Abstract
Inducible nitric-oxide synthase (iNOS) produces biologically stressful levels of nitric oxide (NO) as a potent mediator of cellular cytotoxicity or signaling. Yet, how this nitrosative stress affects iNOS function in vivo is poorly understood. Here we define two specific non-heme iNOS nitrosation sites discovered by combining UV-visible spectroscopy, chemiluminescence, mass spectrometry, and x-ray crystallography. We detected auto-S-nitrosylation during enzymatic turnover by using chemiluminescence. Selective S-nitrosylation of the ZnS4 site, which bridges the dimer interface, promoted a dimer-destabilizing order-to-disorder transition. The nitrosated iNOS crystal structure revealed an unexpected N-NO modification on the pterin cofactor. Furthermore, the structurally defined N-NO moiety is solvent-exposed and available to transfer NO to a partner. We investigated glutathione (GSH) as a potential transnitrosation partner because the intracellular GSH concentration is high and NOS can form S-nitrosoglutathione. Our computational results predicted a GSH binding site adjacent to the N-NO-pterin. Moreover, we detected GSH binding to iNOS with saturation transfer difference NMR spectroscopy. Collectively, these observations resolve previous paradoxes regarding this uncommon pterin cofactor in NOS and suggest means for regulating iNOS activity via N-NO-pterin and S-NO-Cys modifications. The iNOS self-nitrosation characterized here appears appropriate to help control NO production in response to cellular conditions.
Keywords: Crystallography, Nitric Oxide, Nitric-oxide Synthase, Pterin, Radicals, Glutathione, Nitrosation, Nitrosylation, Tetrahydrobiopterin
Introduction
Nitric oxide (NO)4 levels are regulated in vivo at the level of synthesis by three closely related forms of nitric-oxide synthase (NOS): inducible (iNOS), endothelial (eNOS), and neuronal (nNOS) (1). All three NOS enzymes are active only as homodimers and contain two modules: the catalytic oxygenase (NOSox) and the electron-supplying reductase (NOSred) (2). The NOSox dimer contains two catalytic sites at which the substrate l-arginine binds above the heme (3, 4). The symmetrical NOSox dimer interface is stabilized by a bridging ZnS4 cluster and two predominantly buried tetrahydrobiopterin (H4B or (6R)-5,6,7,8-tetrahydro-l-biopterin) cofactors, each hydrogen-bonded to one heme (4–7). Critical differences among the three isozymes include location, regulatory mechanisms, dimer stability, and the amount of NO that is produced (8–10). iNOS rapidly generates large toxic bursts of NO in vivo and has fewer known mechanisms for halting NO production, whereas eNOS and nNOS produce less NO, are calcium-regulated and contain auto-inhibitory elements (9, 11, 12).
Biological signaling by NO is orchestrated by three interconverting redox-active forms: the free radical (NO•), the nitroxide anion (NO−), and the nitrosonium cation (NO+) (13). NO• binds to metal sites in proteins, while NO+ reacts with cysteinyl sulfurs forming S-nitrosylated derivatives (14). S-Nitrosylation is a reversible post-translational modification implicated in protein regulation and signaling within intact cells (15–17). As the major producer of high NO concentrations in vivo, iNOS is a potential target for regulatory auto-S-nitrosylation. The human (and murine) iNOSox dimer contains 18 cysteine residues, 16 of which are conserved in all three isozymes. NOS is S-nitrosylated in arterial mouse preparations, nitrosylated eNOS is evidently generated during turnover (18), and the zinc site has been implicated as a potential S-nitrosylation target (19–21). However, NO-bound nitrosylation sites have not been directly identified in iNOS, and little is known about chemical mechanisms of iNOS nitrosation at the molecular level or their regulatory consequences in the cellular milieu.
To identify nitrosation sites that may act in iNOS regulation, we treated iNOS with NO donors, and characterized the resulting species by spectroscopy, tandem mass spectrometry and x-ray crystallography. Furthermore, we used chemiluminescence to assay iNOS for S-nitrosylation during turnover. We observed spectroscopic evidence for an air-stable, H4B-dependent, NO-modified iNOS. We identified selective S-nitrosylation of the ZnS4 site at low concentrations of exogenous NO donor. By x-ray crystallographic analysis, we also discovered unexpected N-nitrosation (N-NO) of the pterin cofactor. Computational docking placed GSH with the reactive cysteine sulfur adjacent to the N-NO modification site. Saturation transfer difference nuclear magnetic resonance (STD-NMR) spectroscopy indicated that GSH transiently binds iNOS. Our results provide biological insights on pterin cofactor chemical reactivity within iNOS, support a model for iNOS allosteric regulation by multi-site nitrosation, and reveal pterin N-nitrosation as a potential NO signaling mechanism. Significantly, these findings suggest that S-NO and N-NO may regulate NO output from iNOS, the NOS enzyme that typically forms the highest and consequently most toxic levels of NO in vivo.
EXPERIMENTAL PROCEDURES
NOS Expression and Purification
Murine full-length iNOS, Δ65 iNOSox (residues 65–498), and iNOSred (residues 645–1144) were recombinantly expressed and purified by metal chelating and size exclusion chromatography (2). Heme (and iNOS) concentrations were determined using the pyridine hemochromagin method (22).
Synthesis of CysNO and GSNO
Nitrosocysteine (CysNO) (50 mm) was synthesized by mixing equal volumes of 1 m reduced free cysteine (CysH) in 0.5 n HCl with 1 m NaNO2 (15N,18O-NaNO2 for 15N,18O-CysNO), and neutralizing the solution with 40 mm EPPS pH 7.6, 0.5 mm EDTA. Nitrosoglutathione (GSNO) was synthesized similarly.
Saville Reaction
iNOS samples with and without NO donors were assayed for S-nitrosylation using the Saville reaction (23). iNOSox (37.5–50 μm heme), full-length iNOS (14.4–16.5 μm heme), or iNOSred (12.7–13.1 μm monomer) were treated for 5 min with 40-fold molar excess of NO donor (CysNO, GSNO, or papaNONOate ((Z)-1-[N-(3-ammoniopropyl)-N-(n-propyl)amino]diazen-1-ium-1,2-diolate)) or 40 mm EPPS buffer, pH 7.6. Protein samples were spun through G-25 columns to remove unreacted NO donors. One volume of 1% sulfanilamide (SAA), with and without 0.05% HgCl2, was added to each sample and incubated for 15 min. One volume of N-(1-napthyl)ethylenediamine (NNED) was added to both SAA and SAA/HgCl2 samples. After 15 min, NNED/SAA-NO was detected at 540 nm. The absorbance difference between the two samples was plotted on a standard curve to determine the S-NO content.
UV-visible Spectroscopy
All UV-visible measurements were performed on a Hewlett Packard 8453 spectrophotometer. Dimeric iNOSox with H4B bound was diluted in a quartz cuvette open to air to final concentration of 14 μm in 40 mm EPPS at pH 7.6. Freshly prepared CysNO was added aerobically to a final concentration of 500 μm (∼17-fold excess over iNOSox heme). After the resulting 443 nm peak decayed, CysH was added to the cuvette to a final concentration of 500 μm. Experiments were replicated >5 times with iNOSox dimer concentrations ranging from 10–20 μm. Additional experiments were performed using GSNO and GSH in place of CysNO and CysH at the same concentrations. Control experiments were performed in the presence of 10 μm neocuproine to selectively chelate Cu(I) with and without 0.5 mm EDTA to chelate trace amounts of divalent metal ions.
For pterin-free and H4B-analogue experiments, dimeric iNOSox purified in the absence of H4B was diluted to 3 μm and incubated for 1 h with or without 10 μm H4B or other pterin (5-methyl-H4B, 4-amino-H4B, or 2-amino-6-(1,2-dihydroxypropyl)-7,8-dihydro-1H-pteridin-4-one (H2B)). Pterin binding was monitored by shift of the Soret peak in the UV-visible spectrum of the enzyme, indicating a low-spin (∼420 nm) to high-spin FeIII transition (397 nm). CysNO was added to a final concentration of 400 μm. After decay of the 443 nm Soret (FeIII-NO), CysH was added initially at low concentrations (400 μm), and then up to final concentrations of 2 mm. Experiments were also repeated with overnight incubation of pterins and with a higher concentration of 5-methyl-H4B (100 μm).
NO Chemiluminescence
S-NO bonds formed during turnover were detected using a Sievers Chemiluminescence NO analyzer. A reaction mixture containing 2 μm full-length iNOS dimer, 50 μm H4B, 250 μm l-arginine, 20 μm FAD/FMN was prepared, and NADPH was added (final concentration 500 μm) to initiate the reaction. After 5 min, nitrite, nitrate, cofactors, and substrates were removed by rapid microdialysis. 5–10 μl of the dialysate (10–20 pmol of iNOS dimer) was injected into a sealed purge vessel containing I2/75 mm KI/4 ml glacial acetic acid under argon atmosphere at 50 °C. The NO released reacts with ozone to form chemiluminescent N2O*. The chemiluminescent signal generated is directly proportional to moles of NO released and is read out over time as an electrical signal. The integrated signal was compared with standards to determine the amount of S-NO. With these experimental conditions, detection limits for S-NO were ∼1 pmol with a typical working range of 1–100 pmol.
We did the following controls to optimize the assay and confirm the presence of the S-NO signal. Of the purge solutions we tested, heated I2/KI/acetic acid gave the most sensitive, reproducible, and reliable measurements for CysNO, GSNO, and S-NO hemoglobin standard solutions. To confirm that the signal resulted from S-NO, 1% SAA and 0.05% HgCl2 were added to the sample to selectively cleave S-NO bonds prior to injection into the purge vessel, and this abolished the signal. We verified that turnover mixtures, including iNOS, H4B, FAD/FMN, l-arginine, but not NADPH, with 5 mm nitrite added gave no signal after microdialysis. This is in significant excess of the maximal amount of nitrite formed during our turnover. Therefore, our assay was sensitive and specific for S-NO. We verified iNOS activity by injecting samples without the dialysis step. We verified that 2 S-NO per tetramer (24) were detected in samples of prepared S-NO hemoglobin using this experimental set-up.
Tandem Mass Spectrometry
Samples of iNOSox (50 μm heme) were nitrosylated with freshly synthesized CysNO or 15N,18O-CysNO for 15 min with 2-, 5-, and 10-fold excess of CysNO relative to heme. Unreacted CysNO was removed using a Bio-Rad spin column pre-equilibrated with 40 mm EPPS at pH 7.6. Samples were analyzed using tandem mass spectrometry (25, 26). Samples were denatured, and free thiols were alkylated with methyl methanethiosulfonate in dimethylformamide, using published protocols (27). The samples were split into three aliquots, and each aliquot digested into peptide fragments overnight using trypsin, elastase, or chymotrypsin. The peptides formed from the three digests were pooled and analyzed by multidimensional protein identification technology (26) on an LCQ ion trap mass spectrometer. The resulting tandem mass spectra were interpreted, and the S-NO sites identified using SEQUEST (25).
Size Exclusion Chromatography
Samples of iNOSox (+l-Arginine, +H4B) (concentration 160 μm heme) with or without 10 mm GSNO added (60-fold molar excess) were loaded onto a Superdex 200gl 10/300 column using an AKTA FPLC. Samples were eluted at a flow rate of 0.5 ml/min in 40 mm EPPS at pH 7.6. Peaks were identified and integrated using Unicorn 5.11 software. Molecular weight standards were analyzed using the same conditions and used to identify peaks in the iNOSox samples.
Crystallization
15 mg/ml iNOSox in 40 mm EPPS, pH 7.6, was incubated with 50 mm CysNO for 5 h. NOS inhibitor ethyl 4-[(4-methylpyridin-2-yl)amino]piperidine-1-carboxylate (10 μm) was added to the protein mixture to promote dimerization (28). Dimeric nitrosylated iNOSox was crystallized at 4 °C by vapor diffusion in hanging drops containing equal volumes of protein and reservoir solution against a reservoir containing 50 mm MES at pH 5.3, 25–30% Li2SO4, 5% n-octyl-β-d-glucopyranoside.
Structural Characterization
Twenty-four hours following crystallization setup, diffraction data from flash-frozen iNOSox crystals were collected (SSRL beamline 7–1, λ = 0.98 Å), indexed, processed and scaled. X-ray diffraction data were indexed, processed, and scaled using Denzo and Scalepack (29). The structure was solved by molecular replacement (search model PDB code 3E7M). Iterative rounds of refinement (CNS version 1.0 (30)) and manual refitting with Xfit (31) were used to refine the structure.
Automated Docking
AutoDock 4.0 (32) was used to dock GSH to dimeric murine iNOSox. Non-polar hydrogens were added to the iNOSox structure and Kollman charges assigned to each atom using SYBYL. Lennard-Jones parameters for the heme were obtained from AMBER and modified by the AutoDock force field factor. Solvation parameters for heme and pterin were derived from amino acid atoms with chemical similarity. Atomic affinity and electrostatics grid maps (60 × 60 × 60, spacing 0.375 Å) centered on the N-NO-pterin site were created using AutoGrid 4.0. The AutoDock Lamarckian genetic algorithm was used to search the grid maps for low energy solutions. Ten independent dockings, each initiated with 150 randomly oriented GSH molecules, were performed. Results were analyzed using AutoDockTools.
STD-NMR
To assess the interaction of glutathione with iNOSox, STD-NMR (33) experiments were acquired on samples of iNOSox (32 μm heme) that were exchanged into NMR buffer (40 mm K2HPO4/KH2PO4 in D2O, pD 7.6) with and without 1 mm GSH. Selective saturation of protein resonances was achieved using an excitation train composed of 50 ms Gaussian-shaped pulses that were separated by a 4 μs delay. The total saturation time was 3 s, and the on- and off-resonance irradiations were centered at 0.5 ppm and 30 ppm, respectively. The pulse sequence also included a spoil sequence, a T2 filter, and a 3-9-19 water suppression element (34, 35). Identical STD experiments were performed on all samples tested. One-dimensional 1H spectra that included the 3–9-19 water suppression sequence were also collected. All spectra were acquired at 5 °C on a 500 MHz Bruker Avance spectrometer equipped with a 5 mm TXI-HCN triple resonance probe, referenced to 2,2-dimethyl-2-silapentane-5-sulfonate, processed with NMRPipe and viewed in NMRDraw (36).
RESULTS
iNOS Nitrosation by NO Donors
To produce sufficient quantities of nitrosated NOS for study, we treated purified iNOS protein with the NO donor nitrosocysteine (CysNO). We treated full-length iNOS, and the individual iNOSox and iNOSred modules, with CysNO for 5 min, removed unreacted CysNO and quantitated S-nitrosylation by a modified Griess assay (Saville reaction) (23). S-Nitrosylation occurred primarily in iNOSox, with maximally 2–3 S-NO modifications per heme. Background detected in controls lacking iNOS or CysNO were <5% of these S-NO measurements for iNOSox. Alternative NO donors GSNO and papaNONOate produced fewer S-NO modifications in iNOSox (1.3–1.6 per heme).
To probe nitrosylation sites on iNOS, we followed the reaction of iNOSox with CysNO by monitoring spectral changes of the iNOS heme. We observed FeIII-NO formation and discovered evidence for a stable, non-heme, NO derivative of iNOS (Fig. 1). Aerobic reaction of CysNO (or GSNO) with H4B-bound iNOSox rapidly shifted the heme spectrum from a 397 nm Soret peak (resting state, high-spin FeIII) to a 443 nm Soret peak and associated α/β bands at 585 and 549 nm, characteristic of FeIII-NO (37). FeIII-NO could be formed by homolytic cleavage of CysNO catalyzed by trace transition metal ions (supplemental Fig. S1), followed by reaction of NȮ with the ferric heme iron.
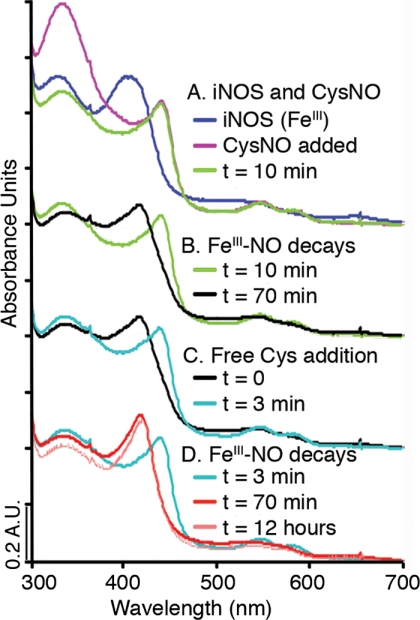
FIGURE 1.
iNOS reactions with NO donor and free thiol, shown by UV-visible spectroscopy. A, reaction of iNOSox (resting state high-spin FeIII, 397 nm, blue) with the NO donor CysNO (purple) forms an FeIII-NO iNOSox derivative (Soret 443 nm, α/β bands 585 nm, 549 nm, green). FeIII-NO is readily distinguishable from FeII-NO spectroscopically by the visible region α/β band absorbances. B, iNOSox FeIII-NO (green) decays to form a low spin Fe species (420 nm, black). C, CysH added to the low spin Fe species (black) re-generates iNOSox FeIII-NO (Soret 443 nm, α/β bands 585 nm, 549 nm, aqua). D, iNOSox FeIII-NO formed by addition of CysH (aqua) decays back to a low spin Fe species (420 nm, red), which is stable for over 12 h (420 nm, pale red).
The high spin FeIII-NO iNOSox signal generated by CysNO treatment (Fig. 1A) gradually decayed (Fig. 1B) to a low spin heme signal (∼420 nm). Surprisingly, subsequent addition of free thiol (reduced CysH or GSH), in the absence of an exogenous NO source, rapidly regenerated the FeIII-NO spectrum (Fig. 1C), which again decayed over time (Fig. 1D). Results with full-length iNOS were similar. Delayed addition of reduced thiols (16 h after the disappearance of FeIII-NO) still regenerated FeIII-NO. Because low spin heme can also be indicative of Fe-NO with changes in the proximal thiolate ligand, we performed a separate control experiment, in which papaNONOate was added to our 420 nm species. FeIII-NO iNOSox rapidly formed when papaNONOate was added to the low spin species, verfiying that NO was not still bound to the heme (supplemental Fig. S2). The spectroscopic data suggest that stable, non-heme, nitrosated adduct(s) of iNOSox can store NO for later release or transfer.
Similar CysNO treatment of iNOSox containing alternative pterins revealed a unique role for H4B in iNOS nitrosation. Although FeIII-NO formation by iNOSox occurred readily regardless of pterin substitution, the time course for FeIII-NO decay was markedly pterin-dependent (supplemental Fig. S3). Notably H4B was the only pterin tested that supported regeneration of FeIII-NO upon addition of reduced CysH to the 420 nm species.
Enzymatic S-Nitrosylation of iNOS
Can NO formed by iNOS during catalysis S-nitrosylate the enzyme? Because of its picomolar sensitivity of NO detection, we used a chemiluminescence NO analyzer to assay for S-nitrosylated iNOS species generated during enzymatic turnover (Fig. 2). Besides full-length iNOS, our catalytic mixture contained only the factors essential for NO synthesis: FAD, FMN, pterin, and substrate l-arginine. We initiated iNOS catalysis by addition of NADPH and also performed controls without NADPH (supplemental Fig. S4). To exclude potential S-nitrosylation targets and minimize breakdown of nitrosylated intermediates, we omitted protein serum albumin, GSH, and other free thiols, which are typically added for sustained activity in NOS turnover assays (38, 39). We created standard plots of S-NO concentration by injecting known amounts of CysNO, GSNO, and S-NO hemoglobin (5–100 pmol of S-NO) into the NO analyzer. We removed any nitrite and nitrate byproducts by using microdialysis. Without this pretreatment of the turnover sample, a large chemiluminescence peak, which we attribute primarily to nitrite, was observed, thus confirming iNOS catalytic activity. From the integrated chemiluminescence signals, we quantitated 5–16 pmol of S-NO, corresponding to molar ratios of 0.6 ± 0.1 S-NO per iNOS dimer (n = 6). These results indicate that iNOS is S-nitrosylated during turnover.
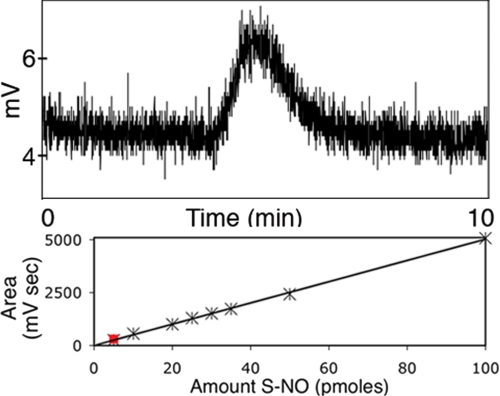
FIGURE 2.
Auto-S-nitrosylation of iNOS during enzymatic turnover, detected photometrically. Following enzymatic turnover, full-length iNOS was micro-dialyzed to remove nitrite/nitrate, excess cofactors, and unreacted substrate. Dialyzed iNOS sample (5 μl, 10 pmol of iNOS dimer) was injected into a chemiluminescence NO analyzer. S-NO bonds were cleaved by acetic acid/KI/I2 in the purge vessel of the NO analyzer, releasing NO gas. NO gas released was reacted with ozone, creating a chemiluminescent species, which was detected photometrically. The integrated area (212 mV s) under the peak (above) corresponds to 5 pmol of S-NO (red square), as determined from a calibration plot (below).
Zinc Site S-Nitrosylation
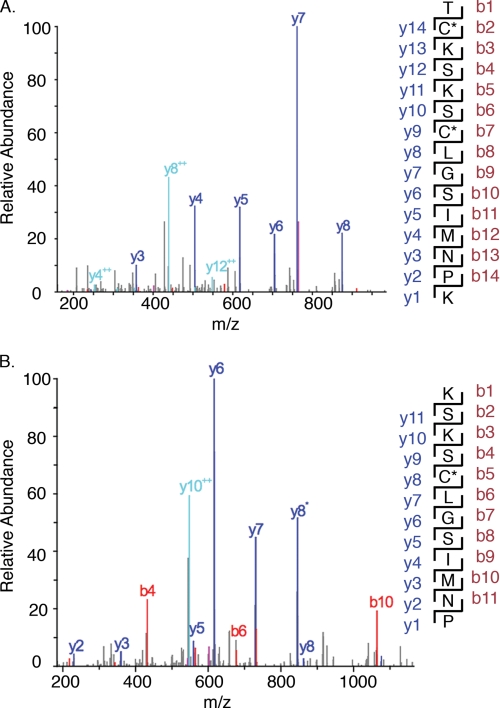
Having demonstrated that iNOS is S-nitrosylated during turnover and by reaction with NO donors, we designed experiments to identify S-nitrosylation target sites in iNOSox. We reacted iNOSox with CysNO (2-, 5-, and 10-fold excess over heme) and analyzed the products by tandem mass spectrometry. By adapting the experimental conditions, we stabilized NO modified peptides for direct mass measurements. In the 5-fold and 10-fold excess CysNO-treated samples, we identified peptides containing S-NO-Cys-104 and S-NO-Cys-109, based on analysis of MS/MS fragmentation patterns (Fig. 3). S-NO modification of both Cys-104 and Cys-109 within the same polypeptide chain was also found. Cys-104 and Cys-109 were confirmed as S-nitrosylation targets in parallel experiments using isotopically 15N,18O-labeled CysNO as the NO donor (supplemental Table S1). At these levels of NO donor, S-nitrosylation was not detected on any other cysteine residues in iNOS. Thus, the ZnS4 cysteine ligands of iNOSox were preferentially targeted for S-nitrosylation.
FIGURE 3.
Tandem mass spectrometry of iNOSoxS-nitrosylated with CysNO. A, mass fragmentation pattern for peptide fragment 103–117 of S-nitrosylated iNOSox. A peptide with mass/charge 1669 was identified as 103TC*KSKSC*LGSIM*NPK117 based on the fragmentation pattern, where C* represents S-NO modification of Cys-104 and Cys-109 and M* represents oxidation of Met-114. B, mass fragmentation pattern for peptide 105–116 of S-nitrosylated iNOSox. Annotations correspond to fragments (mapped at right) generated from the N (red) and C (blue) termini of the peptide, respectively.
To test the effects of nitrosation on the NOS monomer/dimer equilibrium, we prepared samples treated with a large excess of nitrosating agent (60-fold molar excess over heme) and analyzed the oligomeric state by using size exclusion chromatography (supplemental Fig. S5). We found that nitrosation destabilized the iNOSox dimer, producing an almost 2-fold decrease in dimer content.
The iNOSox Crystal Structure Reveals H4B Is a Nitrosation Target
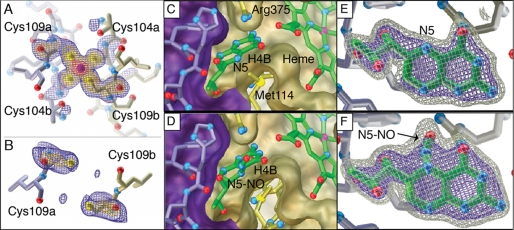
To elucidate the structural effects of nitrosation on the obligate NOS dimer, we grew diffraction quality crystals of iNOSox in the presence of CysNO and a dimer-stabilizing inhibitor (28, 40), and determined the crystal structure (2.2 Å resolution, Rfree 22.9%, Table 1). We found evidence for two distinct types of NO modifications, both at sites that contribute to the dimer interface: not only S-nitrosylation of the ZnS4 site, but also N-nitrosation of the pterin cofactors (Fig. 4).
TABLE 1.
Nitrosated iNOSox data collection and refinement statistics
| PDB code | 3NQS |
| Space group | P6122 |
| Unit cell (Å; o) | 214.1, 214.1, 116.5; 90, 90, 120 |
| Scatterers | 7605 |
| Residues | 2 × (77–99, 109–495) |
| Cofactors | 2 × (heme, N-NO-pterin) |
| Ligands | 2 × (ethyl-4-[(4-methylpyridin-2-yl)amino] piperidine-1-carboxylate) |
| Waters | 598 |
| Resolution (Å) | 20.0-2.2 (2.25-2.2)a |
| Unique refls. | 74686 (6090) |
| Observations | 295816 (17574) |
| % Completeness | 93.9 (74.6) |
| <I/σI> | 21.1 (2.5) |
| Rsym (%)b | 11.5 (41.5) |
| R (%)c | 20.4 |
| Rfree (%)d | 22.9 |
| Avg. B (Å2) | 46.3 |
| Rms bond (Å) | 0.007 |
| Rms angle (°) | 1.2 |
a Values in parentheses are for highest resolution shell.
b Rsym = ΣΣ|Ij − <I>| / ΣΣj Ij.
c R = Σ||Fo| − |Fc|| / Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
d 5% of the reflections were set aside randomly for the Rfree calculation.
FIGURE 4.
iNOSox crystal structures before (top, PDB 3E7M) and after (bottom, PDB 3NQS) nitrosylation, showing the ZnS4 (left), dimer interface (middle), and pterin sites (right). A, in the absence of NO donor, the iNOSox ZnS4 is occupied and ordered as shown by Fo − Fc omit electron density for the zinc and its four cysteine ligands (Cys-104a, Cys-104b, Cys-109a, Cys-109b). B, after CysNO treatment during crystallization, the iNOSox ZnS4 is disrupted and residues 100 to 107 are disordered. Fo − Fc omit electron density observed for Cys-109 from each subunit indicates that the inter-atomic distance between sulfur atoms (5.7 Å) is too long for a ZnS4 cluster or disulfide bond (expected distance S-S is 2.0 Å) formation, but would accommodate two S-NO modifications. C, pterin and D, N-NO pterin (after reaction with CysNO) cofactors (green tubes with atom-colored spheres for oxygen and nitrogen atoms) both hydrogen bond with adjacent heme and are tightly packed between two iNOSox subunits (purple/gold surfaces) at the dimer interface. E, pterin and F, N-NO pterin in Fo − Fc omit electron density contoured at 3σ (blue) and 6σ (gray) are distinguishable by additional protruding electron density for the NO moiety (identified by arrow) covalently attached to position N5.
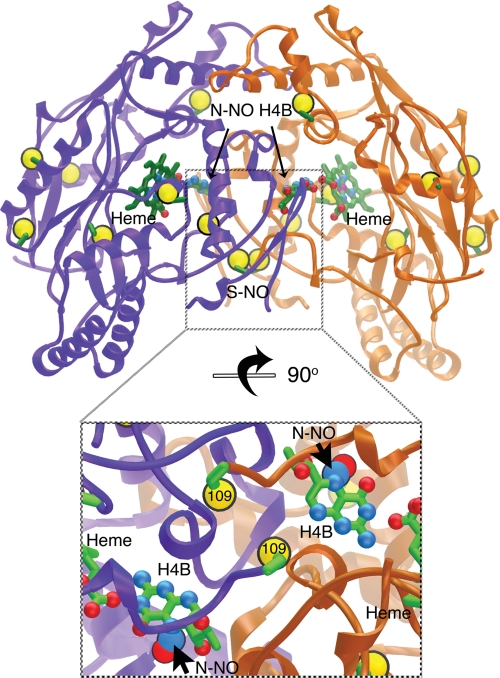
One NO modification target is the dimer-bridging ZnS4, consistent with the selective S-nitrosylation of the zinc ligands Cys-104 and Cys-109 shown in our mass spectrometric analysis. In the absence of the NO donor, the iNOSox zinc site is ordered (Fig. 4A). In the presence of the NO donor, the zinc is lost: no anomalous electron density for zinc is observed and residues 101–108 in the zinc-binding loop are disordered (Fig. 4B). The two Cys-109 sulfur atoms of the iNOSox dimer are too far apart to ligate zinc or form a disulfide bond. Synchrotron radiation breaks S-NO bonds (41); thus, S-NO is not directly observable on Cys-109 or any other cysteine residues in this structure. Yet, S-nitrosylation of iNOSox is evident in the crystal structure because the ZnS4 site is disrupted.
We furthermore discovered novel N-nitrosation of the pterin cofactors in our structure. In the structure of CysNO-treated iNOSox, electron density for the pterin cofactors indicates covalent modification at the N5 positions by a diatomic molecule (Fig. 4F). The additional electron density is absent in the corresponding untreated protein (Fig. 4E) and cannot be accounted for with a water molecule (supplemental Fig. S6). Moreover, we have not observed such additional electron density at the pterin N5 position in any of our other inhibitor-bound crystal structures of iNOSox (4, 6, 40, 42). We attribute the electron density on the pterin cofactor to an N-NO modification because NO was the only reactive diatomic species present during crystallization that is capable of forming a stable pterin adduct. Furthermore, the geometry at the N5 position matches that in small molecule crystal structures of the related heterocylic N-NO-pyrazines (43). No significant structural rearrangements are needed to accommodate N-nitrosation of the pterin. In the crystals, N-NO-pterin may have been formed directly by CysNO or indirectly from iNOSox nitrosation sites, heme-Fe or Cys-104/Cys-109, which are located 13.8 Å and ∼15 Å away from pterin-N5, respectively. The N-NO moiety is solvent-exposed and flanked on one side by the Arg-375 guanidinium group and on the other side by the Met-114 side chain (Fig. 4, C and D). Met-114 (valine in eNOS), the only residue involved in pterin binding that is not conserved among the three isozymes, may have an isozyme-specific role in nitrosation linked to its anti-oxidant properties (44).
Although N-nitrosation of the cofactor within the protein was unexpected, N-nitroso compounds can be readily synthesized. For instance, combining equimolar amounts of acidified tryptophan and nitrite at room temperature yields N-nitrosotryptophan, which exhibits a characteristic yellow color (45). Yet, N-nitrosation in iNOSox occurred uniquely on the pterin cofactor, despite the potential for N-nitrosation of tryptophan residues throughout the protein. Thus N-NO-pterin is a selective chemical modification.
GSH Binding and Release from iNOS
The solvent exposure of the N-NO moiety on pterin led us to investigate the potential for NO transfer to a partner. As intracellular GSH concentrations range from 2 mm to 10 mm (46), and GSNO formation by NOS has been experimentally observed (47), we examined whether NO transfer from pterin to GSH is structurally feasible. We computationally predicted a binding site for GSH on iNOSox, and experimentally determined GSH binding to iNOSox using STD-NMR spectroscopy.
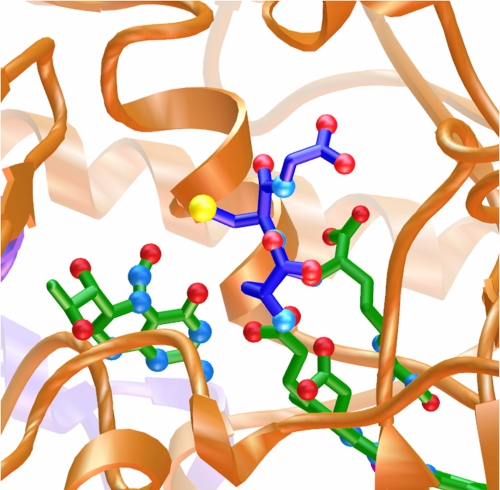
We used AutoDock 4.0 (32) to predict binding orientations of GSH to iNOSox. During the docking, iNOSox was rigid and GSH was flexible with rotatable bonds. AutoDock predicted a binding site near the N-NO-pterin (Fig. 5). In the docked orientation, the GSH reactive thiol is adjacent to the N-NO group (distance < 4 Å), and Arg-375 is positioned to activate acid/base transnitrosation.
FIGURE 5.
A computationally identified GSH-binding site near the N-NO-pterin. Computational docking of GSH (dark blue tubes with atom-colored spheres for oxygen, nitrogen, and sulfur) to iNOSox identified a favorable site near the N-NO-pterin (green tubes at left, oriented with NO up) for transfer of NO to the reactive thiol (yellow) of GSH. Heme and bound N-hydroxyarginine intermediate (green tubes at right) are shown in the context of the iNOSox dimer interface (gold and purple ribbons).
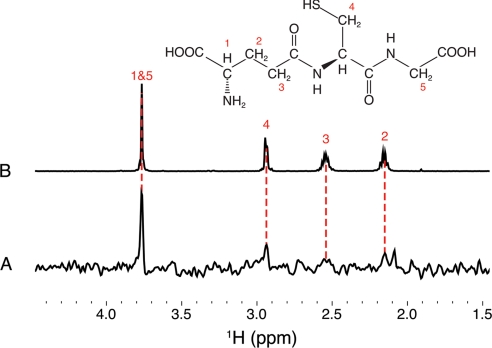
Having computationally identified a putative GSH binding site on iNOS, we designed NMR experiments to test if GSH binds to iNOSox (Fig. 6). The mm GSH concentrations in vivo suggest that a physiologically relevant interaction of GSH with iNOS may be relatively weak (Kd ∼1 mm). So, we chose STD-NMR spectroscopy, a robust technique for detecting specific protein/ligand interactions with dissociation constants between 10−8 and 10−3 m (33). We acquired one-dimensional 1H STD-NMR spectra of iNOSox in the presence and absence of GSH. After the pulse sequence saturates specific resonances of the protein, spin diffusion transfers magnetization from the saturated spins throughout the protein and to any bound ligands. After on- (0.5 ppm) and off-resonance (30 ppm) saturated 1H spectra were collected, the difference spectrum was calculated (supplemental Fig. S7, B and C). To reveal the 1H signals that result from the specific interactions between GSH and iNOSox (Fig. 6A), the iNOSox difference spectrum was then subtracted from the iNOSox:GSH difference spectrum. A clear GSH-associated resonance at 3.8 ppm, and smaller peaks assignable to GSH (2.9, 2.55, and 2.15 ppm) are observed in the double difference spectrum, but not in controls in which either protein or GSH is omitted (supplemental Fig. S7, A and B). These STD-NMR results revealed GSH binding to iNOSox with an affinity of mm or better, consistent with mm biological GSH concentrations.
FIGURE 6.
1H STD-NMR analysis of GSH binding to iNOSox. A, difference spectrum of the iNOSox:GSH STD spectrum minus the iNOSox STD spectrum, showing proton resonances characteristic of GSH at 3.75, 2.9, 2.55, and 2.15 ppm (red dashed lines). B, 1H NMR spectrum of GSH (1 mm) with resonances assigned to numbered protons within the chemical structure (above).
DISCUSSION
NO Interaction Chemistries for iNOS
Nitrosation sites have the potential to regulate iNOS activity, alter product release, and influence the electronic state of NO product. Our results show that iNOS pterin cofactors and Zn site cysteines are nitrosation targets, besides the heme Fe (Fig. 7). Because pterin and ZnS4 are both integral components of the NOS dimer interface, their nitrosation provides a logical means for allosteric regulation (Fig. 8). The heme iron and pterin are interacting (4) redox-active cofactors that exchange electrons (48–50). This interconnectivity could allow pterin modification to tune the heme redox potential, electron transfer between heme and pterin, and electronic state of NO.
FIGURE 7.
Selective iNOSoxS- and N-nitrosation at the dimer interface (subunits colored purple/gold, cysteine sulfurs shown as large gold spheres). The nitrosylated iNOSox crystal structure (top) shows the location of our confirmed S-nitrosylation (S-NO) and N-nitrosation (N-NO-H4B) sites in the interlocked dimer interface, as well as overall distribution of cysteine residues. The inset below shows a close-up view of the S-NO and N-NO sites in the iNOSox dimer interface rotated 90° from the top image.
FIGURE 8.
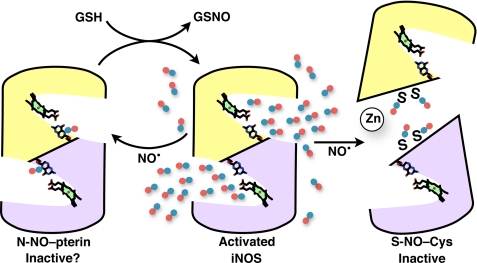
Schematic model for N-NO and S-NO modification and function in iNOS. Activated dimeric iNOS (center panel) synthesizes high levels of NO (blue/red atoms) at the two active-site hemes, from the substrates l-arginine and molecular oxygen. NO nitrosates iNOS, forming N-NO-pterin (left panel) in the active-site funnel and S-NO-Cys (right panel) selectively at ZnS4 bridging the dimer interface. Nitrosation inactivates iNOS by breaking the dimer (right panel). Pterin N-NO modification may inactivate iNOS, if the modified cofactor does not properly transfer electrons to and from the heme. The proposed reaction of GSH and iNOS N-NO-pterin (left panel, top arrow) would form GSNO for signaling and regenerate the H4B cofactor. NOS dimers are shown as gold and purple subunits with heme (green porphyrin) and pterin cofactors represented by atom-colored sticks.
Zinc Site S-Nitrosylation at the NOS Dimer Interface
Our results provide the first direct experimental observation of NO-modified peptides in pterin-bound iNOS. We stabilized the NO modifications by using acidic conditions and low temperature mass spectrometry, whereas other studies depended on indirect chemical markers, mutagenesis or zinc quantitation (21, 51).
NOS enzymes are active only as dimers (2) and therefore can be allosterically regulated by the monomer/dimer equilibrium. A structural zinc site located across the dimer interface stabilizes iNOS in the active dimeric form (5). The three isozymes differ in dimer stability, with iNOSox forming the weakest dimer (dimer stability: nNOSox > eNOSox > iNOSox) (10). Of the 18 reactive cysteine residues in the iNOSox dimer, we found that S-nitrosylation selectively targets the conserved ZnS4 site Cys residues at low levels of NO donor (5–10 fold excess over heme). This striking selectivity is characteristic of biological S-NO signaling (52).
Destabilization of the iNOS dimer by ZnS4 site S-nitrosylation (supplemental Fig. S5) provides a feasible allosteric auto-inhibition mechanism: at high NO concentrations, nitrosation limits iNOS activity by shifting the enzyme from active dimer toward inactive monomer. Such an allosteric regulatory mechanism is supported by dimer dissociation in iNOS (53) and eNOS (20) upon treatment with NO donors. In contrast, nNOS truncation mutants that lack the ZnS4 site remain dimeric (54), suggesting this regulatory mechanism is isoform-specific. For its physiological roles in immune function, iNOS generates large bursts of toxic nitrogen oxides (55). Unlike eNOS and nNOS, iNOS is not regulated by calcium levels and auto-inhibitory elements (8, 9). Nitrosylation of the structural ZnS4 site leading to dissociation of the obligate dimer may therefore be an important alternative regulatory mechanism for controlling this NOS isozyme.
Pterin Nitrosation Chemistry
Our structural identification of NO bound at the pterin N5 position after treatment with CysNO is surprising, but consistent with pterin radical formation in NOS. During catalysis, H4B transfers an electron to the heme iron, forming a pterin radical localized to the N5 position (50), which is solvent-exposed in the active-site funnel (4). Of the pterins we tested in our spectroscopic experiments (supplemental Fig. S3), only H4B supported thiol-dependent release of non-heme NO to reform iNOSox FeIII-NO. H4B is unique among these pterins in that it both supports NO synthesis and has an open site (6, 56) for the N-NO modification that we observed in our crystal structure.
How might N-NO-H4B form during catalysis? N-NO-pterin is the logical product of NO radical with the N5-centered-pterin radical of NOS (Fig. 8). In a currently accepted NOS mechanism, release of NO radical is coupled to reduction of pterin radical (48, 49), such that these two radicals would not coexist during a single turnover. However, NO could react with the pterin radical if the process becomes uncoupled or at high NO concentrations, as may occur during inflammatory responses. Also, NOS enzymes have been shown to sequester NO at non-heme sites near the active site (57, 58), which would then be available for reaction with a subsequent pterin radical. Moreover, Schmidt and co-workers (59) showed that NOS catalysis leads to H4B loss or modification that is distinct from oxidation and dependent on a NOS-derived reactive nitrogen species. Bacterial NOS synthesizes nitrated Trp in the pterin binding site via a proposed N-NO intermediate (60). Thus, the H4B binding site appears to have evolved for nitrosative chemistry.
N-NO species are well characterized chemically, yet just beginning to be understood biologically (45, 61). For example, N-NO-Trp-containing dipeptides are vasoactive, and N-NO-Trp melatonin reacts with GSH to form GSNO (62). Significantly, the redox-active pterin in iNOS is solvent-exposed at the N5 position, leading us to test if a partner may interact there.
Functional Consequences of N-NO-pterin
N-NO-pterin has the potential to regulate NOS activity, transfer NO, or both. Given the stringent redox and electronic requirements of H4B in NOS, N-NO-pterin may inhibit catalysis directly by preventing pterin electron transfer to the heme (Fig. 8). Alternatively, N-NO-pterin may allosterically control NOS by altering the monomer/dimer equilibrium. Thus, the ability of H4B to participate in nitrosative chemistry may be an additional reason why NOS utilizes this cofactor.
Nitrosative chemistry favors NO transfer from N-NO to S-NO (62). S-Nitrosothiols are prevalent in vivo and mediate biological NO signals (15, 17, 27, 63). Furthermore, iNOS binds and S-nitrosylates at least one protein target (52). NOS has been shown to form GSNO (47, 64), and iNOS requires GSH to achieve maximal sustained activity (38, 39, 65). Therefore we investigated GSH binding at a site near the N-NO-pterin. Computational docking analysis found that GSH can be accommodated near pterin with the reactive sulfur adjacent to the exposed N-NO modification. Our NMR binding experiments demonstrate that GSH interacts with iNOSox. Transnitrosation from N-NO-pterin to GSH could regenerate H4B and release GSNO (Fig. 8).
GSNO has been identified as an active agent for NO signaling. Importantly, GSNO is produced by iNOS in cells (66) and is the species responsible for killing mycobacteria in vivo (67). Significantly, GSNO is more stable and less prone to opportunistic side reactions than NO (68, 69), and thus allows for NOS action at a distance.
General Implications
Structures of proteins controlling reactive oxygen species are potential master keys to understanding cancer, aging, and degenerative diseases (70), as these proteins and their interactions regulate a balance for signaling, repair, or cytotoxic outcomes at the cellular and subcellular level. The N- and S-nitrosation sites that we identified in iNOS are linked to dimer stability and hence activity. Consequently these nitrosation sites relate directly to the role of NOS as such a master key. Structural identification of N-NO-pterin and direct mass spectrometric characterizations of S-NO are unequivocal. These findings have general implications for the interpretation of both in vitro and in vivo experiments: temporal and functional relationships among S-nitrosation, N-nitrosation, and NOS activities merit targeted study.
Together our results demonstrate unprecedented nitrosative chemistry in NOS. These discovered S-NO and N-NO sites underscore the remarkable functional versatility of NOS enzymes: their intrinsic structural chemistry may dynamically regulate responses to physiological or pathophysiological contingencies.
Supplementary Material
Acknowledgments
We thank Stanford Synchrotron Radiation Lightsource (SSRL) for beamline facilities, Dennis J. Stuehr for providing protein, James R. Williamson for critical discussions and access to the NMR facilities, and Michael E. Pique for creating schematic figure with “Pages” software.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-HL58883 (to E. D. G.) and RR011823 (to J. R. Y.) and the Skaggs Institute for Chemical Biology (to R. J. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
The atomic coordinates and structure factors (code 3NQS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- NO
- nitric oxide
- iNOS
- inducible NOS
- GSH
- reduced glutathione
- NOS
- nitric-oxide synthase
- eNOS
- endothelial NOS
- nNOS
- neuronal NOS
- NOSox
- NOS oxygenase
- NOSred
- NOS reductase
- H4B
- (6R)-5,6,7,8-tetrahydro-l-biopterin
- STD
- saturation transfer difference
- NMR
- nuclear magnetic resonance
- CysNO
- nitrosocysteine
- CysH
- reduced free cysteine
- GSNO
- nitrosoglutathione
- papaNONOate
- (Z)-1-[N-(3-ammoniopropyl)-N-(n-propyl)amino]diazen-1-ium-1,2-diolate
- SAA
- sulfanilamide
- NNED
- N-(1-napthyl)ethylenediamine
- H2B
- 2-amino-6-(1,2-dihydroxypropyl)-7,8-dihydro-1H-pteridin-4-one
- Rms
- root mean square.
REFERENCES
- 1.Griffith O. W., Stuehr D. J. (1995) Annu. Rev. Physiol. 57, 707–736 [DOI] [PubMed] [Google Scholar]
- 2.Stuehr D. J. (1996) Methods Enzymol. 268, 324–333 [DOI] [PubMed] [Google Scholar]
- 3.Crane B. R., Arvai A. S., Gachhui R., Wu C., Ghosh D. K., Getzoff E. D., Stuehr D. J., Tainer J. A. (1997) Science 278, 425–431 [DOI] [PubMed] [Google Scholar]
- 4.Crane B. R., Arvai A. S., Ghosh D. K., Wu C., Getzoff E. D., Stuehr D. J., Tainer J. A. (1998) Science 279, 2121–2126 [DOI] [PubMed] [Google Scholar]
- 5.Crane B. R., Rosenfeld R. J., Arvai A. S., Ghosh D. K., Ghosh S., Tainer J. A., Stuehr D. J., Getzoff E. D. (1999) EMBO J. 18, 6271–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crane B. R., Arvai A. S., Ghosh S., Getzoff E. D., Stuehr D. J., Tainer J. A. (2000) Biochemistry 39, 4608–4621 [DOI] [PubMed] [Google Scholar]
- 7.Aoyagi M., Arvai A. S., Ghosh S., Stuehr D. J., Tainer J. A., Getzoff E. D. (2001) Biochemistry 40, 12826–12832 [DOI] [PubMed] [Google Scholar]
- 8.Michel T., Feron O. (1997) J. Clin. Invest. 100, 2146–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcin E. D., Bruns C. M., Lloyd S. J., Hosfield D. J., Tiso M., Gachhui R., Stuehr D. J., Tainer J. A., Getzoff E. D. (2004) J. Biol. Chem. 279, 37918–37927 [DOI] [PubMed] [Google Scholar]
- 10.Panda K., Rosenfeld R. J., Ghosh S., Meade A. L., Getzoff E. D., Stuehr D. J. (2002) J. Biol. Chem. 277, 31020–31030 [DOI] [PubMed] [Google Scholar]
- 11.Cho H. J., Xie Q. W., Calaycay J., Mumford R. A., Swiderek K. M., Lee T. D., Nathan C. (1992) J. Exp. Med. 176, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoyagi M., Arvai A. S., Tainer J. A., Getzoff E. D. (2003) EMBO J. 22, 766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnelle D. R., Stamler J. S. (1995) Arch. Biochem. Biophys. 318, 279–285 [DOI] [PubMed] [Google Scholar]
- 14.Stamler J. S., Singel D. J., Loscalzo J. (1992) Science 258, 1898–1902 [DOI] [PubMed] [Google Scholar]
- 15.Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 16.Derakhshan B., Hao G., Gross S. S. (2007) Cardiovasc. Res. 75, 210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gow A. J., Chen Q., Hess D. T., Day B. J., Ischiropoulos H., Stamler J. S. (2002) J. Biol. Chem. 277, 9637–9640 [DOI] [PubMed] [Google Scholar]
- 18.Erwin P. A., Lin A. J., Golan D. E., Michel T. (2005) J. Biol. Chem. 280, 19888–19894 [DOI] [PubMed] [Google Scholar]
- 19.Erwin P. A., Mitchell D. A., Sartoretto J., Marletta M. A., Michel T. (2006) J. Biol. Chem. 281, 151–157 [DOI] [PubMed] [Google Scholar]
- 20.Ravi K., Brennan L. A., Levic S., Ross P. A., Black S. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2619–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell D. A., Erwin P. A., Michel T., Marletta M. A. (2005) Biochemistry 44, 4636–4647 [DOI] [PubMed] [Google Scholar]
- 22.Berry E. A., Trumpower B. L. (1987) Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 23.Saville B. (1958) Analyst 83, 670–672 [Google Scholar]
- 24.Jia L., Bonaventura C., Bonaventura J., Stamler J. S. (1996) Nature 380, 221–226 [DOI] [PubMed] [Google Scholar]
- 25.Eng J., McCormack A., Yates J. R., 3rd. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 26.Washburn M. P., Wolters D., Yates J. R., 3rd. (2001) Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 27.Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Nat. Cell Biol. 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 28.Connolly S., Aberg A., Arvai A., Beaton H. G., Cheshire D. R., Cook A. R., Cooper S., Cox D., Hamley P., Mallinder P., Millichip I., Nicholls D. J., Rosenfeld R. J., St-Gallay S. A., Tainer J., Tinker A. C., Wallace A. V. (2004) J. Med. Chem. 47, 3320–3323 [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–336 [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 32.Morris G., Huey R., Lindstrom W., Sanner M., Belew R., Goodsell D., Olson A. (2009) J. Comp. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayer M., Meyer B. (1999) Angew. Chem. Int. Ed. 38, 1784–1788 [DOI] [PubMed] [Google Scholar]
- 34.Piotto M., Saudek V., Sklenár V. (1992) J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 35.Sklenar V., Piotto M., Leppik R., Saudek V. (1993) J. Magn. Reson. Ser. A 102, 241–245 [Google Scholar]
- 36.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37.Hurshman A. R., Marletta M. A. (1995) Biochemistry 34, 5627–5634 [DOI] [PubMed] [Google Scholar]
- 38.Hofmann H., Schmidt H. H. (1995) Biochemistry 34, 13443–13452 [DOI] [PubMed] [Google Scholar]
- 39.Stuehr D. J., Kwon N. S., Nathan C. F. (1990) Biochem. Biophys. Res. Commun. 168, 558–565 [DOI] [PubMed] [Google Scholar]
- 40.Garcin E. D., Arvai A. S., Rosenfeld R. J., Kroeger M. D., Crane B. R., Andersson G., Andrews G., Hamley P. J., Mallinder P. R., Nicholls D. J., St-Gallay S. A., Tinker A. C., Gensmantel N. P., Mete A., Cheshire D. R., Connolly S., Stuehr D. J., Aberg A., Wallace A. V., Tainer J. A., Getzoff E. D. (2008) Nat. Chem. Biol. 4, 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiter E. R., Rodríguez M. M., Weichsel A., Montfort W. R., Bonaventura J. (2007) J. Biol. Chem. 282, 19773–19780 [DOI] [PubMed] [Google Scholar]
- 42.Rosenfeld R. J., Garcin E. D., Panda K., Andersson G., Aberg A., Wallace A. V., Morris G. M., Olson A. J., Stuehr D. J., Tainer J. A., Getzoff E. D. (2002) Biochemistry 41, 13915–13925 [DOI] [PubMed] [Google Scholar]
- 43.Lowe-Ma C. K., Fischer J. W., Willer R. L. (1990) Acta Crystallogr. C 46, 1853–1859 [Google Scholar]
- 44.Levine R. L., Berlett B. S., Moskovitz J., Mosoni L., Stadtman E. R. (1999) Mech. Ageing Dev. 107, 323–332 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y. Y., Xu A. M., Nomen M., Walsh M., Keaney J. F., Jr., Loscalzo J. (1996) J. Biol. Chem. 271, 14271–14279 [PubMed] [Google Scholar]
- 46.Meister A., Anderson M. E. (1983) Annu. Rev. Biochem. 52, 711–760 [DOI] [PubMed] [Google Scholar]
- 47.Mayer B., Pfeiffer S., Schrammel A., Koesling D., Schmidt K., Brunner F. (1998) J. Biol. Chem. 273, 3264–3270 [DOI] [PubMed] [Google Scholar]
- 48.Woodward J. J., Nejatyjahromy Y., Britt R. D., Marletta M. A. (2010) J. Am. Chem. Soc. 132, 5105–5113 [DOI] [PubMed] [Google Scholar]
- 49.Wei C. C., Wang Z. Q., Hemann C., Hille R., Stuehr D. J. (2003) J. Biol. Chem. 278, 46668–46673 [DOI] [PubMed] [Google Scholar]
- 50.Hurshman A. R., Krebs C., Edmondson D. E., Huynh B. H., Marletta M. A. (1999) Biochemistry 38, 15689–15696 [DOI] [PubMed] [Google Scholar]
- 51.Li D., Hayden E. Y., Panda K., Stuehr D. J., Deng H., Rousseau D. L., Yeh S. R. (2006) J. Biol. Chem. 281, 8197–8204 [DOI] [PubMed] [Google Scholar]
- 52.Kim S. F., Huri D. A., Snyder S. H. (2005) Science 310, 1966–1970 [DOI] [PubMed] [Google Scholar]
- 53.Chen Y., Panda K., Stuehr D. J. (2002) Biochemistry 41, 4618–4625 [DOI] [PubMed] [Google Scholar]
- 54.Panda K., Adak S., Aulak K. S., Santolini J., McDonald J. F., Stuehr D. J. (2003) J. Biol. Chem. 278, 37122–37131 [DOI] [PubMed] [Google Scholar]
- 55.MacMicking J., Xie Q. W., Nathan C. (1997) Annu. Rev. Immunol. 15, 323–350 [DOI] [PubMed] [Google Scholar]
- 56.Wei C. C., Wang Z. Q., Arvai A. S., Hemann C., Hille R., Getzoff E. D., Stuehr D. J. (2003) Biochemistry 42, 1969–1977 [DOI] [PubMed] [Google Scholar]
- 57.Pant K., Crane B. R. (2006) Biochemistry 45, 2537–2544 [DOI] [PubMed] [Google Scholar]
- 58.Slama-Schwok A., Négrerie M., Berka V., Lambry J. C., Tsai A. L., Vos M. H., Martin J. L. (2002) J. Biol. Chem. 277, 7581–7586 [DOI] [PubMed] [Google Scholar]
- 59.Reif A., Fröhlich L. G., Kotsonis P., Frey A., Bömmel H. M., Wink D. A., Pfleiderer W., Schmidt H. H. (1999) J. Biol. Chem. 274, 24921–24929 [DOI] [PubMed] [Google Scholar]
- 60.Kers J. A., Wach M. J., Krasnoff S. B., Widom J., Cameron K. D., Bukhalid R. A., Gibson D. M., Crane B. R., Loria R. (2004) Nature 429, 79–82 [DOI] [PubMed] [Google Scholar]
- 61.Kirsch M., Korth H. G. (2007) Org. Biomol. Chem. 5, 3889–3894 [DOI] [PubMed] [Google Scholar]
- 62.Sonnenschein K., de Groot H., Kirsch M. (2004) J. Biol. Chem. 279, 45433–45440 [DOI] [PubMed] [Google Scholar]
- 63.Foster M. W., McMahon T. J., Stamler J. S. (2003) Trends. Mol. Med. 9, 160–168 [DOI] [PubMed] [Google Scholar]
- 64.Gorren A. C., Mayer B. (2002) Curr. Drug Metab. 3, 133–157 [DOI] [PubMed] [Google Scholar]
- 65.Harbrecht B. G., Di Silvio M., Chough V., Kim Y. M., Simmons R. L., Billiar T. R. (1997) Ann. Surg. 225, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dijkers P. F., O'Farrell P. H. (2009) Mol. Biol. Cell 20, 4083–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venketaraman V., Dayaram Y. K., Amin A. G., Ngo R., Green R. M., Talaue M. T., Mann J., Connell N. D. (2003) Infect. Immun. 71, 1864–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ignarro L. J., Lippton H., Edwards J. C., Baricos W. H., Hyman A. L., Kadowitz P. J., Gruetter C. A. (1981) J. Pharmacol. Exp. Ther. 218, 739–749 [PubMed] [Google Scholar]
- 69.Stamler J. S., Simon D. I., Osborne J. A., Mullins M. E., Jaraki O., Michel T., Singel D. J., Loscalzo J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perry J. J., Fan L., Tainer J. A. (2007) Neuroscience 145, 1280–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.