Abstract
Matriptase, a membrane-tethered serine protease, plays essential roles in epidermal differentiation and barrier function, largely mediated via its activation of prostasin, a glycosylphosphatidylinositol-anchored serine protease. Matriptase activity is tightly regulated by its inhibitor hepatocyte growth factor activator inhibitor-1 (HAI-1) such that free active matriptase is only briefly available to act on its substrates. In the current study we provide evidence for how matriptase activates prostasin under this tight control by HAI-1. When primary human keratinocytes are induced to differentiate in a skin organotypic culture model, both matriptase and prostasin are constitutively activated and then inhibited by HAI-1. These processes also occur in HaCaT human keratinocytes when matriptase activation is induced by exposure of the cells to a pH 6.0 buffer. Using this acid-inducible activation system we demonstrate that prostatin activation is suppressed by matriptase knockdown and by blocking matriptase activation with sodium chloride, suggesting that prostatin activation is dependent on matriptase in this system. Kinetics studies further reveal that the timing of autoactivation of matriptase, prostasin activation, and inhibition of both enzymes by HAI-1 binding are closely correlated. These data suggest that, during epidermal differentiation, the matriptase-prostasin proteolytic cascade is tightly regulated by two mechanisms: 1) prostasin activation temporally coupled to matriptase autoactivation and 2) HAI-1 rapidly inhibiting not only active matriptase but also active prostasin, resulting in an extremely brief window of opportunity for both active matriptase and active prostasin to act on their substrates.
Keywords: Cell Surface Enzymes, Protease, Protease Inhibitor, Proteolytic Enzymes, Serine Protease
Introduction
Matriptase, a type II transmembrane serine protease, plays essential roles in epidermal differentiation and barrier function (1, 2). Mutations of matriptase are associated with autosomal recessive ichthyosis and hypotrichosis (3). Four mutations have been identified in ST14, the gene that encodes for matriptase (3–5). Targeted deletion of matriptase in mice also causes severe epidermal defects with impairment of desquamation, lipid matrix formation, and profilaggrin processing. These defects lead to compromised epidermal barrier function and postnatal death (2). The severe epidermal defects appear to result from the lack of activation of the glycosylphosphatidylinositol-anchored serine protease prostasin by matriptase, because both matriptase-deficient and prostasin-deficient mice share an almost identical pattern of epidermal defects. Furthermore, lack of prostasin activation was observed in matriptase-deficient mouse skin (6, 7), suggesting that much of the function of matriptase in the epidermis is manifested through the activation of prostasin.
Hepatocyte growth factor activator inhibitor-1 (HAI-1)2 is a transmembrane Kunitz-type serine protease inhibitor (8). HAI-1 can inhibit matriptase in a competitive and reversible manner, consistent with the characteristics of Kunitz-type serine protease inhibitors (9). What is unusual, however, is its high efficiency of matriptase inhibition by HAI-1. As a consequence of this efficiency, free active matriptase is rapidly inactivated by HAI-1 binding and can only act on its downstream substrates for extremely short time periods after activation (10–12). This unusually efficient inhibition results not only from the high inhibitory potency of HAI-1 (13) and the subcellular co-localization of enzyme and inhibitor (11) but also from several uncommon biochemical mechanisms. For example, most matriptase-expressing epithelial cells express HAI-1 in significantly higher levels compared with the levels of matriptase, typically in excess of 10-fold molar excess.3 In the absence of HAI-1, newly synthesized matriptase accumulates within the endoplasmic reticulum/Golgi, which results in very low levels of matriptase being expressed (11). HAI-1 may also participate in matriptase zymogen activation, because matriptase activation is significantly reduced when point mutations are introduced into the LDL receptor class A domain of HAI-1, or this domain is deleted (14). This unusual relationship between matriptase and HAI-1 appears to result in two unique consequences: 1) the very short-lived availability of free active matriptase, due to the direct and rapid access of HAI-1 to the newly generated active matriptase, and 2) a paradoxical reduction, rather than increase in matriptase activity in the absence of HAI-1. The short-lived availability of active matriptase suggests that activation of the downstream matriptase substrate must be coupled with matriptase's own activation. The paradoxical decrease in matriptase function associated with HAI-1 deficiency appears to be consistent with animal studies in which the epidermal defects seen in HAI-1-deficient mice resemble those in matriptase hypomorphic mice (15, 16).
Although data from genetic manipulation models and studies of tissue distribution clearly demonstrate a close functional linkage between matriptase and prostasin and their essential roles in epidermal function, the details of how and when the activation of prostasin by active matriptase occurs, remain largely unknown. In the current study, we examine the expression and activation state of matriptase and prostasin during epidermal differentiation using an organotypic skin culture model and dissect the molecular events associated with the activation and inactivation of these enzymes. Our results demonstrate that matriptase and prostasin are rapidly and constitutively activated throughout the course of epidermal differentiation, consistent with their roles in skin function. More importantly, although HAI-1 rapidly inhibits active matriptase, immediately after matriptase zymogen activation, the active matriptase apparently has no difficulty activating prostasin in the brief window of opportunity, prior to its inactivation by HAI-1. Unexpectedly, active prostasin appears to follow the same fate as active matriptase and is rapidly inactivated by binding with HAI-1. These data suggest that epidermal keratinocytes regulate matriptase-initiated cell surface proteolysis with unprecedentedly tight control to safeguard themselves from the proteolytic activities of matriptase and prostasin.
EXPERIMENTAL PROCEDURES
Antibodies
Two types of matriptase antibodies were used: the mouse monoclonal antibodies (mAbs) M32 and M24 and rat mAb 21-9 recognize both latent and activated forms of matriptase (17–19); mAb M69 recognizes an epitope present only on activated matriptase and is, therefore, able to distinguish activated matriptase from latent matriptase (9, 20). Human HAI-1 protein was detected using the HAI-1 mAb M19 (17). Two prostasin antibodies were used: a prostasin polyclonal antibody, described previously (21, 22), which is able to recognize both activated prostasin in complex with HAI-1 and uncomplexed prostasin, which is presumably the latent form of the enzyme. Interestingly, this antibody appears to recognize the prostasin-HAI-1 complex better than latent prostasin. The second antibody was a prostasin mAb (BD Biosciences, Franklin Lakes, NJ) that only recognizes prostasin after heat-denaturing treatment, which results in the dissociation of the prostasin-HAI-1 complex.
Lentiviral Particle Preparation and Cell Transduction for the Delivery of shRNAs
Matriptase-targeting and non-targeting control small hairpin RNAs (shRNA) in the vector PLKO.1-puro were purchased from Open Biosystems (Huntsville, AL). The shRNA constructs were packaged into lentiviral particles using 293T cells using standard methods. Briefly, on day 1, 293T cells were switched to fresh media without antibiotics 3 h prior to transfection. Ten micrograms of the shRNA plasmids were mixed with 5 μg of vesicular stomatitis virus glycoprotein, 5 μg of d89, 50 μl of CaCl2, and water to a total volume of 500 μl. The mixture was then mixed with 500 μl of 2× Hebs buffer (dextrose 2.0 g/liter, HEPES, 10 g/liter, KCI 0.74 g/liter, NaCl 16 g/liter, Na2HPO4.2H2O 0.27 g/liter and incubated at room temperature for 20 min. The resultant mixture was then added dropwise onto a 100-mm dish of 293T cells, and the cells were incubated overnight at 37 °C, with 5% CO2. The following day, the transfection media were replaced with fresh media, and the cultures were again incubated overnight. On the third day, conditioned media were harvested from the plates and filtered through a 22-μm polyethersulfone syringe-driven filter unit.
Cell Culture and Media
The immortalized human keratinocyte line HaCaT was cultured in DMEM (Cellgro, Manassas, VA) supplemented with 10% FBS (Gemini, West Sacramento, CA), 100 units/ml penicillin, and 100 μg/ml streptomycin. Primary human keratinocytes isolated from neonatal foreskin tissue were cultured in keratinocyte serum-free media (Invitrogen) supplemented with 25 μg/ml bovine pituitary extract and 1.5 ng/ml of recombinant epidermal growth factor. Cultures were maintained in a humidified 5% CO2 atmosphere at 37 °C.
Organotypic Culture
Organotypic raft cultures were generated based on the method described previously (23, 24). Collagen (type I rat tail, BD Biosciences, Bedford, MA) plugs with 105 J2–3T3 fibroblasts per milliliter were prepared in 6-well cell culture inserts (0.4-μm pore size, PET track-etched membrane, BD Biosciences), and incubated for 1 day. A total of 106 primary human keratinocytes was plated on top of each plug in keratinocyte serum-free media supplemented with 25 μg/ml bovine pituitary extract and 1.5 ng/ml recombinant epidermal growth factor. After 2 days, primary human keratinocyte differentiation was initiated by lifting the organotypic cultures to the air-liquid interface and changing the culture medium to keratinocyte serum-free media supplemented with 25 μg/ml bovine pituitary extract, 1.5 ng/ml recombinant epidermal growth factor. The medium was changed every second day for the next 8 days.
Acid-induced Matriptase Activation in HaCaT Cells
Matriptase activation was induced by exposing HaCaT cells to a 0.15 m phosphate buffer (PB), pH 6.0, for 30 min at room temperature, as described previously (18). For the activation kinetics assay, the cells were incubated with PB for various times as indicated in the figure. Cell lysates were harvested and subjected to immunoblot analysis.
Immunoblot Analysis
Proteins for immunoblot analysis were prepared from the organotypic cultures and regular cell cultures by washing the cells three times with phosphate-buffered saline (PBS, Cellgro), followed by dissolving cells in 1% Triton X-100 in PBS at 4°C for 5 min. Insoluble debris was removed by centrifugation. The protein concentrations in these lysates were determined using the BCA protein assay reagents (Pierce) according to the manufacturer's instructions. Samples containing the same amount of total protein were mixed with 5× sample buffer and resolved by 7.5% SDS-PAGE and then transferred to nitrocellulose membrane (Pall Corp., Pensacola, FL). The membranes were probed with the appropriate antibodies and an HRP-conjugated secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD), before signal detection with Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences).
Prostasin-HAI-1 Complex Purification
HaCaT cells were treated with PB for 30 min at room temperature to activate prostasin and induce prostasin-HAI-1 complex formation. After activation, the cells were washed three times with PBS and lysed with 1% Triton X-100 in PBS. The cell lysates were centrifuged to remove insoluble debris and then dialyzed against 20 mm Tris-HCl, pH 8.0, containing 1% Triton X-100 (buffer A). The dialyzed sample was loaded onto a DEAE-Sepharose Fast Flow column (Amersham Biosciences) pre-equilibrated with buffer A. The DEAE column was washed with 5 column volumes of buffer A, and proteins were eluted with a linear gradient of 0 to 0.5 m NaCl in buffer A. The eluted fractions containing the prostasin-HAI-1 complex were pooled and loaded onto a HAI-1 mAb M19 immunoaffinity column pre-equilibrated with buffer A. The column was washed with buffer A, and bound protein was then eluted with 0.1 n glycine-HCl, pH 2.4. The eluate fractions were neutralized immediately after collection with 2 m Trizma Base.
Mass Spectrometry Analysis and Identification of Prostasin
The protein bands from SDS-PAGE gels stained with Protoblue (National Diagnostics, Atlanta, GE) were excised, washed, destained, and trypsinized overnight at 37 °C using standard protocols after dithiothreitol reduction and iodoacetamide alkylation. Analysis of the tryptic peptides derived from the protein samples was performed by liquid chromatography/mass spectrometry (LS/MS) using services provided by Prottech Inc. (Norristown, PA).
RESULTS
Activation of Matriptase Starts Early, and Is Maintained throughout, in Vitro Human Epidermal Differentiation
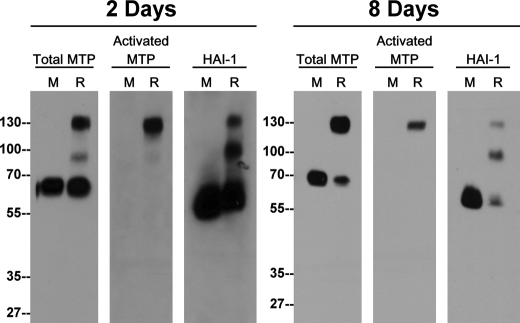
Epidermal differentiation involves a host of proteases and protease inhibitors of which matriptase is an essential player in the development of skin barrier function. Here we used a three-dimensional organotypic skin cultures to examine the functional status of matriptase during epidermal differentiation. When cultured as a monolayer, primary human keratinocytes express matriptase in its mature latent form, yielding a 70-kDa protein band on immunoblots using matriptase mAb M24 (Fig. 1, Total MTP, lanes M). The cells are devoid of activated matriptase as shown by the lack of a band recognized by the mAb M69, which can specifically distinguish active matriptase from latent matriptase (Fig. 1, Activated MTP, lanes M). The cognate inhibitor of matriptase, HAI-1, is also expressed by human keratinocytes in monolayer culture, detectable in its un-complexed form at 55-kDa detected by HAI-1 mAb M19 (Fig. 1, HAI-1, lanes M).
FIGURE 1.
Detection of a novel HAI-1 complex in differentiating human keratinocytes. Primary human keratinocytes were grown in monolayer (M) or in a skin organotypic culture system (R). Cells were harvested on the second or eighth days after they were lifted to the air-liquid interface and analyzed by immunoblot for total matriptase (Total MTP), activated matriptase (activated MTP), and HAI-1.
To study how matriptase is regulated during epidermal differentiation, epidermal organotypic cultures were established using primary human keratinocytes grown on collagen lattices prepared with fibroblasts. Two days after being lifted to the air-liquid interface, significant activation of matriptase occurs, as indicated by the appearance of a 120-kDa (activated) matriptase-HAI-1 complex, which can be detected by all three mAbs: M24, M69, and M19 (Fig. 1, 2 days, lanes R). These data suggest that matriptase activation occurs at an early stage of epidermal differentiation. Keratinocytes appear to employ HAI-1 to inhibit matriptase in a fashion identical to that of other types of epithelial cell, with the active matriptase being rapidly inactivated by the formation of a 120-kDa matriptase-HAI-1 complex. In addition to this species, however, we noticed that differentiating keratinocytes also form an 85-kDa HAI-1 complex (Fig. 1, 2 days, HAI-1, lane R). The level of the 85-kDa HAI-1 complex detectable in the lysates appeared to be comparable to that of the 120-kDa matriptase-HAI-1 complex in epidermal keratinocytes, and yet an 85-kDa HAI-1 complex has not been noticed or reported in other epithelial systems, such as 184 A1N4 mammary epithelial cells suggesting that this species is not present in other epithelial systems (18). Because HAI-1 only binds and forms stable complexes with active but not latent serine proteases, the appearance of the 85-kDa HAI-1 complex suggests that a second serine protease may be being activated and then bound by HAI-1 in differentiating keratinocytes. The matriptase-HAI-1 complex and 85-kDa HAI-1 complex were also detected in raft culture lysates prepared on the 8th day of culture (Fig. 1, 8 Days), suggesting that the activation of matriptase and the putative second serine protease may be maintained during the course of epidermal differentiation.
Acid Exposure Induces Simultaneous Activation of Matriptase and a Putative Serine Protease in HaCaT Cells
The appearance of the 85-kDa HAI-1 complex throughout the course of keratinocyte differentiation in the organotypic skin culture model might be related to activation of matriptase in a keratinocyte-selective manner. Previously we discovered using other epithelial cells that robust matriptase activation can rapidly be induced by simply exposing the cells to mildly acidic conditions (12, 18). We, therefore, tested if this would occur in keratinocytes by exposing HaCaT cells to a pH 6.0 buffer and found that acid exposure of these cells resulted in the rapid conversion of the 70-kDa matriptase zymogen to the 120-kDa matriptase-HAI-1 complex (Fig. 2, Total MTP and Activated MTP, compare lanes 3 to lanes 1), suggesting that keratinocytes indeed share this activation mechanism with other epithelial cells. In addition to the 120-kDa matriptase-HAI-1 complex, the 85-kDa HAI-1 complex was also detected in these cells in response to the mild acid exposure (Fig. 2, HAI-1, lane 3).
FIGURE 2.
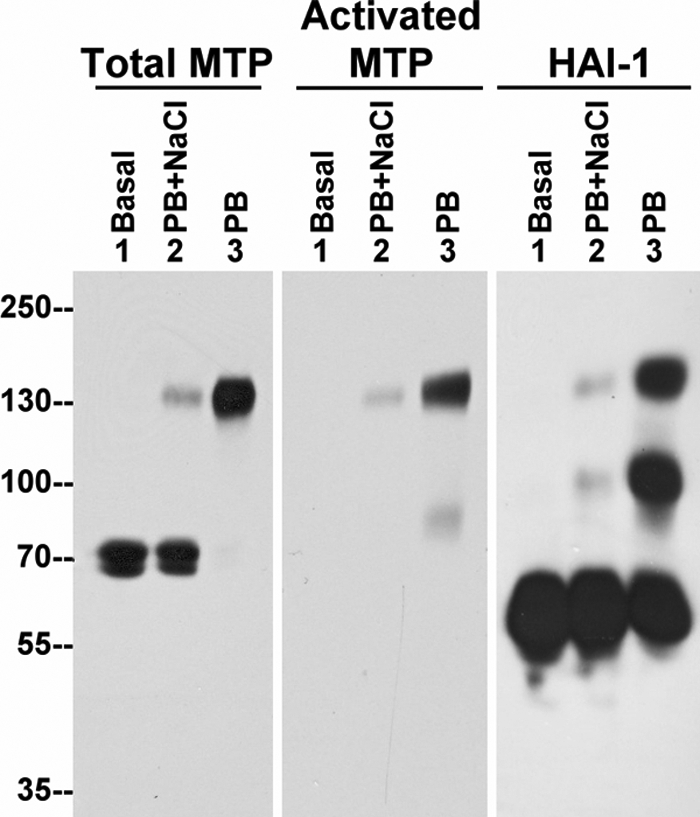
Acid-induced activation of matriptase is associated with the formation of a novel 85-kDa HAI-1 complex in immortalized human keratinocytes. HaCaT cells were incubated with basal media (Basal, lane 1) or phosphate buffer (PB), pH 6.0, with 0.15 m NaCl (PB+NaCl, lane 2) or PB, pH 6.0, alone (PB, lane 3) for 30 min. Cell lysates were extracted and subjected to immunoblot analyses for total matriptase (Total MTP), activated matriptase (Activated MTP), and HAI-1.
Acid-driven matriptase activation is thought to result from an unusual feature of the matriptase zymogen. The intrinsic activity of the zymogen form matriptase is greatly enhanced at a mildly acidic pH, and this may contribute to matriptase autoactivation (25). Furthermore, matriptase zymogen activity can also be inhibited by elevated concentrations of sodium chloride and so can the autoactivation of matriptase in 184 A1N4 mammary epithelial cells (12, 25). The HaCaT cells also appear to exhibit the NaCl-mediated inhibition of acid-induced matriptase activation, because the levels of the 120-kDa matriptase-HAI-1 complex were significantly reduced in the presence of increased NaCl (Fig. 2, compare lanes 2 to lanes 3). Interestingly, sodium chloride also significantly suppressed formation of the 85-kDa HAI-1 complex (Fig. 2, HAI-1, compare lane 2 to lane 3). These data suggest that formation of the 85-kDa HAI-1 complex may be an event common to keratinocytes and that its appearance is associated with matriptase activation.
Purification of the 85-kDa HAI-1 Complex and Identification of Prostasin as a Component of the Complex
To identify the putative serine protease that binds to HAI-1 to form the 85-kDa complex, the complex was isolated from cell lysates prepared from acid-exposed HaCaT cells using a combination of DEAE chromatography, immunodepletion to remove the 120-kDa matriptase-HAI-1 complex using immobilized matriptase mAb 21-9, and immunoaffinity chromatography using immobilized HAI-1 mAb M19. Following these procedures, the 85-kDa HAI-1 complex was visualized by Protoblue protein staining of an SDS gel under non-boiled and non-reducing conditions (Fig. 3A, lane 1, a). Heat-denaturing the sample resulted in dissociation of the 85-kDa HAI-1 complex, and was accompanied by the appearance of bands at 55 and 35 kDa (Fig. 3A, lane 2, b and c, respectively). The heat-sensitive nature of the 85-kDa complex is consistent with the non-covalent interactions between serine proteases and Kunitz-type serine protease inhibitors. In-gel trypsin digestion and MS-based protein identification revealed that the 85-kDa protein band contained HAI-1, as expected, and the serine protease prostasin (Fig. 3B). The constituents of 85-kDa prostasin-HAI-1 complex were further confirmed by MS-based protein identification of the bands yielded by heat denaturation of the 85-kDa complex with the 35-kDa band being confirmed as prostasin and the 55-kDa band as HAI-1 (Fig. 3B). Immunoblot analysis using a prostasin antibody also confirmed that the 85-kDa HAI-1 complex contained prostasin under non-boiled and non-reducing conditions (Fig. 3C, lane 1), and that the 35-kDa protein band released from heat-denaturing was indeed prostasin (Fig. 3C, lane 2). These data and those described above suggest that keratinocytes simultaneously activate matriptase and prostasin throughout the course of epidermal differentiation, and in response to acid exposure, and that both active matriptase and active prostasin are rapidly inhibited by HAI-1.
FIGURE 3.
Identification of prostatin as a component of the novel 85-kDa HAI-1 complex. A, partially purified 85-kDa HAI-1 complex was analyzed by SDS-PAGE under either non-reducing and non-boiled conditions (NRNB) or under non-reducing and boiled conditions (NR, B), and the protein bands were visualized by staining with Protoblue. B, the protein bands indicated by a, b, and c in A were subjected to proteomic protein identification by MS/MS. Among the tryptic peptides obtained from protein band a, 12 peptides matched to HAI-1 and 6 peptides matched to prostasin. Thirteen peptides obtained from band b matched to HAI-1, and three from band c matched to prostasin. C, the purified 85-kDa HAI-1 complex was analyzed by immunoblot for prostasin under either non-reducing and non-boiled conditions (NRNB) or non-reducing and boiled conditions (NR, B) using a prostasin polyclonal antibody.
Prostasin Activation Is Dependent on Matriptase
We next set out to determine if the activation of prostasin in keratinocytes simply occurs at the same time as matriptase activation, or if its activation is coupled to, and dependent on matriptase activation. As shown in Fig. 2, using the HAI-1 mAb, the formation of the prostasin-HAI-1 complex can be induced by exposing HaCaT cells to a pH 6.0 buffer, and NaCl can suppress complex formation. Using a prostasin polyclonal antibody, we confirmed that the 85-kDa complex was not detected in HaCaT keratinocytes before acid exposure (Fig. 4, lane 1). In response to the acid exposure, the appearance of the 85-kDa complex can be clearly detected by the prostasin polyclonal antibody (Fig. 4, lane 3), and its appearance is suppressed by the inclusion of NaCl in the pH 6.0 buffer (Fig. 4, lane 2), recapitulating the data generated with the HAI-1 mAb shown in Fig. 2. Because HAI-1 only binds to active serine proteases, formation of the prostasin-HAI-1 complex should be at the cost of latent prostasin, in a situation similar to the disappearance of 70-kDa latent matriptase and the concomitant appearance of 120-kDa matriptase-HAI-1 complex (Fig. 2). The prostasin polyclonal antibody used in the current study appears to be less sensitive to latent prostasin than to prostasin-HAI-1 complex. Latent prostasin with a size close to the 35-kDa protein marker was clearly detected in HaCaT cells only after exposing the nitrocellulose membrane to x-ray film for a longer time (Fig. 4, lane 1 in low panel). Nevertheless, the conversion of prostasin zymogen to active prostasin and inactivation of active prostasin by the formation of the 85-kDa HAI-1 complex results in the levels of latent prostasin being significantly decreased in parallel with the formation of the 85-kDa complex (Fig. 4, low panel, compare lane 3 with lane 1). The reduction of zymogen activation caused by the NaCl (Fig. 4, lane 2 in low panel) results in latent prostasin being detected at intermediate levels (Fig. 4, lower panel) consistent with the partial suppression of the appearance of the 85-kDa complex (Fig. 4, lower and upper panel). These data clearly demonstrate that prostasin undergoes activation and HAI-1 complexation in keratinocytes in response to acid exposure and that both matriptase and prostasin activation are tightly coupled.
FIGURE 4.
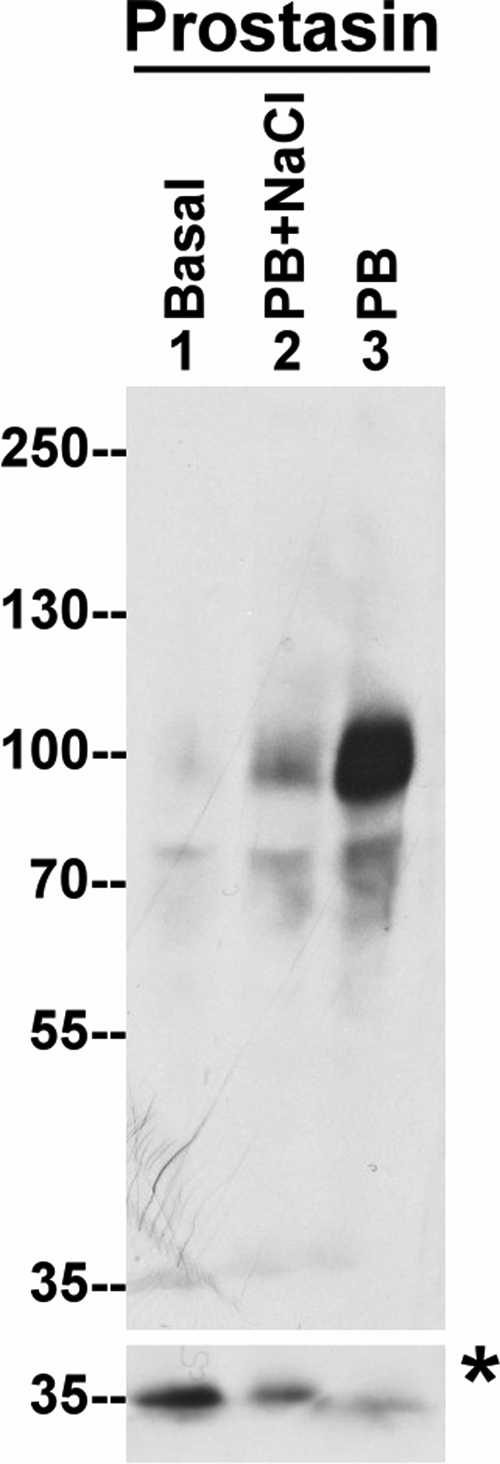
Acid exposure of HaCaT keratinocytes results in the activation of prostasin associated with matriptase activation. HaCaT cells were incubated with basal media (Basal, lane 1) or phosphate buffer, pH 6.0, with 0.15 m NaCl (PB+NaCl, lane 2) or phosphate buffer, pH 6.0 (PB, lane 3) for 30 min. Cell lysates were extracted and subjected to immunoblot analyses for prostasin using a prostasin polyclonal antibody. The lower panel (marked with an asterisk) containing proteins near the 35-kDa protein marker shows a longer exposure of the x-film to show the levels of latent prostasin.
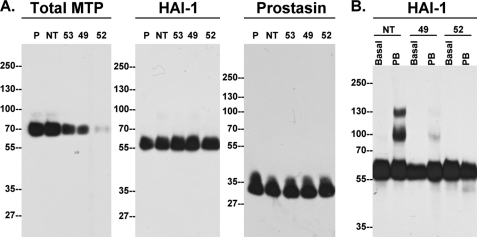
Matriptase has been shown to become active through an autoactivation process (14), and prostasin activation does not occur in the skin of matriptase-deficient mice (6). This suggests that matriptase may serve as an upstream activator of prostasin. To test this hypothesis we first generated three HaCaT clones in which the level of matriptase expression had been suppressed to various degrees by transduction with lentiviruses carrying matriptase-targeting shRNAs, or non-targeting controls (Fig. 5A, Total MTP, 49, 52, 53, and NT). The levels of HAI-1 and prostasin were not altered in these clones when compared with the levels in the parental and non-target control cells (Fig. 5A, HAI-1 and Prostasin). For better detection of prostasin, a commercially available prostasin monoclonal antibody was used, which only detects the protease after heat-denaturation of the sample prior to SDS-PAGE, which results in dissociation of the prostasin-HAI-1 complex. When matriptase activation was induced by acid exposure of the cells, formation of the 85-kDa prostasin-HAI-1 complex along with 120-kDa matriptase-HAI-1 complex was clearly observed in the non-target control clone (Fig. 5B, NT, compare lane 2 to lane 1), but was barely detectible in the matriptase knockdown clones 49 and 52 (Fig. 5B, 49 and 52). These data clearly demonstrate that prostasin activation depends on matriptase proteolytic action, providing key evidence for a cascaded linkage between matriptase and prostasin activation.
FIGURE 5.
Prostasin activation is dependent on matriptase. A, generation of matriptase knockdown HaCaT cells. HaCaT cells were infected with lentiviruses bearing three different matriptase-targeting shRNAs and a non-targeting shRNA. Stable clones were selected with puromycin, and cell lysates were analyzed by immunoblot for total matriptase (Total MTP), HAI-1, and prostasin. Total matriptase and HAI-1 were detected under non-reducing and non-boiled conditions. Prostasin was detected under reduced and boiled conditions using a prostasin monoclonal antibody. B, prostasin activation is dependent on matriptase. The matriptase knockdown clones (49 and 52) and the non-target control (NT) cells were either exposed to basal medium (Basal) or phosphate buffer, pH 6.0 (PB), for 30 min to induce activation of matriptase and prostasin. Formation of the prostasin-HAI-1 complex, as an indication of prostasin activation, was analyzed by immunoblot using HAI-1 mAb M19.
Matriptase and Prostasin Activation Kinetics in HaCaT Cells
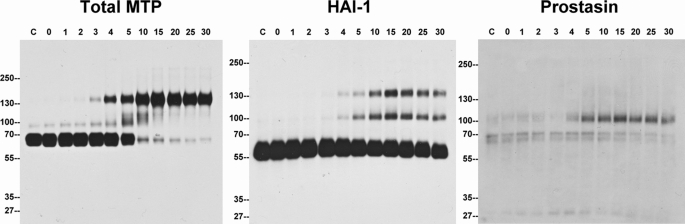
We further established the “lockstep” relationship between matriptase activation and prostasin activation by comparing their respective activation kinetics in response to acid exposure. As rapidly as 3–4 min after starting acid exposure, matriptase began to become activated, with the active enzyme being immediately inhibited by binding to HAI-1 with the appearance of the 120-kDa matriptase-HAI-1 complex detected by both matriptase and HAI-1 mAbs (Fig. 6, Total MTP and HAI-1). Matriptase activation reached a plateau after ∼10–15 min of acid exposure as there was almost no latent matriptase left at this point (Fig. 6, Total MTP). The appearance of the 85-kDa prostasin-HAI-1 complex followed similar kinetics to the appearance of the 120-kDa matriptase-HAI-1 complex demonstrated using the HAI-1 mAb M19 (Fig. 6, HAI-1). The kinetics of prostasin-HAI-1 complex formation can also be followed using the prostasin polyclonal antibody (Fig. 6, Prostasin). These kinetics studies demonstrate that matriptase activation, prostasin activation, and inhibition of both proteases by HAI-1 occur at almost the same time in human keratinocytes. These data provide evidence that, despite the extremely tight control by HAI-1 and the short period in which free active matriptase is present, it is able to activate its downstream substrate, prostasin.
FIGURE 6.
Activation and HAI-1 binding kinetics of the matriptase-prostasin proteolytic cascade. HaCaT cells were treated with either basal medium (C) or phosphate buffer, pH 6.0, and harvested at indicated time points (minutes). Cell lysates were prepared by lysis with 1% Triton X-100 in PBS and followed by immunoblot analyses for total matriptase (Total MTP), HAI-1, and prostasin.
DISCUSSION
In the present study we investigate how a matriptase-initiated cell surface proteolytic cascade is regulated throughout the course of epidermal differentiation. The proteolytic cascade is constitutively activated from an early stage of epidermal differentiation, and the active enzymes generated are immediately inhibited by binding to HAI-1. The constitutive activation of the system followed by the immediate HAI-1-mediated inhibition of both matriptase and prostasin suggests that the proteolytic cascade not only plays an important role in epidermal differentiation but that its activity must also be under tight control. Our data clearly demonstrate that matriptase is able to act on its substrate even in the presence of overwhelming levels of HAI-1. The rapid inhibition of active prostasin by HAI-1 further indicates that the tight regulation of the matriptase-prostasin cascade lies not only at the level of the initiator protease matriptase but also at the secondary protease prostasin. With such a tightly regulatory mechanism that only allows the first two proteases of the cascade to have an extremely limited opportunity to act on their downstream substrates, it is reasonable to suppose that deregulation of the cascade at either one of the two proteases or at their cognate inhibitor HAI-1 would affect the entire protease cascade, and in turn derail epidermal differentiation.
Although several protein substrates have been identified for matriptase based on the cleavage preferences of the enzyme, prostasin is the first matriptase substrate for whose activation can be demonstrated during the course of matriptase's own activation and in the context of rapid HAI-1-mediated inhibition of active matriptase. The functional linkage between matriptase and prostasin activation is consistent with the observation that prostasin activation does not occur in the skin of matriptase-deficient mice (6). The very similar epidermal defects that are observed in matriptase-deficient and prostasin-deficient mice strongly suggest that prostasin may be the predominant substrate of matriptase in this tissue functional context, and that the lack of prostasin activation may be responsible for the majority of the defects associated with matriptase deletion. Despite their close relationship in skin biology, matriptase and prostasin may not function in tandem in other epithelial cells, or at least their functional linkage in other epithelial cells is not as straightforward as in keratinocytes. During the development and progression of squamous cells carcinoma induced by initiation with 7,12-dimethylbenzanthracene and promotion with phorbol 12-myristate 13-acetate, matriptase and prostasin are eventually expressed in separate parts of the tumors (26). In polarized epithelial cells grown in moist environments, matriptase is targeted to basolateral plasma membrane and prostasin, a glycosylphosphatidylinositol-anchored protein, is likely targeted to the apical plasma membrane (27). Although a glycosylphosphatidylinositol-anchored protein might be targeted to the apical surface after transiently moving to the basolateral plasma membrane, it is conceivable that the different subcellular localizations of matriptase and prostasin in polarized epithelial cells renders the ability of matriptase to activate prostatin less relevant. Different mechanisms are likely employed by other epithelial cells to activate prostasin, if matriptase is not the activator. Furthermore, the rapid inhibition of prostasin by HAI-1 also may not be a universal mechanism. Prostasin was initially isolated from the seminal fluid as an active protease, but not in an HAI-1 complex (28). The epidermis is a specialized epithelium designed to encounter the terrestrial environment, and although keratinocytes resemble other epithelial types, keratinocytes undergo progressive remodeling of cell morphology and tissue structure during epidermal differentiation. These cellular remodeling events may alter the subcellular distribution of matriptase and prostasin and allow matriptase and prostasin to team up to form a relatively keratinocyte-selective cell surface protease cascade.
The epithelial sodium channel (ENaC), a membrane-bound ion channel, is located in the apical membrane of polarized epithelial cells (29). Both prostasin and matriptase have been demonstrated to activate ENaC, resulting in their alternative names as mouse channel activating proteases 1 and 2 (mCAP 1 and mCAP 3, respectively) (30). Because matriptase is targeted to the basolateral membrane in polarized epithelial cells where it is activated and immediately inhibited by HAI-1 binding (31), it is of interest to determine whether or how matriptase could activate the apically localized ENaC. In contrast to matriptase, prostasin is co-localized with ENaC at the apical plasma membrane, and this co-localization makes prostasin a more likely candidate protease activator for ENaC in polarized epithelial cells. In contrast to some non-keratinocyte epithelial cells in which the inhibition of prostasin by HAI-1 seems less effective, active prostasin in keratinocytes much like active matriptase is a short-lived species. It remains to be determined whether prostasin is still an effective activator of ENaC in keratinocytes (32).
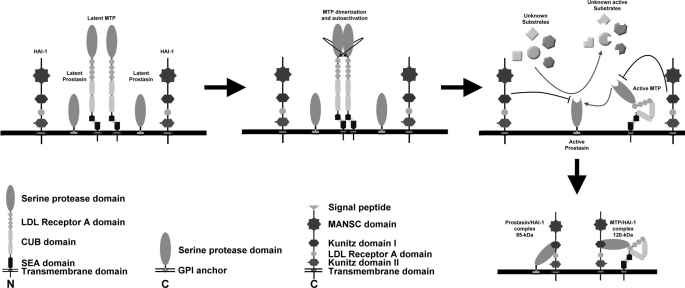
In conclusion, the matriptase-prostasin proteolytic cascade, a relatively keratinocyte-selective, and tightly regulated cell surface proteolysis system, is essential for epidermal differentiation. Both matriptase and prostasin zymogens and their cognate inhibitor HAI-1 are likely co-localized and anchored at the cell membrane. In response to activation stimuli, such as acid exposure or signals of epidermal tissue differentiation, the matriptase zymogen undergoes autoactivation mediated via the formation a homodimer that generates active matriptase that in turn rapidly activates prostasin. Both active matriptase and active prostasin likely also rapidly activate downstream substrates before HAI-1 moves in and inhibits both proteases and suppresses the activity of the proteolytic cascade. Although HAI-1 contains two Kunitz domains, Kunitz domain 1 at the N terminus is likely responsible for the inhibition (33). We summarize these molecular events associated with activation and inactivation of the protease cascade in Fig. 7. The unprecedentedly tight control of this cell surface proteolytic cascade allows keratinocytes to undergo differentiation through a precisely programmed mechanism.
FIGURE 7.
The model for the regulation of matriptase-prostasin proteolytic cascade. A schematic model of the activation and inactivation of matriptase and prostasin is presented. Both matriptase and prostasin are synthesized as single chain zymogens and anchored on cell membrane. Matriptase undergoes autoactivation via the interaction of two zymogen molecules to generate active matriptase. Active matriptase subsequently activate prostasin and the downstream substrates. Both active matriptase and active prostasin are short-lived due to the rapid HAI-1-mediated inhibition.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA-104944 (to C.-Y. L.) and R01-CA-123223 (to M. D. J. and C.-Y. L.). This work was also supported by the Florida Biomedical Research Program (Grant 06NIR-03 to L.-M. C.) and the Susan G. Komen for the Cure (Grant BCTR0707538 to K. X. C.).
H. Xu, I.-C. Tseng, F.-P. Chou, J.-K. Wang, M. D. Johnson, and C.-Y. Lin, manuscript submitted for publication.
- HAI-1
- hepatocyte growth factor activator inhibitor-1
- ENaC
- epithelial sodium channel.
REFERENCES
- 1.List K., Haudenschild C. C., Szabo R., Chen W., Wahl S. M., Swaim W., Engelholm L. H., Behrendt N., Bugge T. H. (2002) Oncogene 21, 3765–3779 [DOI] [PubMed] [Google Scholar]
- 2.List K., Szabo R., Wertz P. W., Segre J., Haudenschild C. C., Kim S. Y., Bugge T. H. (2003) J. Cell. Biol. 163, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basel-Vanagaite L., Attia R., Ishida-Yamamoto A., Rainshtein L., Ben Amitai D., Lurie R., Pasmanik-Chor M., Indelman M., Zvulunov A., Saban S., Magal N., Sprecher E., Shohat M. (2007) Am. J. Hum. Genet. 80, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avrahami L., Maas S., Pasmanik-Chor M., Rainshtein L., Magal N., Smitt J., van Marle J., Shohat M., Basel-Vanagaite L. (2008) Clin. Genet. 74, 47–53 [DOI] [PubMed] [Google Scholar]
- 5.Alef T., Torres S., Hausser I., Metze D., Türsen U., Lestringant G. G., Hennies H. C. (2009) J. Invest Dermatol. 129, 862–869 [DOI] [PubMed] [Google Scholar]
- 6.Netzel-Arnett S., Currie B. M., Szabo R., Lin C. Y., Chen L. M., Chai K. X., Antalis T. M., Bugge T. H., List K. (2006) J. Biol. Chem. 281, 32941–32945 [DOI] [PubMed] [Google Scholar]
- 7.Leyvraz C., Charles R. P., Rubera I., Guitard M., Rotman S., Breiden B., Sandhoff K., Hummler E. (2005) J. Cell. Biol. 170, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimomura T., Denda K., Kitamura A., Kawaguchi T., Kito M., Kondo J., Kagaya S., Qin L., Takata H., Miyazawa K., Kitamura N. (1997) J. Biol. Chem. 272, 6370–6376 [DOI] [PubMed] [Google Scholar]
- 9.Benaud C., Dickson R. B., Lin C. Y. (2001) Eur. J. Biochem. 268, 1439–1447 [DOI] [PubMed] [Google Scholar]
- 10.Lee M. S., Kiyomiya K., Benaud C., Dickson R. B., Lin C. Y. (2005) Am. J. Physiol. Cell. Physiol. 288, C932–C941 [DOI] [PubMed] [Google Scholar]
- 11.Oberst M. D., Chen L. Y., Kiyomiya K., Williams C. A., Lee M. S., Johnson M. D., Dickson R. B., Lin C. Y. (2005) Am. J. Physiol. Cell. Physiol. 289, C462–C470 [DOI] [PubMed] [Google Scholar]
- 12.Lee M. S., Tseng I. C., Wang Y., Kiyomiya K., Johnson M. D., Dickson R. B., Lin C. Y. (2007) Am. J. Physiol. Cell. Physiol. 293, C95–105 [DOI] [PubMed] [Google Scholar]
- 13.Kirchhofer D., Peek M., Li W., Stamos J., Eigenbrot C., Kadkhodayan S., Elliott J. M., Corpuz R. T., Lazarus R. A., Moran P. (2003) J. Biol. Chem. 278, 36341–36349 [DOI] [PubMed] [Google Scholar]
- 14.Oberst M. D., Williams C. A., Dickson R. B., Johnson M. D., Lin C. Y. (2003) J. Biol. Chem. 278, 26773–26779 [DOI] [PubMed] [Google Scholar]
- 15.Nagaike K., Kawaguchi M., Takeda N., Fukushima T., Sawaguchi A., Kohama K., Setoyama M., Kataoka H. (2008) Am. J. Pathol. 173, 1464–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.List K., Currie B., Scharschmidt T. C., Szabo R., Shireman J., Molinolo A., Cravatt B. F., Segre J., Bugge T. H. (2007) J. Biol. Chem. 282, 36714–36723 [DOI] [PubMed] [Google Scholar]
- 17.Lin C. Y., Anders J., Johnson M., Dickson R. B. (1999) J. Biol. Chem. 274, 18237–18242 [DOI] [PubMed] [Google Scholar]
- 18.Tseng I. C., Xu H., Chou F. P., Li G., Vazzano A. P., Kao J. P., Johnson M. D., Lin C. Y. (2010) J. Biol. Chem. 285, 3261–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C. Y., Wang J. K., Torri J., Dou L., Sang Q. A., Dickson R. B. (1997) J. Biol. Chem. 272, 9147–9152 [PubMed] [Google Scholar]
- 20.Benaud C., Oberst M., Hobson J. P., Spiegel S., Dickson R. B., Lin C. Y. (2002) J. Biol. Chem. 277, 10539–10546 [DOI] [PubMed] [Google Scholar]
- 21.Chen M., Fu Y. Y., Lin C. Y., Chen L. M., Chai K. X. (2007) Biochim. Biophys. Acta. 1773, 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M., Chen L. M., Lin C. Y., Chai K. X. (2007) Biochim. Biophys. Acta [Google Scholar]
- 23.Stark H. J., Baur M., Breitkreutz D., Mirancea N., Fusenig N. E. (1999) J. Invest Dermatol. 112, 681–691 [DOI] [PubMed] [Google Scholar]
- 24.Stark H. J., Szabowski A., Fusenig N. E., Maas-Szabowski N. (2004) Biol. Proced. Online. 6, 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inouye K., Yasumoto M., Tsuzuki S., Mochida S., Fushiki T. (2010) J. Biochem. 147, 485–492 [DOI] [PubMed] [Google Scholar]
- 26.List K., Hobson J. P., Molinolo A., Bugge T. H. (2007) J. Cell. Physiol. 213, 237–245 [DOI] [PubMed] [Google Scholar]
- 27.Paladino S., Pocard T., Catino M. A., Zurzolo C. (2006) J. Cell. Biol. 172, 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J. X., Chao L., Chao J. (1994) J. Biol. Chem. 269, 18843–18848 [PubMed] [Google Scholar]
- 29.Myerburg M. M., Harvey P. R., Heidrich E. M., Pilewski J. M., Butterworth M. (2010) Am. J. Respir. Cell. Mol. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreasen D., Vuagniaux G., Fowler-Jaeger N., Hummler E., Rossier B. C. (2006) J. Am. Soc. Nephrol. 17, 968–976 [DOI] [PubMed] [Google Scholar]
- 31.Wang J. K., Lee M. S., Tseng I. C., Chou F. P., Chen Y. W., Fulton A., Lee H. S., Chen C. J., Johnson M. D., Lin C. Y. (2009) Am. J. Physiol. Cell. Physiol. 297, C459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ovaere P., Lippens S., Vandenabeele P., Declercq W. (2009) Trends Biochem. Sci. 34, 453–463 [DOI] [PubMed] [Google Scholar]
- 33.Fan B., Wu T. D., Li W., Kirchhofer D. (2005) J. Biol. Chem. 280, 34513–34520 [DOI] [PubMed] [Google Scholar]