Abstract
BCR-ABL is a causative tyrosine kinase (TK) of chronic myelogenous leukemia (CML). In CML patients, although myeloid cells are remarkably proliferating, erythroid cells are rather decreased and anemia is commonly observed. This phenotype is quite different from that observed in polycythemia vera (PV) caused by JAK2 V617F, whereas both oncogenic TKs activate common downstream molecules at the level of hematopoietic stem cells (HSCs). To clarify this mechanism, we investigated the effects of BCR-ABL and JAK2 V617F on erythropoiesis. Enforced expression of BCR-ABL but not of JAK2 V617F in murine LSK (Lineage−Sca-1hiCD117hi) cells inhibited the development of erythroid cells. Among several signaling molecules downstream of BCR-ABL, an active mutant of N-Ras (N-RasE12) but not of STAT5 or phosphatidylinositol 3-kinase (PI3-K) inhibited erythropoiesis, while N-RasE12 enhanced the development of myeloid cells. BCR-ABL activated Ras signal more intensely than JAK2 V617F, and inhibition of Ras by manumycin A, a farnesyltransferase inhibitor, ameliorated erythroid colony formation of CML cells. As for the mechanisms of Ras-induced suppression of erythropoiesis, we found that GATA-1, an erythroid-specific transcription factor, blocked Ras-mediated mitogenic signaling at the level of MEK through the direct interaction. Furthermore, enforced expression of N-RasE12 in LSK cells derived from p53-, p16INK4a/p19ARF-, and p21CIP1/WAF1-null/wild-type mice revealed that suppressed erythroid cell growth by N-RasE12 was restored only by p21CIP1/WAF1 deficiency, indicating that a cyclin-dependent kinase (CDK) inhibitor, p21CIP1/WAF1, plays crucial roles in Ras-induced suppression of erythropoiesis. These data would, at least partly, explain why respective oncogenic TKs cause different disease phenotypes.
Keywords: Cell Cycle, Erythropoeisis, MAP Kinases (MAPKs), Ras, Tumor Suppressor, BCR-ABL, CML, GATA-1, JAK2 V617F, p21CIP1/WAF1
Introduction
Oncogenic tyrosine kinases (TKs)2 such as BCR-ABL, FLT3-ITD, and JAK2 V617F are known to confer growth and/or survival advantage on hematopoietic cells, thereby causing hematologic malignancies (1–3). These gene alterations are supposed to occur at the hematopoietic stem cell (HSC) level (3, 4). Although these oncogenic TKs activate common downstream pathways including Ras/Raf/MEK/ERK, PI3-K/Akt, and STAT (1, 2, 5), their disease phenotypes are quite different: BCR-ABL is a causative gene of chronic myelogenous leukemia (CML) (1), FLT3-ITD of acute myeloid leukemia (AML) (2), and JAK2 V617F of myeloproliferative neoplasms including polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (PMF) (3). In patients with chronic-phase CML, anemia is a common feature in contrast to the marked leukocytosis in the peripheral blood. Also, bone marrow (BM) examination shows that erythroid islands are reduced in number and size despite the increased cellularity due to the granulocytic proliferation (6). This disease phenotype is totally different from that of PV, in which JAK2 V617F causes erythrocytosis together with the mild leukocytosis and thrombocytosis. Furthermore, in blast-phase CML, blast lineages are generally myeloid or lymphoid, and erythroid crisis is a rare incidence with a frequency no more than 5% (7, 8). These data suggest that, in contrast to the trilinear promoting activities of JAK2 V617F, BCR-ABL might not support the development of erythroid cells.
BCR-ABL activates several downstream pathways including Ras/Raf/MEK/ERK, STAT5, and PI3-K/Akt pathways (1, 4). Among them, we have previously shown that Ras plays crucial roles in the growth and survival of BCR-ABL-positive K562 cells, while STAT5 and PI3-K pathways contribute to their growth and survival to the only limited extent (9). In addition, although the role of STAT5 in BCR-ABL-mediated leukemogenesis remains controversial (10, 11), another group also reported that transformation of murine BM cells by BCR-ABL is blocked by dominant-negative Ras (12). Furthermore, Ras signaling was shown to be indispensable for the pathogenesis of CML in a murine BM transplantation model (13). Therefore, the activated Ras is considered to be essential for the pathogenesis of CML, and is also speculated to principally determine the disease phenotype of CML, that is, prominent proliferation of myeloid cells accompanied by the suppressed erythropoiesis.
Ras is constitutively activated by various oncogenic TKs or mutations of Ras itself in various malignant tumors. Although oncogenic (or constitutively activated) Ras was originally shown to transmit mitogenic and survival signals through Raf/MEK/ERK (14), recent studies have demonstrated that, like other oncogenic stimuli, it also induces growth inhibition/arrest in normal cells to prevent their malignant transformation. In general, this biological phenomenon is called “cellular senescence” and observed in various types of non-hematopoietic cells (15, 16). In addition, excessive Ras signaling was reported to inhibit erythropoiesis (17, 18), indicating the presence of a similar cellular response in hematopoietic cells. So far, oncogenic Ras has been shown to cause senescence through several signaling pathways other than Raf/MEK/ERK (15, 19–21). Also, several cell cycle regulatory molecules such as p53, p16INK4a, p19ARF and p21CIP1/WAF1, have been shown to play central roles in oncogene-induced senescence (15, 19, 21).
In this report, we found that BCR-ABL but not JAK2 V617F, and among their downstream molecules, Ras but not STAT5 or PI3-K suppress erythropoiesis from murine LSK cells. As for this mechanism, we found that an erythroid-lineage specific transcription factor, GATA-1, blocks Ras-dependent growth and survival by inhibiting MEK1 activity through the direct interaction. Furthermore, we showed that a cyclin-dependent kinase (CDK) inhibitor, p21CIP1/WAF1, plays crucial roles in Ras-induced suppression of erythropoiesis using p21CIP1/WAF1-deficient hematopoietic cells.
EXPERIMENTAL PROCEDURES
Cytokines and Reagents
Recombinant human thrombopoietin (rhTPO) and recombinant murine interleukin-3 (rmIL-3) were provided by Kyowa Hakko Kirin (Tokyo, Japan). Recombinant human erythropoietin (rhEPO) and murine stem cell factor (rmSCF) were purchased from R & D Systems (Minneapolis, MN). Manumycin A was purchased from Merck KGaA (Darmstadt, Germany).
Plasmid Constructs and cDNAs
Expression vectors for GATA-1/ERT (G1ERT) and wild-type (WT) GATA-1 were described previously (22). Active forms of N-Ras (N-RasE12) (23) and STAT5A (1*6 STAT5A) (24), and membrane-targeted PI3-K catalytic subunit (p110CAAX) (25) were subcloned into pMYs-IRES-EGFP, a retrovirus expression vector, which was kindly provided by Dr. T. Kitamura (University of Tokyo, Tokyo, Japan). pMSCV-IRES-GFP-p210-BCR-ABL is a generous gift from Dr. C. J. Eaves (Terry Fox Laboratory, Vancouver, BC, Canada) (26). The cDNA of JAK2 V617F was kindly provided by Dr. K. Shimoda (University of Miyazaki, Miyazaki, Japan) (27) and was subcloned into pMSCV-IRES-GFP.
Cell Lines and Cultures
A murine IL-3-dependent hematopoietic cell line, Ba/F3, was maintained in RPMI (nacalai tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS) (Equitech-Bio, Kerrville, TX) and 0.3 ng/ml rmIL-3. NIH3T3 and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; nacalai tesque) supplemented with 10% FBS.
Preparation of Stable Transformants from Ba/F3
We introduced G1ERT into Ba/F3 cells by electroporation (250 V and 950 microfarads) and selected stably transfected clones by the culture with G-418 (1.0 mg/ml; Wako Pure Chemical Industries, Osaka, Japan). We further introduced pMYs-IRES-EGFP-N-RasE12 and obtained doubly transfected clones by sorting GFP-positive cells with BD FACSAria Cell-Sorting System (BD Biosciences, San Jose, CA). Their IL-3-independent growth and cell cycle were analyzed with or without the activation of GATA-1 by 4-hydroxytamoxifen (4-HT; Sigma-Aldrich). DNA contents of the cells were evaluated by staining with propidium iodide.
Luciferase Assays
Luciferase assays were performed with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) as previously described (22). As for assays using Ba/F3 cells, transfection was performed with Amaxa Nucleofector technology (Lonza, Cologne, Germany), followed by the measurement of luciferase activities after 24 h.
Immunoblotting and Coimmunoprecipitation Analyses
Preparation of cell lysates, immunoprecipitation, gel electrophoresis, and immunoblotting were performed according to the methods described previously (22, 28). Antibodies (Abs) and reagents were supplied by the manufacturers described in supplemental methods.
Glutathione S-transferase (GST) Pull-down Assays
GST pull-down assays were performed as previously reported (22).
Animals
The congenic C57BL/6J mice were purchased from Clea Japan, Inc. (Tokyo, Japan). B6.129-Cdkn2atm1Rdp (p16INK4a/p19ARF-null) mice and p53-null mice were kindly provided by Technology Transfer Center National Cancer Institute (Rockville, MD) and Dr. N. Nishimoto (Wakayama Medical University, Wakayama, Japan), respectively. B6.129S2-Cdkn1atm1Tyj/J (p21CIP1/WAF1-null) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). The experimental designs of this study were approved by the Institutional Animal Care and Use Committee at Osaka University Graduate School of Medicine.
Separation of Murine Hematopoietic Progenitors
Murine BM cells were flushed from both femora and tibiae, and progenitors were concentrated by anti-mouse CD117 MicroBeads and autoMACS Pro Separator (Miltenyi Biotec, Bergisch Gladbach, Germany). To isolate LSK (Lineage−Sca-1hiCD117hi) cells, selected progenitors were stained with phycoerythrin-conjugated (PE-conjugated) monoclonal Abs against murine lineage markers (CD3e (145–2C11), CD45R/B220 (RA3–6B2), Gr-1 (RB6–8C5), CD11b (M1/70), and TER-119 (TER-119)), fluorescein isothiocyanate-conjugated (FITC-conjugated) anti-Sca-1 Ab (E13–161.7), and allophycocyanin-conjugated (APC-conjugated) anti-CD117 Ab (2B8), and isolated by FACSAria. All Abs were purchased from BD Biosciences.
Preparation of Retrovirus Particles
Preparation of retrovirus particles was performed as described previously (29) (see supplemental methods).
Retrovirus Transfection into Murine BM Progenitors
Isolated LSK cells were precultured overnight in DMEM supplemented with 10% FBS, rmSCF (100 ng/ml), and rhTPO (100 ng/ml). Then, the cells were seeded on 24-well tissue plates coated with RetroNectin (TaKaRa Bio Inc., Shiga, Japan), infected with each viral supernatant by spinoculation, and cultured in the same medium containing 10% FBS, protamine sulfate (10 μg/ml; Sigma-Aldrich), rmSCF (50 ng/ml), and rhTPO (50 ng/ml). After 48 h of culture, retrovirus-transduced GFP+ cells were sorted with FACSAria and were subjected to colony assays or stromal coculture.
Colony Assays
Cells were plated at the indicated density in methylcellulose medium (MethoCult; Stem Cell Technologies, Vancouver, BC, Canada) supplemented with the indicated growth factors. Cells were incubated with 5% CO2 at 37 °C, and the numbers of colonies were counted after the indicated days.
Stromal Coculture
A murine BM stromal cell line, MS-5, was cultured in minimum essential medium (MEM) α (Invitrogen, Carlsbad, CA) with 10% FBS and prepared in 24-well tissue plates 1 day before the seeding. The sorted GFP+ progenitors were seeded (1.5 × 103 cells/well) on the monolayer of MS-5 and cocultured in 2 ml of MEMα supplemented with 10% FBS, rmSCF (50 ng/ml), and rhEPO (3 units/ml). Five days after the initiation of coculture, hematopoietic cells were harvested and stained with PE-conjugated anti-CD45 (30-F11) Ab, and APC-conjugated anti-CD11b (M1/70) or anti-TER-119 (TER-119) Ab (all of them from BD Biosciences). To evaluate the phosphorylation status of ERK1/2, we used BD Phosflow technology (BD Biosciences). The harvested cells were further incubated in DMEM containing 2% FBS without cytokines for 4 h, then fixed, permeabilized, and stained with Alexa Fluor®647-conjugated anti-ERK1/2 (pT202/pY204) Ab (BD Biosciences) according to the manufacturer's recommendation.
Flow Cytometric Analyses
Flow cytometric analyses were performed using BD FACSCanto II (BD Biosciences). The data analyses were done with BD FACSDiva software (BD Biosciences) or FlowJo software (TreeStar, Ashland, OR).
Immunofluorescence Microscopy
5 × 104 of the transduced cells were cytospun onto microscope slides, fixed in 2% paraformaldehyde, and permeabilized in 1% Nonidet P-40 in PBS. After the incubation in blocking buffer (1 mg/ml of γ-globulin in PBS), the slides were incubated with a monoclonal Ab against p16INK4a (F-12) or p19ARF (5-C3–1) (both from Santa Cruz Biotechnology, Santa Cruz, CA). The slides were then incubated with an Alexa Fluor®546-conjugated secondary antibody (goat anti-mouse IgG for p16INK4a, or goat anti-rat IgG for p19ARF), followed by the staining of nuclei with Hoechst 33342 (all from Invitrogen). The slides were mounted in Fluoromount (Diagnostic BioSystems, Pleasanton, CA) before viewing on a LSM 5 PASCAL microscope (Carl Zeiss, Oberkochen, Germany).
Semiquantitative RT-PCR
Total RNA was isolated from 5 × 103 of the transduced cells using RNeasy Mini Kit (Qiagen, Hilden, Germany) and converted to cDNA by SuperScript III First Strand Synthesis System (Invitrogen). PCR was performed using Ampli Taq Gold (Applied Biosystems, Carlsbad, CA) with primers described in supplemental Table S1.
Real-time RT-PCR
Quantitative real-time RT-PCR was performed using FastStart Universal SYBR Green Master (Roche Diagnostics GmbH, Mannheim, Germany) and PRISM 7900HT (Applied Biosystems). Amplified signals were normalized to the levels of hypoxanthine phosphoribosyl transferase (HPRT). The primer sequences are described in supplemental Table S1.
BM Samples from CML Patients
BM samples were obtained from three patients with newly diagnosed chronic-phase CML. CD34+ cells were separated using the MACS immunomagnetic separation system, and were subjected to colony assays. All BM samples were obtained after receiving written informed consent in accordance with the Declaration of Helsinki, and this study protocol was approved by the institutional review board of Osaka University Hospital.
Statistical Methods
Statistical analyses were carried out by standard Student t tests. Error bars used throughout indicate S.D.
RESULTS
BCR-ABL but Not JAK2 V617F Inhibits the Development of Erythroid Cells
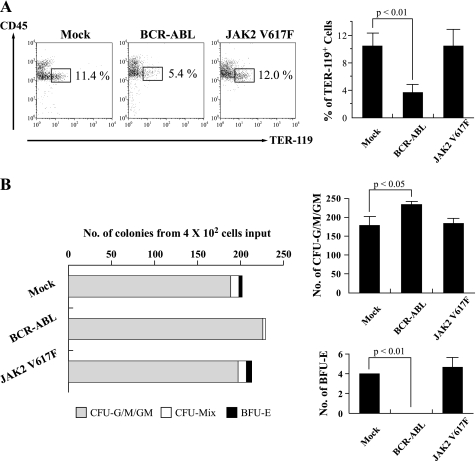
To examine the effects of BCR-ABL on erythropoiesis, we first introduced p210-BCR-ABL into murine LSK cells using the retrovirus vector harboring GFP as a reporter gene. After 48 h, GFP+ cells were sorted and cocultured with a murine BM stromal cell line, MS-5, in the presence of rmSCF and rhEPO for 5 days. As compared with mock-transduced cells, the proportion of CD45lowTER-119+ erythroid cells was reduced in BCR-ABL-transduced cells significantly (Fig. 1A). We also examined the effects of JAK2 V617F on erythropoiesis with the same strategy and found that JAK2 V617F did not reduce the proportion of erythoroid cells. In colony assays, BCR-ABL significantly decreased the number of burst-forming units-erythroid (BFU-E), while it increased the number of myeloid colonies (Fig. 1B). On the other hand, JAK2 V617F did not reduce the number of BFU-E. These data indicate that BCR-ABL but not JAK2 V617F inhibits the development of erythroid cells from murine hematopoietic progenitors.
FIGURE 1.
Effects of oncogenic TKs on proliferation of erythroid cells. A, after infection of retrovirus expressing Mock, BCR-ABL, or JAK2 V617F into murine LSK cells, GFP+ cells were sorted and cocultured with MS-5 in the presence of rmSCF and rhEPO. After 5-day cultures, expression of CD45 and TER-119 was analyzed by flow cytometry (left panels). The proportions of CD45lowTER-119+ cells are shown in the right bar graph (n = 3). B, respective retrovirus-transduced LSK cells were seeded at a density of 2.0 × 102 cells/35-mm dish in methylcellulose medium containing rmSCF, rmIL-3, rmIL-6, rhTPO, and rhEPO. Colony numbers were counted after 9 days. Representative colony numbers (left) and myeloid/erythroid colony numbers (right, n = 3) are shown. BFU-E, burst-forming units-erythroid; CFU-G/M/GM, colony-forming unit-granulocyte/macrophage/ granulocyte-macrophage.
Oncogenic Ras Inhibits Erythropoiesis Downstream of BCR-ABL
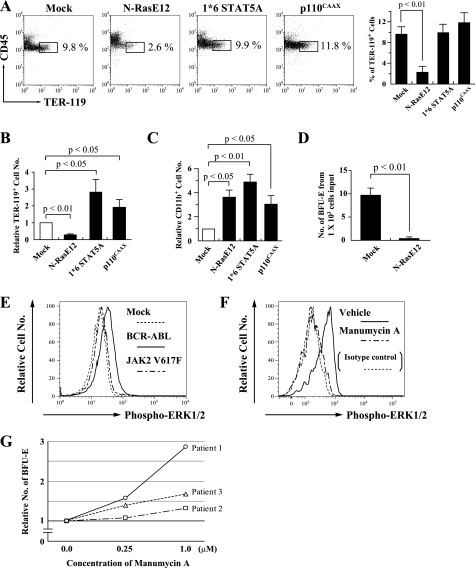
BCR-ABL activates mainly Ras/Raf/MEK/ERK, JAK2/STAT5, and PI3-K/Akt pathways. Next, to examine the roles of these pathways in erythropoiesis, we transduced LSK cells with an active form of each signal transduction molecule: N-RasE12 for an active form of N-Ras, 1*6 STAT5A for STAT5, and p110CAAX for PI3-K. Compared with Mock, 1*6 STAT5A and p110CAAX increased total erythroid cell numbers by 2.8- and 1.9-fold, respectively (both, p < 0.05), while the proportion of erythroid cells was scarcely influenced by both molecules due to the increase in total cell numbers (Fig. 2, A and B). In contrast, N-RasE12 remarkably reduced not only the frequency (Fig. 2A) but also the number of erythroid cells (0.28-fold) (Fig. 2B), while it significantly increased the number of CD11b+-myeloid cells (Fig. 2C). We also performed colony assays using N-RasE12- or Mock-transduced LSK cells. As shown in Fig. 2D, N-RasE12 significantly reduced the number of BFU-E (average colony numbers from 1.0 × 103 cells input: Mock-transduced cells, 9.7; N-Ras-transduced cells, 0.33) (p < 0.01).
FIGURE 2.
Roles of downstream molecules of oncogenic TKs in erythropoiesis. A–C, LSK cells each transfected with the indicated gene were cocultured with MS-5 in the medium containing rmSCF and rhEPO. After 5 days, expression of CD45 and TER-119 was analyzed by flow cytometry (A, left panels) and the proportions of CD45lowTER-119+ cells are shown (A, right bar graph, n = 3). Numbers of the TER-119+ cells were calculated by multiplication of the frequencies and total cell numbers. Relative numbers to Mock are shown (B). Relative CD11b+ myeloid cell numbers are shown (C). D, retrovirus-infected LSK cells were seeded at a density of 5.0 × 102 cells/dish in methylcellulose medium containing rmSCF, rmIL-3, and rhEPO. The numbers of BFU-E were counted after 8 days (n = 3). E, LSK cells, each transfected with Mock, BCR-ABL, or JAK2 V617F, were further incubated without cytokines after the coculture with MS-5, and the phosphorylation status of ERK1/2 was analyzed using Phosflow technology. F, after 5-h incubation of CML patients blood mononuclear cells with manumycin A (7 μm) or vehicle, the phosphorylation status of ERK1/2 was analyzed. G, CD34+ cells were separated from BM samples of three CML patients, and seeded in methylcellulose medium containing rhSCF, rhIL-3, and rhEPO, with manumycin A at the indicated concentrations or vehicle. The numbers of BFU-E were counted after 9 days, and shown as relative numbers to vehicle in each patient.
BCR-ABL Activates Ras Signal More Intensely than JAK2 V617F
Next, we tried to clarify why JAK2 V617F did not suppress erythropoiesis, because it has been reported to activate Ras as well as BCR-ABL (5). For this purpose, we introduced JAK2 V617F and BCR-ABL into murine LSK cells, cocultured them with MS-5, and evaluated the Ras activity by expediently measuring the phosphorylation status of ERK1/2 after 4-h starvation of cytokines. As shown in Fig. 2E, ERK1/2 was more intensely phosphorylated (activated) in cells transduced with BCR-ABL than in those with JAK2 V617F. We also examined the phosphorylation status of ERK1/2 in CML patients' blood cells treated with manumycin A, a potent farnesyltransferase inhibitor which selectively suppresses Ras, or vehicle only. As shown in Fig. 2F, phosphorylation of ERK was reduced by Ras inhibition, indicating that BCR-ABL activates ERK through the activation of Ras. These data indicate that different growth status of erythroid cells between these TKs might result from the preferential activation of Ras signal by BCR-ABL.
Suppression of Ras Signal Ameliorates the Inhibition of Erythropoiesis Caused by BCR-ABL
Furthermore, to make sure that suppressed erythropoiesis caused by BCR-ABL is due to the activation of Ras signal, we examined the effects of Ras-inhibition on erythroid colony formation of BCR-ABL expressing cells. CD34+ cells were separated from BM samples of three patients with newly diagnosed chronic-phase CML. They were then cultured in methylcellulose medium containing rhSCF, rhIL-3, and rhEPO, with or without manumycin A. Complete blockage of Ras signal by supplement of sufficient dose (10 μm) of manumycin A eradicated erythroid colony formation (data not shown). However, as shown in Fig. 2G, the number of erythroid colonies was restored by low doses of manumycin A in all three patients, though there was some difference in degree. This result, actually in primary CML cells, supports our model that, although Ras is indispensable for erythroid cell survival, excessive Ras signal downstream of BCR-ABL rather inhibits erythroid cell proliferation.
GATA-1 Inhibits Ras-dependent Cell Proliferation and Survival
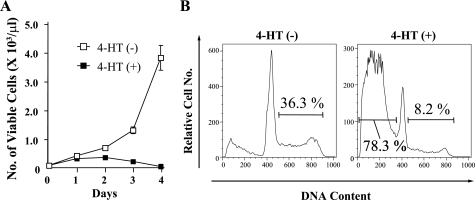
As described above, oncogenic Ras signaling promoted the proliferation of myeloid cells, but inhibited that of erythroid cells. To elucidate the mechanisms underlying the different responses to oncogenic Ras between the two lineages, we examined the effects of GATA-1, which is expressed in erythroid cells but not in myeloid cells, on Ras signal. For this purpose, we transduced N-RasE12 and G1ERT, a chimera gene consisting of full-length GATA-1 and the mutated ligand-binding domain of estrogen receptor, into Ba/F3 cells, which was named Ba/F3/N-RasE12/G1ERT. G1ERT reveals GATA-1 activity in response to 4-HT as previously reported (22). As shown in Fig. 3A, N-RasE12 enabled this clone to proliferate and survive independently of IL-3. However, when GATA-1 activity was induced by 4-HT treatment, N-RasE12-dependent cell growth was completely suppressed (Fig. 3A). In agreement with this result, the proportion of growing cells in S-G2/M phase was reduced by 4-HT treatment from 36% to 8% in DNA contents analysis (Fig. 3B). Furthermore, 4-HT treatment induced apoptosis in 78% of cells, which was detected as a subdiploid fraction. From these results, we speculated that GATA-1 might inhibit oncogenic Ras activities, which transmit proliferation and survival signals.
FIGURE 3.
Inhibition of Ras-dependent cell proliferation and survival by GATA-1. A, Ba/F3/N-RasE12/G1ERT cells were seeded at a density of 100/μl and cultured in RPMI supplemented with 1% FBS without IL-3 in the presence or absence of 1 μm 4-HT. Total numbers of viable cells were counted by trypan blue dye exclusion method on the indicated days. The results are shown as means ± S.D. of triplicate cultures. B, after 48 h of culture, DNA contents of 4-HT-treated or untreated cells were examined by propidium iodide staining. The proportions of cells in S-G2/M phase and subdiploid fraction are shown, respectively.
GATA-1 Suppresses MEK Activity
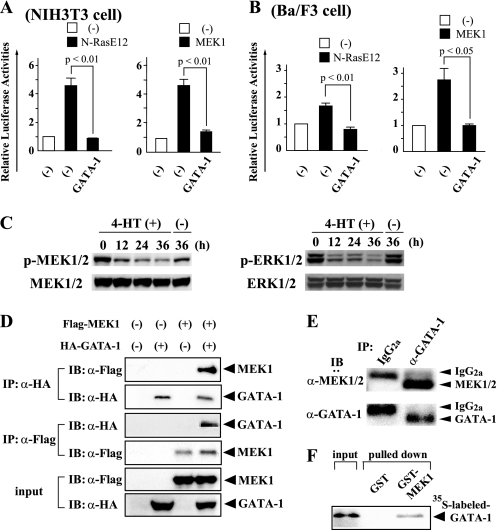
Ras signal is known to be transmitted to the nucleus through Raf, MEK, and ERK in this order. To identify which molecule was inhibited by GATA-1 in this pathway, we performed luciferase assays using a reporter gene for ERK (3 × AP-1-Luc) in NIH3T3 and Ba/F3 cells. As shown in Fig. 4, A and B, GATA-1 significantly reduced the N-Ras- and MEK1-induced AP-1-luciferase activities almost to the baseline levels (white boxes), which indicates that GATA-1 inhibits Ras signal at the level or downstream of MEK. Next, we examined the phosphorylation status of MEK1/2 and ERK1/2 in Ba/F3/N-RasE12/G1ERT cells by immunoblot analysis. As shown in Fig. 4C, both MEK1/2 and ERK1/2 were phosphorylated by N-RasE12 even under the culture without IL-3, which was suppressed by 4-HT in a time-dependent manner. This result implies that GATA-1 suppresses Ras signal at the level or upstream of MEK. Together with the results from luciferase assays, it was speculated that GATA-1 would inhibit MEK activity.
FIGURE 4.
GATA-1 blocks the Ras/Raf/MEK/ERK pathway through its direct interaction with MEK1. A, NIH3T3 cells (2 × 105 cells seeded in 60-mm dish) were transfected with the indicated expression vectors and the reporter gene (3 × AP-1-Luc) together with pRL-CMV. After 12 h, the cells were serum-deprived for 24 h, then lysed, and subjected to the measurement of the firefly and Renilla luciferase activities. The relative firefly luciferase activities normalized by the Renilla luciferase activities are shown as means ± S.D. of three separate experiments. B, Ba/F3 cells (2 × 106 cells) were transfected with the same vectors as Fig. 4A using Amaxa Nucleofector technology. After 24 h of culture, the cells were lysed and subjected to the measurement of the luciferase activities. C, Ba/F3/N-RasE12/G1ERT cells cultured in RPMI supplemented with 1% FBS were treated with 1 μm 4-HT or vehicle. Total cellular lysates were prepared at the indicated time and subjected to immunoblotting with the indicated Abs. The filters were reprobed with corresponding Abs to confirm that the equal amounts of the proteins were loaded. D, coimmunoprecipitation analyses were performed using 293T cells transfected with HA-tagged GATA-1 and/or Flag-tagged MEK1 as indicated. IP, immunoprecipitation; IB, immunoblotting; α, anti. E, total cellular lysate was prepared from murine BM CD71+ cells. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies. F. The in vitro binding between GATA-1 and MEK1 was examined by GST pull-down assays. 35S-labeled GATA-1 was incubated with GST-MEK1 bound to glutathione-Sepharose beads, and the binding complex was separated by gel electrophoresis and subjected to autoradiography.
GATA-1 Blocks the Ras Signal through Its Direct Interaction with MEK1
To clarify how GATA-1 inhibits MEK activities, we examined the interaction between GATA-1 and MEK1. First, we transfected 293T cells with hemagglutinin-tagged (HA-tagged) GATA-1 and/or Flag-tagged MEK1. Total cellular lysates were prepared after 36 h, and GATA-1 was immunoprecipitated with the anti-HA Ab and MEK1 with the anti-Flag Ab. As shown in Fig. 4D, immunoblotting with the anti-Flag Ab showed that MEK1 was coimmunoprecipitated with GATA-1 only when both molecules were cotransduced. Also, immunoblotting with the anti-HA Ab showed that GATA-1 was coimmunoprecipitated with MEK1.
Next, to examine whether endogenous GATA-1 and MEK interact in primary erythroid cells, we performed a coimmunoprecipitation analysis using murine BM erythroid cells: Cells positive for CD71 (transferrin receptor), which is expressed at high levels on erythroid progenitors, were purified using the MACS immunomagnetic separation system. Total cellular lysate was prepared and subjected to immunoprecipitation with an anti-GATA-1 Ab or rat isotype IgG. Fig. 4E shows that MEK is coimmunoprecipitated with GATA-1, indicating that these molecules actually interact with each other in primary erythroid cells.
Finally, we investigated whether MEK1 directly binds to GATA-1 in vitro by GST pull-down assays. After verifying the quality and quantity of GST-MEK1 fusion protein by Coomassie Brilliant Blue staining (data not shown), we analyzed the binding between GST-MEK1 and in vitro-translated GATA-1. As shown in Fig. 4F, GST-MEK1 but not GST alone, bound to 35S-labeled GATA-1 in vitro.
Together with the results of Fig. 4, A–C, we proved the following two facts: GATA-1 inhibits MEK activation; GATA-1 and MEK interact with each other in primary erythroid progenitors. From these facts, we speculated that GATA-1 blocks Ras signal at least partly through the direct interaction with MEK1.
Oncogenic Ras Induces Suppression of Erythropoiesis through the Induction of p21CIP1/WAF1
In addition to the functions to deliver mitogenic and anti-apoptotic signals (14), Ras paradoxically causes growth arrest (senescence) in normal cells through several cell cycle regulatory molecules such as p53, p16INK4a, p19ARF, and p21CIP1/WAF1 (15, 21). Among them, p53 is a tumor-suppressor and acts as a pivotal regulator of these responses (15, 16, 19, 21). p19ARF is a splicing variant of p16INK4a and inhibits the function of H/MDM2, which promotes degradation of p53 (30). p16INK4a is a member of the INK4 family of CDK inhibitors, which causes cell cycle arrest at G1 phase by inhibiting CDK4/6 activities (30). Meanwhile, p21CIP1/WAF1 is a member of the Cip/Kip family of CDK inhibitors and also induces G1 arrest by inhibiting CDK2 activities. In this report, we next examined their roles in N-RasE12-induced suppression of erythropoiesis.
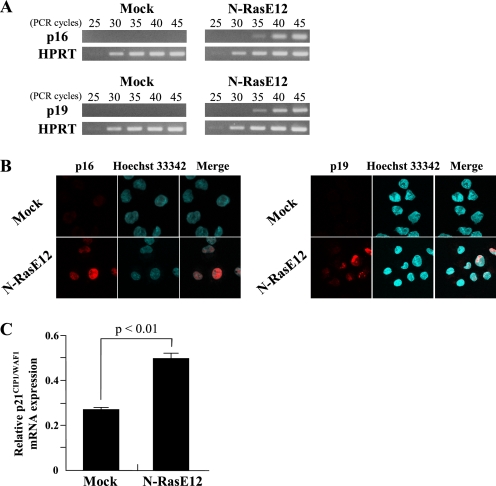
At first, we examined the effects of N-RasE12 on the expression of p16INK4a, p19ARF, and p21CIP1/WAF1 by semiquantitative/real-time RT-PCR analyses or immunofluorescence. As shown in Fig. 5A and B, the expression of p16INK4a and p19ARF was induced in N-RasE12-transduced LSK cells both in mRNA and protein levels. Also, the expression of p21CIP1/WAF1 was increased by nearly 2-fold in N-RasE12-transduced LSK cells compared with mock-transduced LSK cells (Fig. 5C), suggesting that the up-regulated p16INK4a, p19ARF, and/or p21CIP1/WAF1 might be involved in N-RasE12-induced suppression of erythropoiesis.
FIGURE 5.
Increase in expression levels of p16INK4a, p19ARF, and p21CIP1/WAF1 by oncogenic Ras. A–C, LSK cells transfected with Mock or N-RasE12 were cultured with rmSCF, rmIL-3, and rhEPO for 2 days. Total RNA was isolated from GFP+ cells, and the expression levels of p16INK4a and p19ARF were analyzed by semiquantitative RT-PCR (A). Immunofluorescence staining of p16INK4a and p19ARF localizations (red) in Hoechst 33342-stained nuclei of GFP+ cells are shown (magnification, 630×) (B). The expression levels of p21CIP1/WAF1 were analyzed by real-time RT-PCR. The results are normalized to the levels of HPRT gene and shown as means ± S.D. (n = 3) (C).
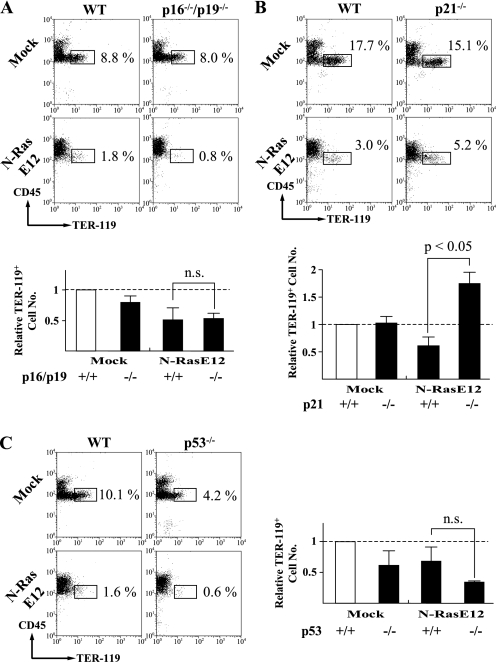
To further analyze the roles of these molecules, we next introduced N-RasE12 into LSK cells isolated from p16INK4a/p19ARF double knock-out (KO) mice, cocultured them with MS-5, and examined the development of erythroid cells by flow cytometry. As shown in Fig. 6A, the frequency of CD45lowTER-119+ erythroid cells was a little lower in N-RasE12-transduced double KO cells than in N-RasE12-transduced WT cells (WT 1.8% versus double KO 0.8%) (upper panels). In addition, although the number of these erythroid cells was slightly restored in N-RasE12-transduced double KO cells compared with N-RasE12-transduced WT cells (lower graph), this difference was not significant.
FIGURE 6.
p21CIP1/WAF1 but not p53 or p16INK4a/p19ARF mediates oncogenic Ras-induced suppression of erythropoiesis. A–C, LSK cells were isolated from BM of the indicated mice. After retrovirus infection, GFP+ cells were sorted and cocultured with MS-5 in the presence of rmSCF and rhEPO. The expression of CD45 and TER-119 was analyzed after 5 days. Bar graphs represent the relative TER-119+ cell numbers normalized to mock-transduced WT cells (dashed lines). n.s., not significant.
We also introduced N-RasE12 into LSK cells isolated from p21CIP1/WAF1-null mice. As observed in the other experiments, N-RasE12 reduced the proportion of CD45lowTER-119+ erythroid cells both in WT and p21CIP1/WAF1-null LSK cells (Fig. 6B, upper panels). However, p21CIP1/WAF1 deficiency partially, but significantly, restored the proportion of this fraction from 3.0 to 5.2%. In addition, surprisingly, N-RasE12 increased the number of erythroid cells in p21CIP1/WAF1-null LSK cells compared with mock-transduced LSK cells (Fig. 6C, lower graph), indicating that p21CIP1/WAF1 is a major regulator of N-RasE12-induced suppression of erythropoiesis.
Because the expression of p21CIP1/WAF1 is regulated in p53-dependent and independent manners (31–33), we finally investigated the roles of p53 in N-RasE12-induced suppression of erythropoiesis with the similar experiment. As shown in Fig. 6C, the proportion and number of erythroid cells in mock-transduced LSK cells were reduced by p53 deficiency. In addition, p53 deficiency did not cancel the inhibition of erythroid cell development by N-RasE12. Together, these results indicate that N-RasE12 inhibits erythropoiesis through p21CIP1/WAF1 in a p53-independent manner.
DISCUSSION
We here found that BCR-ABL suppresses erythroid cell proliferation. This finding is largely consistent with clinical features of CML, in which anemia is commonly observed and erythroid blast crisis is a rare event. Also, we found that constitutively activated Ras, but not PI3-K or STAT5, inhibits erythropoiesis and that a farnesyltransferase inhibitor, manumycin A, restores erythroid colony formation of CML patients BM cells at relatively low concentrations. These results strongly indicate that Ras is a negative regulator of erythropoiesis downstream of BCR-ABL. So far, functions of Ras in normal erythropoiesis are controversial. It was reported that Ras signaling was essential for development of erythroid progenitors (34, 35). In contrast, H-Ras−/−, N-Ras−/−, and double KO (H-Ras−/− N-Ras−/−) mice had no apparent hematopoietic abnormality, indicating that Ras is dispensable for normal erythropoiesis (36, 37). Regarding the roles of oncogenic Ras in erythropoiesis, it was shown that oncogenic H-Ras blocks terminal erythroid differentiation (38), and that enforced expression of an active mutant of N-Ras in primitive hematopoietic cells inhibits proliferation of erythroid cells (17, 18). Our results indicate that the excessive Ras signal would inhibit erythropoiesis, though Ras signal might be to some extent necessary for erythroid cell survival. Ras is mutated in a significant proportion of cases with acute myeloid leukemia and myelodysplastic syndromes (39), or constitutively activated by various oncogenic TKs, including FLT3-ITD (2), c-KIT D816V (40), and TEL-PDGFRB (41). So, anemia observed in these hematologic malignancies also might be, at least partly, attributed to the constitutively activated Ras signal. However, in this study, JAK2 V617F slightly enhanced erythropoiesis as observed in patients with PV, whereas its downstream pathways including Ras, PI3-K, and STAT5 are common to BCR-ABL (1, 5). As for this difference, we here found that JAK2 V617F does not activate Ras signal so strongly as BCR-ABL. Also, it was speculated that JAK2 V617F would utilize mainly STAT5 to promote erythropoiesis in PV patients. Although Ras has some isoforms, we focused on N-Ras, because, in myeloid malignancies, N-Ras mutations are more frequent than K-Ras, whereas H-Ras mutations are rare (39, 42–44). It is predictable that activated N-Ras has stronger leukemogenic potential than activated H-Ras or K-Ras.
In contrast to the negative role of oncogenic Ras in erythropoiesis, Ras activation prominently enhanced the development of myeloid cells from LSK cells as observed in CML patients. To clarify the mechanism through which the active form of Ras plays different roles in the growth of hematopoietic cells according to the cell lineages (i.e. inhibition of erythropoiesis but promotion of myelopoiesis), we examined the role of GATA-1, which is a transcription factor mainly expressed in erythroid and megakaryocytic cells but not in myeloid cells. Ras-induced suppression of erythropoiesis can be considered to result from inhibition of proliferation of already committed erythroid progenitors, and blockage of commitment into erythroid lineage from HSCs. In this study, we found that GATA-1 inhibits MEK activity and suppresses the Ras-dependent proliferation of GATA-1-posistive cells. GATA-1 is necessary in the post-commitment stages of erythroid and megakaryocytic development, and is highly expressed after the commitment into megakaryocyte-erythrocyte progenitors (MEPs), but is scarcely expressed in HSCs (45). So, it is unlike that the interaction between GATA-1 and MEK1 is associated with the lineage determination of HSCs. On the other hand, recent reports showed that suppression of erythroid cell development by H-, K-, and N-Ras occurs at later stages of differentiation (18, 38, 46). These data are consistent with our result that GATA-1 interacts with MEK1, thereby inhibiting Ras-mediated mitogenic signals.
However, this result raises a question where these molecules interact together in the cells because GATA-1 is located in the nucleus and MEK is in the cytoplasm (47). As an explanation it was previously reported that MEK contains a nuclear export signal in its N-terminal domain, indicating that MEK is translocated to the nucleus upon mitogenic stimulation and then goes back to the cytoplasm after transduction of its signal (48). So, GATA-1 is supposed to interact with MEK1 in the nucleus, thereby inhibiting its activity. This hypothesis that GATA-1 would inhibit MEK activities is also contradictory to the fact that platelet counts are often elevated in CML patients, because MEK has been shown to be important for the maturation (polyploidization) of megakaryocytes, in which GATA-1 is highly expressed as well as in erythroid cells. Regarding this issue, Jacquel et al. reported that PMA-induced megakaryocytic maturation is only partly dependent on the MEK/ERK pathway and suggested the involvement of other pathways such as Jun N-terminal kinase (JNK) and protein kinase C (PKC) in CML cells (49). Alternatively, it is also possible that the interaction between GATA-1 and MEK might be inhibited in megakaryocytes due to the presence of some nuclear protein(s) specific for this lineage. However, further studies are required to clarify how megakaryocytes develop and platelets are effectively produced in CML patients.
Among various signaling molecules downstream of Ras, the Raf/MEK/ERK pathway mainly promotes cell growth and prevents apoptosis of hematopoietic cells (14). On the other hand, oncogenic stimuli including constitutively activated Ras, also cause growth inhibition (senescence) that acts as a fail-safe mechanism against malignant transformation (15, 16, 21). Although the mechanism of Ras-induced senescence is not fully understood, recent findings have unveiled several MEK/ERK-independent pathways (19). These pathways regulate the function of two main tumor-suppressor molecules, p53 and retinoblastoma protein (pRb) (50). Downstream of oncogenic Ras, p38-regulated/activated protein kinase (PRAK), a substrate of p38 mitogen-activated protein kinase (p38 MAPK), activates p53 by direct phosphorylation (20). Ras/Raf stabilizes p53 independently of MEK through the up-regulation of p19ARF (21). The PI3-K pathway also stabilizes p53 through the inhibition of H/MDM2 (19). So, we speculated that N-RasE12 might induce growth arrest in erythroid cells even if MEK activities are blocked by GATA-1.
Ras-induced senescence is executed by CDK inhibitors such as p16INK4a and p21CIP1/WAF1, and a tumor-suppressor, p19ARF, which consequently activate both p53 and pRb pathways. Among these molecules, we here found that p21CIP1/WAF1 is a major player of Ras-induced suppression of erythropoiesis (may well be called nearly equal to senescence). Although p21CIP1/WAF1 is a transcriptional target of p53 (51), p53 deficiency did not cancel Ras-induced suppression of erythropoiesis. So, p53-independent expression of p21CIP1/WAF1 was supposed to be important for Ras-induced suppression of erythropoiesis. Because Darley et al. (18) previously showed that oncogenic N-Ras conferred developmental abnormalities on human erythroid cells through the activation of PKC, one of the reported activators of p21CIP1/WAF1 (52), PKC may be a candidate molecule involved in Ras-induced expression of p21CIP1/WAF1 and consequent suppression of erythropoiesis.
Mutation and/or deletion of the p53 gene and the INK4a/ARF locus are frequently observed in CML blast phase (1), but to our knowledge, there is no report demonstrating the inactivation of the p21CIP1/WAF1 gene. So, our findings that p21CIP1/WAF1 but not p53 or p16INK4a/p19ARF is the major regulator of Ras-induced suppression of erythropoiesis are again consistent with the clinical features that anemia is continued and erythroid transformation is a rare event in blast-phase CML (7, 8). Furthermore, because loss-of-function mutations of the p21CIP1/WAF1 gene are rare in most of the hematologic malignancies, anemia observed in these diseases might be attributable to p21CIP1/WAF1.
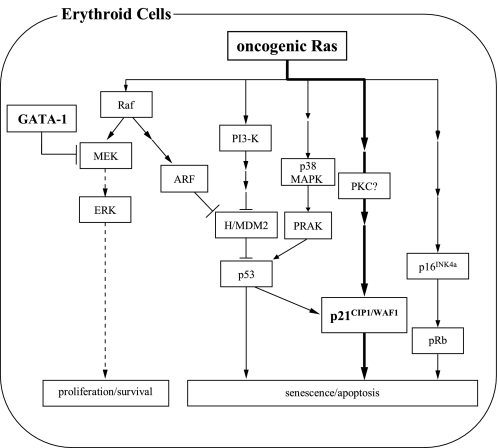
In conclusion, we here show that BCR-ABL but not JAK2 V617F inhibits erythropoiesis through the Ras signal. We also identified p21CIP1/WAF1 as a central regulator of Ras-induced suppression of erythropoiesis. Ras transmits both growth promoting and inhibitory signals, and then induces proliferation or senescence dependently on their balance. In erythroid but not in myeloid progenitors, the growth promoting signal is inhibited at the level of MEK by GATA-1, which would lead to the relative dominance of the growth inhibitory signal mediated by p21CIP1/WAF1 (Fig. 7). These mechanisms would explain why oncogenic Ras simultaneously reveals conflicting effects according to the cell lineage, i.e. growth promotion in myeloid cells and growth inhibition in erythroid cells. This model may be also useful to understand the mechanism of anemia caused by other oncogenic TKs.
FIGURE 7.
A proposed model for oncogenic Ras-induced suppression of erythropoiesis. Oncogenic Ras simultaneously activates several downstream molecules including Raf, PI3-K, and p38 MAPK. The Raf/MEK/ERK pathway mainly transduces proliferation and survival signals, while the remaining pathways commonly induce growth arrest (senescence) through cell cycle regulatory molecules such as p16INK4a, p19ARF, p21CIP1/WAF1, and p53. So, oncogenic Ras is supposed to induce proliferation or senescence dependently on the balance between these two signals. In this study, we found that GATA-1 inhibits mitogenic signal from Ras through its interaction with MEK1 in erythroid cells, which resulted in their growth inhibition due to the dominance of senescence-inducing signals. In addition, we found that p21CIP1/WAF1 is a crucial regulator of oncogenic Ras-induced senescence of erythroid cells.
Supplementary Material
Acknowledgments
We thank Dr. Connie J. Eaves for providing the vector expressing p210-BCR-ABL, Dr. Kazuya Shimoda for providing the plasmid encoding JAK2 V617F, and Dr. Hiroyuki Miyoshi for providing 293gp cells.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Methods.
- TK
- tyrosine kinase
- CML
- chronic myelogenous leukemia
- PV
- polycythemia vera
- HSC
- hematopoietic stem cell
- LSK
- Lineage−Sca-1hiCD117hi
- BM
- bone marrow
- CDK
- cyclin-dependent kinase
- rh
- recombinant human
- TPO
- thrombopoietin
- rm
- recombinant murine
- EPO
- erythropoietin
- SCF
- stem cell factor
- G1ERT
- GATA-1/ERT
- 4-HT
- 4-hydroxytamoxifen
- Ab
- antibody
- HPRT
- hypoxanthine phosphoribosyl transferase
- pRb
- retinoblastoma protein
- PRAK
- p38-regulated/activated protein kinase.
REFERENCES
- 1.Quintás-Cardama A., Cortes J. (2009) Blood 113, 1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Small D. (2006) Hematology Am. Soc. Hematol. Educ. Program, 178–184 [DOI] [PubMed] [Google Scholar]
- 3.Levine R. L., Gilliland D. G. (2008) Blood 112, 2190–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren R. (2005) Nat. Rev. Cancer 5, 172–183 [DOI] [PubMed] [Google Scholar]
- 5.Zeuner A., Pedini F., Signore M., Ruscio G., Messina C., Tafuri A., Girelli G., Peschle C., De Maria R. (2006) Blood 107, 3495–3502 [DOI] [PubMed] [Google Scholar]
- 6.Thiele J., Kvasnicka H. M., Schmitt-Graeff A., Zirbes T. K., Birnbaum F., Kressmann C., Melguizo-Grahmann M., Frackenpohl H., Sprungmann C., Leder L. D., Diehl V., Zankovich R., Schaefer H. E., Niederle N., Fischer R. (2000) Leuk. Lymphoma 36, 295–308 [DOI] [PubMed] [Google Scholar]
- 7.Saikia T., Advani S., Dasgupta A., Ramakrishnan G., Nair C., Gladstone B., Kumar M. S., Badrinath Y., Dhond S. (1988) Leuk. Res. 12, 499–506 [DOI] [PubMed] [Google Scholar]
- 8.Griffin J. D., Todd R. F., 3rd, Ritz J., Nadler L. M., Canellos G. P., Rosenthal D., Gallivan M., Beveridge R. P., Weinstein H., Karp D., Schlossman S. F. (1983) Blood 61, 85–91 [PubMed] [Google Scholar]
- 9.Sonoyama J., Matsumura I., Ezoe S., Satoh Y., Zhang X., Kataoka Y., Takai E., Mizuki M., Machii T., Wakao H., Kanakura Y. (2002) J. Biol. Chem. 277, 8076–8082 [DOI] [PubMed] [Google Scholar]
- 10.Sexl V., Piekorz R., Moriggl R., Rohrer J., Brown M. P., Bunting K. D., Rothammer K., Roussel M. F., Ihle J. N. (2000) Blood 96, 2277–2283 [PubMed] [Google Scholar]
- 11.Hoelbl A., Kovacic B., Kerenyi M. A., Simma O., Warsch W., Cui Y., Beug H., Hennighausen L., Moriggl R., Sexl V. (2006) Blood 107, 4898–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawyers C. L., McLaughlin J., Witte O. N. (1995) J. Exp. Med. 181, 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baum K. J., Ren R. (2008) J. Hematol. Oncol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platanias L. C. (2003) Blood 101, 4667–4679 [DOI] [PubMed] [Google Scholar]
- 15.Campisi J. (2005) Cell 120, 513–522 [DOI] [PubMed] [Google Scholar]
- 16.Braig M., Schmitt C. A. (2006) Cancer Res. 66, 2881–2884 [DOI] [PubMed] [Google Scholar]
- 17.Darley R. L., Hoy T. G., Baines P., Padua R. A., Burnett A. K. (1997) J. Exp. Med. 185, 1337–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darley R. L., Pearn L., Omidvar N., Sweeney M., Fisher J., Phillips S., Hoy T., Burnett A. K. (2002) Blood 100, 4185–4192 [DOI] [PubMed] [Google Scholar]
- 19.Yaswen P., Campisi J. (2007) Cell 128, 233–234 [DOI] [PubMed] [Google Scholar]
- 20.Sun P., Yoshizuka N., New L., Moser B. A., Li Y., Liao R., Xie C., Chen J., Deng Q., Yamout M., Dong M. Q., Frangou C. G., Yates J. R., 3rd, Wright P. E., Han J. (2007) Cell 128, 295–308 [DOI] [PubMed] [Google Scholar]
- 21.Wahl G. M., Carr A. M. (2001) Nat. Cell Biol. 3, E277–286 [DOI] [PubMed] [Google Scholar]
- 22.Ezoe S., Matsumura I., Gale K., Satoh Y., Ishikawa J., Mizuki M., Takahashi S., Minegishi N., Nakajima K., Yamamoto M., Enver T., Kanakura Y. (2005) J. Biol. Chem. 280, 13163–13170 [DOI] [PubMed] [Google Scholar]
- 23.Delgado M. D., Vaqué J. P., Arozarena I., López-Ilasaca M. A., Martínez C., Crespo P., León J. (2000) Oncogene 19, 783–790 [DOI] [PubMed] [Google Scholar]
- 24.Onishi M., Nosaka T., Misawa K., Mui A. L., Gorman D., McMahon M., Miyajima A., Kitamura T. (1998) Mol. Cell. Biol. 18, 3871–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egawa K., Sharma P. M., Nakashima N., Huang Y., Huver E., Boss G. R., Olefsky J. M. (1999) J. Biol. Chem. 274, 14306–14314 [DOI] [PubMed] [Google Scholar]
- 26.Jiang X., Ng E., Yip C., Eisterer W., Chalandon Y., Stuible M., Eaves A., Eaves C. J. (2002) Blood 100, 3731–3740 [DOI] [PubMed] [Google Scholar]
- 27.Shide K., Shimoda H. K., Kumano T., Karube K., Kameda T., Takenaka K., Oku S., Abe H., Katayose K. S., Kubuki Y., Kusumoto K., Hasuike S., Tahara Y., Nagata K., Matsuda T., Ohshima K., Harada M., Shimoda K. (2008) Leukemia 22, 87–95 [DOI] [PubMed] [Google Scholar]
- 28.Matsumura I., Kanakura Y., Kato T., Ikeda H., Horikawa Y., Ishikawa J., Kitayama H., Nishiura T., Tomiyama Y., Miyazaki H., Matsuzawa Y. (1996) Blood 88, 3074–3082 [PubMed] [Google Scholar]
- 29.Fukushima K., Matsumura I., Ezoe S., Tokunaga M., Yasumi M., Satoh Y., Shibayama H., Tanaka H., Iwama A., Kanakura Y. (2009) J. Biol. Chem. 284, 7719–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roussel M. F. (1999) Oncogene 18, 5311–5317 [DOI] [PubMed] [Google Scholar]
- 31.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 32.Macleod K. F., Sherry N., Hannon G., Beach D., Tokino T., Kinzler K., Vogelstein B., Jacks T. (1995) Genes Dev. 9, 935–944 [DOI] [PubMed] [Google Scholar]
- 33.Parker S. B., Eichele G., Zhang P., Rawls A., Sands A. T., Bradley A., Olson E. N., Harper J. W., Elledge S. J. (1995) Science 267, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 34.Khalaf W. F., White H., Wenning M. J., Orazi A., Kapur R., Ingram D. A. (2005) Blood 105, 3538–3541 [DOI] [PubMed] [Google Scholar]
- 35.Sui X., Krantz S. B., You M., Zhao Z. (1998) Blood 92, 1142–1149 [PubMed] [Google Scholar]
- 36.Esteban L. M., Vicario-Abejón C., Fernández-Salguero P., Fernández-Medarde A., Swaminathan N., Yienger K., Lopez E., Malumbres M., McKay R., Ward J. M., Pellicer A., Santos E. (2001) Mol. Cell. Biol. 21, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umanoff H., Edelmann W., Pellicer A., Kucherlapati R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1709–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Socolovsky M., Gross A. W., Lodish H. F. (2003) Blood 102, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 39.MacKenzie K. L., Dolnikov A., Millington M., Shounan Y., Symonds G. (1999) Blood 93, 2043–2056 [PubMed] [Google Scholar]
- 40.Metcalfe D. D. (2008) Blood 112, 946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheadon H., Welham M. J. (2003) Blood 102, 1480–1489 [DOI] [PubMed] [Google Scholar]
- 42.Döhner K., Döhner H. (2008) Haematologica 93, 976–982 [DOI] [PubMed] [Google Scholar]
- 43.Neubauer A., Greenberg P., Negrin R., Ginzton N., Liu E. (1994) Leukemia 8, 638–641 [PubMed] [Google Scholar]
- 44.Flotho C., Valcamonica S., Mach-Pascual S., Schmahl G., Corral L., Ritterbach J., Hasle H., Aricò M., Biondi A., Niemeyer C. M. (1999) Leukemia 13, 32–37 [DOI] [PubMed] [Google Scholar]
- 45.Akashi K., Traver D., Miyamoto T., Weissman I. L. (2000) Nature 404, 193–197 [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., Liu Y., Beard C., Tuveson D. A., Jaenisch R., Jacks T. E., Lodish H. F. (2007) Blood 109, 5238–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenormand P., Sardet C., Pagès G., L'Allemain G., Brunet A., Pouysségur J. (1993) J. Cell Biol. 122, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaaro H., Rubinfeld H., Hanoch T., Seger R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3742–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacquel A., Herrant M., Defamie V., Belhacene N., Colosetti P., Marchetti S., Legros L., Deckert M., Mari B., Cassuto J. P., Hofman P., Auberger P. (2006) Oncogene 25, 781–794 [DOI] [PubMed] [Google Scholar]
- 50.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 51.Deng Y., Chan S. S., Chang S. (2008) Nat. Rev. Cancer 8, 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biggs J. R., Kudlow J. E., Kraft A. S. (1996) J. Biol. Chem. 271, 901–906 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.