Abstract
SET domain lysine methyltransferases (KMTs) methylate specific lysine residues in histone and non-histone substrates. These enzymes also display product specificity by catalyzing distinct degrees of methylation of the lysine ϵ-amino group. To elucidate the molecular mechanism underlying this specificity, we have characterized the Y245A and Y305F mutants of the human KMT SET7/9 (also known as KMT7) that alter its product specificity from a monomethyltransferase to a di- and a trimethyltransferase, respectively. Crystal structures of these mutants in complex with peptides bearing unmodified, mono-, di-, and trimethylated lysines illustrate the roles of active site water molecules in aligning the lysine ϵ-amino group for methyl transfer with S-adenosylmethionine. Displacement or dissociation of these solvent molecules enlarges the diameter of the active site, accommodating the increasing size of the methylated ϵ-amino group during successive methyl transfer reactions. Together, these results furnish new insights into the roles of active site water molecules in modulating lysine multiple methylation by SET domain KMTs and provide the first molecular snapshots of the mono-, di-, and trimethyl transfer reactions catalyzed by these enzymes.
Keywords: Enzyme Mechanisms, Histone Methylation, Protein Methylation, S-Adenosylmethionine, X-ray Crystallography, Chromatin Modification, Isothermal Titration Calorimetry, SET Domain Methyltransferase, TAF10, Transcription
Introduction
SET domain enzymes represent a family of S-adenosylmethionine (AdoMet)3-dependent methyltransferases that catalyze the site-specific methylation of protein lysyl residues in a host of proteins, including histones, transcription factors, chromatin-modifying enzymes, ribosomal subunits, and other substrates (1–3). In many instances, these modifications serve to recruit effector proteins that recognize methyl-lysyl residues in a sequence-dependent fashion (4). In addition, SET domain KMTs exhibit product specificity, defined as their ability to catalyze mono-, di-, or trimethylation of the lysine ϵ-amino group. This specificity is biologically relevant because many methyl-lysine-binding proteins can discriminate among different degrees of lysine methylation (4). Thus, both the site and degree of lysine methylation are critical to recognition by effector proteins.
Structural and functional studies have identified a Phe/Tyr switch in the active site of SET domain KMTs that governs their respective product specificities (5, 6). According to this model, KMTs that possess a tyrosine in the Phe/Tyr switch site are limited to catalyzing lysine monomethylation, whereas enzymes that possess a phenylalanine or another hydrophobic residue in this position display di- or trimethyltransferase activity. Mutational analysis of various SET domain KMTs, including DIM-5 (KMT1) (6), SET7/9 (6), G9A (KMT1C) (5), and SET8 (also known as PR-SET7 and KMT5A) (7, 8), has demonstrated that substitutions in the Phe/Tyr switch result in predictable changes in product specificity. Several models have been proposed to explain the mechanism by which the Phe/Tyr switch site governs this specificity, including variations in the diameter of the active site due to the size of Phe/Tyr switch residue and steric hindrance by the tyrosine hydroxyl group (6, 9–11). However, our recent studies of the Phe/Tyr switch mutant Y334F in the human histone H4 Lys-20 (H4K20) methyltransferase SET8 indicate that the Phe/Tyr switch regulates product specificity via a more subtle mechanism (8). Specifically, the switch modulates the binding of an active site water molecule that in turn regulates the transition from monomethylation to multiple methylation.
Among the KMTs that have been structurally characterized, SET7/9 has emerged as an archetypal model for studying the catalytic mechanism and product specificity of the SET domain family. Although initially isolated as a histone H3 Lys-4 (H3K4)-specific methyltransferase, this KMT has been shown to regulate the functions of numerous non-histone substrates through site-specific methylation (12–21). Early structural and functional studies of SET7/9 identified two active site mutants, Y245A and Y305F, which change its product specificity. The Phe/Tyr switch mutant Y305F alters SET7/9 product specificity from a mono- to dimethyltransferase (6), whereas the Y245A substitution converts the enzyme into a trimethyltransferase with weak monomethyltransferase activity (11). These mutants have been the subjects of numerous molecular modeling simulations that have led to various models to explain their distinct product specificities (22–26). However, the lack of structural data for the SET7/9 Y245A and Y305F mutants in complex with cognate methylated peptides has hindered our understanding of the mechanisms that define the respective product specificities of these mutants. Moreover, these structures would yield a framework for visualizing the mono-, di-, and trimethylation reactions catalyzed by SET domain KMTs.
To gain insight into the molecular basis of their product specificities, we have determined high resolution crystal structures of the SET7/9 Y245A and Y305F mutants in complex with peptides of the TATA box-binding protein-associated factor TAF10 bearing the Lys-189 methylation site in unmodified (K189), monomethylated (K189me1), dimethylated (K189me2), and trimethylated (K189me3) states. The structures and accompanying biochemical data support a model whereby changes in the occupancy or position of water molecules in the active site are critical in establishing the product specificities of the SET7/9 Y245A and Y305F mutants. Together, our results provide new insights into the mechanisms that govern SET domain product specificity and provide stepwise snapshots of the lysine mono-, di-, and trimethyl transfer reactions catalyzed by KMTs.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of the SET7/9 Mutants
The Y245A and Y305F mutants were introduced into the pHIS2 SET7/9 expression vector encoding residues 110–366 (27) via QuikChange site-directed mutagenesis (Stratagene) and were verified by dideoxy DNA sequencing. The plasmids encoding wild type (WT) SET7/9 and the Y245A and Y305F mutants were transformed into Rosetta2 DE3 cells (Novagen) and were expressed as described previously (27, 28). In the course of characterizing WT SET7/9, we observed that the enzyme co-purified with AdoMet or another contaminant that resulted in technical difficulties in the isothermal titration calorimetry (ITC) experiments and co-crystallization trials with the TAF10 peptides. To overcome this problem, a denaturation and refolding step was inserted in the purification scheme. The denaturation and refolding protocol involved adding 6 m guanidine HCl, 50 mm sodium phosphate, pH 7.5, and 500 mm NaCl to the protein while it was immobilized on a nickel-Sepharose column (GE Healthcare). The column was washed with this buffer, followed by a solution of 6 m urea, 50 mm sodium phosphate, pH 7.5, 150 mm NaCl, and 10 mm 2-mercaptoethanol to remove the cofactor from the denatured enzyme. A reverse gradient from 6 to 0 m urea was then performed in the same buffer to refold the protein, which was subsequently eluted from the column using a linear gradient of 0–500 mm imidazole in 50 mm sodium phosphate, pH 7.5, 500 NaCl, and 10 mm 2-mercaptoethanol. The refolded protein was digested with tobacco etch virus protease (29) during dialysis against 20 mm Tris, pH 7.5, 150 mm NaCl, and 5 mm 2-mercaptoethanol and then purified using a Superdex 200 gel filtration column (GE Healthcare). Protein concentration was determined by its absorbance at 280 nm.
Synthetic Peptides
The TAF10 peptides bearing K189, K189me1, K189me2, and K189me3 (sequence, acetyl-SKSK189DRKYTL-amide) and a biotinylated TAF10 peptide (sequence, acetyl-SKSK189DRKYTLT(K-EZLink-S-S-biotin)-amide) were synthesized and purified by New England Peptide, Inc. Peptide concentrations were measured using the absorbance of their tyrosine residue at 274 nm.
Crystallization and Data Collection
Crystals were produced by hanging drop vapor diffusion by mixing the crystallization solution in a 1:1 ratio with 10–20 mg/ml SET7/9, 1 mm S-adenosylhomocysteine (AdoHcy), and 1.0–3.0 mm unmodified or methylated TAF10-K189 peptide in 20 mm Tris, pH 8.0, 100 mm NaCl, and 2 mm tris(2-carboxyethyl)phosphine. Crystals were obtained at 20 °C in either 1.8–2.0 m (NH4)2SO4, with 0.1 m BisTris, pH 6.2–6.6, or in 0.95–1.1 m sodium citrate with 100 mm imidazole pH 8.0–8.4. In both crystallization conditions, the final pH values were between pH 8.0 and 9.0. Crystals in the (NH4)2SO4 condition were typically flash-frozen in the mother liquor containing 25–30% glycerol, and the crystals in the citrate condition were frozen in 1.6 m sodium citrate. Data were collected at the Advanced Photon Source beamlines 21-IDG (LS-CAT) and 23-IDD (GM/CA-CAT). Images were indexed, integrated, and scaled using HKL2000 (30). Structures of the mutants were solved by molecular replacement using MOLREP (31) with the coordinates of a previously reported SET7/9 ternary complex used as the search model (Protein Data Bank code 2F69). Successive rounds of model building and refinement were carried out using Coot (32) and REFMAC (33), respectively. The geometry of the models were verified by MolProbity (34). Simulated annealing omit maps were calculated using CNS (35) with the peptide and cofactor removed to eliminate model bias in the active site. Structural figures were rendered using PyMOL (Schrödinger, LLC.).
Fluorescent Methyltransferase Assay
A coupled fluorescent methyltransferase assay was used to measure the kinetic parameters of WT SET7/9 and the Y245A and Y305F mutants as reported previously, with the exception that 50–150 nm enzyme, 100 μm AdoMet, and varying concentrations of TAF10 peptide substrate were used (27, 36). Assays were performed in triplicate, and a homocysteine calibration curve was used to calculate the initial velocities. Kinetic parameters were calculated by plotting the velocities versus peptide concentration and by fitting the Michaelis-Menten equation to the data via nonlinear regression using Prism 5.0 (GraphPad). In cases where the Km value was beyond the measurable range of the assay, the kcat/Km value was determined as described previously (7).
Calorimetry Experiments
ITC was performed at 20 °C using a MicroCal VP-ITC calorimeter (GE Healthcare) with 0.12 mm protein and 4 mm AdoHcy in 20 mm sodium phosphate, pH 7, and 100 mm NaCl with 1.5 mm peptide as the injectant. Data were processed, and equilibrium dissociation constants (KD) and curve fitting errors were calculated from the binding isotherms using Origin 7.0 (OriginLab Corp.). WT SET7/9 and the Y245A and Y305F mutants displayed ligand:protein binding stoichiometries (N values) between 0.8 and 1.0, demonstrating that WT SET7/9 and its mutants were properly refolded due to their ability to bind peptides in an ∼1:1 molar ratio.
TLC Product Analysis
Methyltransferase assays were performed in triplicate at 37 °C with the biotinylated TAF10 peptide (0.5 mm) for 30 min in the presence of WT SET7/9 (3 pmol), Y305F mutant (6 pmol), or the Y245A mutant (100 pmol). Assays contained 100 mm HEPES, pH 8.0, 50 mm NaCl, 1.0 mm [methyl-3H]AdoMet (diluted to a specific activity of 0.2 μCi/nmol with purified AdoMet (37)), 15 μm Sulfolobus solfataricus AdoHcy hydrolase (36), and 2 units of adenosine deaminase (Roche Applied Science) in a final volume of 20 μl. The reactions were terminated by addition of an equal volume of 200 mm MES, pH 5.1, prior to addition of a 2-fold molar excess of immobilized avidin resin (UltraLink; Pierce). Biotinylated peptides were allowed to bind at room temperature for 30 min, and the resin was then collected by centrifugation (∼9000 × g). The resin was washed three times with 300 mm NaCl, and the peptide was eluted overnight from the avidin resin by cleavage of the disulfide bond in the linker of the peptide using 10 mm tris(2-carboxyethyl)phosphine (Thermo Scientific). The resin was incubated with additional 10 mm tris(2-carboxyethyl)phosphine the following day until the radiolabel was essentially removed from the resin. The recovered peptides were hydrolyzed by incubation with 6 m HCl at 110 °C for 24 h. Subsequent steps in measuring the radiolabel incorporated into the mono-, di-, and trimethyl-lysine products were performed as reported previously (8).
RESULTS
Functional Analysis of the SET7/9 Y305F Mutant
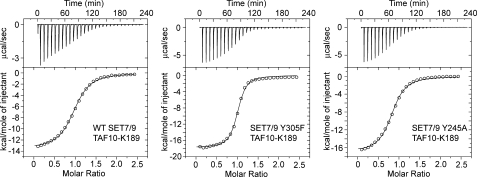
Prior studies of SET7/9 by Zhang et al. (6) reported that mutation of the Phe/Tyr switch residue Tyr-305 to a phenylalanine alters its product specificity from a mono- to dimethyltransferase. We verified these findings by demonstrating that WT SET7/9 monomethylated the TAF10-K189 peptide, whereas the Y305F mutant mono- and dimethylated this substrate, as demonstrated by mass spectrometry (data not shown). We next examined whether the Y305F substitution altered the affinity of SET7/9 for the TAF10-K189 peptides using ITC (Fig. 1). A comparison of the KD values revealed that SET7/9 Y305F bound the TAF10-K189 and TAF10-K189me1 peptides ∼4- and 6-fold more tightly, respectively, than the WT enzyme, whereas this mutant displayed a substantially diminished affinity for the TAF10-K189me2 peptide (Table 1). Although the WT enzyme and the Y305F mutant exhibited discernable differences in their affinities for the unmodified and monomethylated peptides, these variations are modest and cannot account for their distinct product specificities, suggesting that a kinetic effect during methylation may be responsible.
FIGURE 1.
ITC analysis of WT SET7/9 and the Y245A and Y305F mutants. Representative titrations and binding isotherms for the calorimetry experiments are shown for the unmodified TAF10-K189 peptide titrated into WT SET7/9 (left plot), SET7/9 Y305F (middle plot), and SET7/9 Y245A (right plot). In each panel, the ITC titration experiment (upper panel) is shown with the binding isotherms (lower panel) fit to a single binding site model.
TABLE 1.
Analysis of the binding affinity of WT SET7/9 and its catalytic mutants for unmodified and methylated TAF10 peptides
| SET7/9 | TAF10 peptide | KDa |
|---|---|---|
| μm | ||
| WT | K189 | 4.9 ± 0.20 |
| WT | K189me1 | 4.0 ± 0.36 |
| Y305F | K189 | 1.3 ± 0.10 |
| Y305F | K189me1 | 0.62 ± 0.065 |
| Y305Fb | K189me2 | >70 |
| Y245A | K189 | 4.0 ± 0.25 |
| Y245A | K189me1 | 3.3 ± 0.10 |
| Y245A | K189me2 | 5.8 ± 0.22 |
| Y245A | K189me3 | 11 ± 0.28 |
a Curve fitting errors were calculated from the binding isotherms.
b An estimate of the affinity is reported due to weak peptide binding.
To investigate this possibility, we characterized the kinetic parameters of WT SET7/9 and the Y305F mutant using the TAF10 peptides as substrates. Both enzymes methylated the unmodified peptide with comparable kcat and Km values (Table 2). In analyzing the kinetic parameters for the methylation of the monomethylated peptide by SET7/9 Y305F, we found that this substrate displayed an elevated Km value that was beyond the measurable range of the assay due to its limited solubility. In this case, we measured the catalytic efficiency (kcat/Km) for the methylation of this peptide and found that it was methylated >15-fold less efficiently than the unmodified peptide by SET7/9 Y305F. Given the fact that the Y305F mutant exhibited a higher binding affinity for the TAF10-K189me1 peptide than the WT enzyme (Table 1), the kinetic data suggest that a step in the reaction pathway following substrate binding limits the catalytic efficiency of this mutant.
TABLE 2.
Kinetic parameters of WT SET7/9 and the Y305F and Y245A mutants
| Enzyme | TAF10 peptide substrate | Kma | kcata | kcat/Kma |
|---|---|---|---|---|
| μm | min−1 | μm−1min−1 × 103 | ||
| WT | K189 | 160 ± 17 | 17 ± 0.62 | 110 ± 17 |
| Y305F | K189 | 88 ± 5.0 | 17 ± 0.30 | 190 ± 11 |
| Y305Fb | K189me1 | 11 ± 0.50 | ||
| Y245A | K189 | 200 ± 35 | 0.53 ± 0.04 | 2.6 ± 0.47 |
| Y245A | K189me1 | 210 ± 23 | 5.9 ± 0.23 | 28 ± 3.3 |
| Y245A | K189me2 | 400 ± 29 | 6.5 ± 0.16 | 15 ± 1.2 |
a Curve fitting errors were calculated from the hyperbolic fits for the Michaelis-Menten equation.
b Km and kcat were not determined because the Km value was beyond the measurable range; therefore, the kcat/Km value is reported.
We next examined whether the Y305F mutant dimethylated the TAF10-K189 peptide via a processive or a distributive mechanism. In a processive mechanism, the methyl-lysine substrate would remain bound to the enzyme during successive methyl transfer reactions; thus, the concentration of an intermediate, such as monomethyl-lysine, cannot exceed the enzyme concentration during the assay. In a distributive mechanism, the intermediates are released into solution where they accumulate prior to the next round of methylation, resulting in an intermediate concentration that is greater than that of the enzyme. Using a radiometric TLC assay and a biotinylated TAF10 peptide, we quantified the amounts of monomethylated products generated by the WT SET7/9 and the Y305F mutant (Table 3). The data demonstrate that comparable amounts of monomethyl-lysine were generated when the quantity of enzyme used is taken into account, in agreement with their similar turnover numbers for the TAF10-K189 peptide (Table 2). The Y305F mutant also produced small but measurable quantities of radiolabeled dimethyl-lysine product that were substantially smaller than the amount of monomethyl-lysine generated. Therefore, the TLC data are consistent with a distributive mechanism for dimethylation by the Y305F mutant because the amount of monomethyl-lysine produced exceeded the quantity of enzyme used in the assay.
TABLE 3.
Product analysis of WT SET7/9 and the Y305F and Y245A mutants
| Enzyme | Quantity of enzyme | Measured product | Amount of product formeda |
|---|---|---|---|
| nmol | nmol | ||
| WT | 0.003 | Kme1 | 0.65 ± 0.07 |
| Y305F | 0.006 | Kme1 | 1.5 ± 0.49 |
| Kme2 | 0.033 ± 0.009 | ||
| Y245A | 0.100 | Kme1 | 0.80 ± 0.22 |
| Kme2 | 0.39 ± 0.021 | ||
| Kme3 | 0.076 ± 0.019 |
a Standard deviation was calculated from triplicate measurements.
Structures of WT SET7/9 and the Y305F Mutant in Complex with Unmodified and Methylated TAF10 Peptides
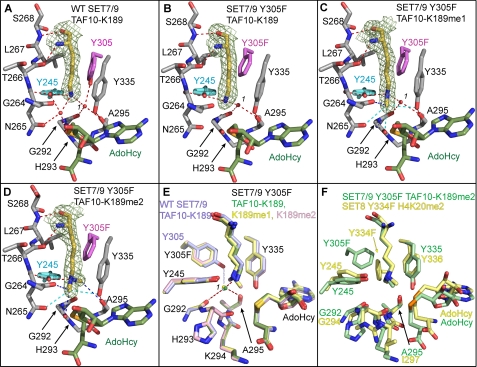
To determine the mechanism by which the Y305F substitution alters the product specificity of SET7/9, we determined the crystal structures of this mutant bound to AdoHcy and TAF10-K189, TAF10-K189me1, and TAF10-K189me2 peptides and compared these to the structures of the WT SET7/9·AdoHcy· TAF10-K189 complex (supplemental Table 1). The structures of these complexes were determined to 1.85 Å or higher resolution, permitting unambiguous modeling of the K189 side chains in the active site of the enzyme based on simulated annealing omit maps (Fig. 2). The ternary complexes of the WT and the Y305F mutant superimpose with overall root mean square differences of less than 0.3 Å for all aligned atoms, indicating that neither the Y305F mutation nor the binding of the various TAF10-K189 peptides results in substantial changes in its overall structure.
FIGURE 2.
Crystal structures of WT SET7/9 and the Y305F mutant in complex with AdoHcy and unmodified and methylated TAF10 peptides. A, active site of WT SET7/9 bound to AdoHcy and TAF10-K189 and the active site of SET7/9 Y305F bound to AdoHcy and TAF10-K189 (B), TAF10-K189me1 (C), and TAF10-K189me2 peptides (D). A–D, the K189 residue in the TAF10 peptide is depicted with yellow carbon atoms with the corresponding Fo – Fc simulated-annealing omit maps (contoured at 2.5 σ) shown. AdoHcy is colored with green carbon atoms, and SET7/9 carbon atoms are colored gray, with the exception of Tyr-305/Y305F (magenta) and Tyr-245 (cyan). Gly-264 to Ser-268 are shown, and the main chain atoms of Gly-292 to Ala-295 are depicted for clarity. Red dashed lines indicate conventional hydrogen bonds, and blue dashed lines indicate CH–O hydrogen bonds. Cyan dashed lines indicate weak CH–O hydrogen bonds between 3.5 and 3.7 Å in length. The water molecule in the solvent binding pocket is numbered 1. E, overlay of the active sites of the WT enzyme (light blue carbon atoms) and the Y305F mutant (green, yellow, and pink carbon atoms correspond to K189, K189me1, and K189me2, respectively). Red dashed lines indicate coordination of the waters in the structures of the Y305F mutant, and the light blue dashed line indicates hydrogen bonding that only occurs in the WT enzyme. F, superimposition of the active sites of the SET7/9 Y305F·AdoHcy·TAF10-K189me2 and SET8 Y334F·AdoHcy·H4K20me2 (Protein Data Bank code 3F9X) complexes shown with green and yellow carbon atoms, respectively.
An inspection of the active sites of the SET7/9 WT and Y305F complexes illustrates the binding modes of the unmodified and methylated forms of K189 in the TAF10 peptides (Fig. 2, A–D). The K189 side chain binds in an extended all trans conformation in a deep pocket, termed the lysine binding channel, that is composed of Tyr-245, Thr-266, Leu-267, Ser-268, Tyr-305, Tyr-335, and Tyr-337 (not shown for clarity) (Fig. 2A). These residues interact with the aliphatic portion of the K189 side chain primarily through van der Waals contacts. The lysine binding channel connects to the AdoMet-binding site on the opposite face of the catalytic domain via an oxygen-lined methyl transfer pore (38). During catalysis, the methyl group of the cofactor is positioned within the methyl transfer pore for the SN2 reaction with the ϵ-amino group of the lysine or methyl-lysine substrate (see below).
To lower the activation barrier for this reaction, the lysine ϵ-amine nucleophile is aligned for methyl transfer through a hydrogen bond network within the active site. In the WT enzyme, the K189 ϵ-amino group hydrogen bonds to the hydroxyl group of Tyr-245 as well as to two water molecules (Fig. 2A). One of the water molecules (termed water 1), is coordinated in a solvent pocket, through hydrogen bonds to the carbonyl oxygens of Gly-292 and Ala-295 and to the hydroxyl group of the Phe/Tyr switch residue Tyr-305. This solvent pocket is structurally conserved in SET domain KMTs and has an important role in defining product specificity through the adjacent Phe/Tyr switch residue, as shown in our prior studies of the human H4K20 methyltransferase SET8 (8). The other water molecule is bound within the methyl transfer pore between the lysine substrate and the thioether sulfur atom of AdoHcy through hydrogen bonds to Tyr-245, Asn-265, and His-293 in SET7/9 and the TAF10-K189 ϵ-amino group. This water is not observed in other structures of SET7/9 ternary complexes and may represent the approximate position that the AdoMet methyl group occupies in the methyl transfer pore in the Michaelis complex.
In structures of the Y305F ternary complexes, the K189, K189me1, and K189me2 side chains also adopt extended trans side chain geometries within the lysine binding channel that are stabilized via hydrogen bonding to Tyr-245 in the enzyme (Fig. 2, B–D). The orientations of the K189me1 and K189me2 side chains are further maintained through carbon-oxygen (CH–O) hydrogen bonding between the methyl groups and oxygen atoms within the vicinity of the methyl transfer pore, as reported previously in other SET domain KMT structures (8, 10, 38). A superimposition of the SET7/9 WT and Y305F complexes underscores the similarity of the lysyl binding conformations (Fig. 2E). However, there are notable differences in the hydrogen bond patterns and occupancy of water 1 within the solvent pocket in the Y305F mutant compared with the WT enzyme. Specifically, the Y305F substitution results in the loss of one hydrogen bond to water 1 in the structures of the TAF10-K189 and TAF10-K189me1 complexes (Fig. 2, B and C). In contrast, water 1 is absent in TAF10-K189me2 complex, and the vacated solvent pocket is occupied by one of the methyl groups of the dimethyl ϵ-amine (Fig. 2D). This methyl group forms a 3.5-Å CH–O hydrogen bond to the carbonyl oxygen of Ala-295, further stabilizing the binding of the dimethyl-lysine side chain. A homologous dimethyl-lysine-binding mode and CH–O hydrogen bond was observed in our prior structural studies of the SET8 Y334F Phe/Tyr switch mutant that confers an analogous change in product specificity from a mono- to a dimethyltransferase (8). A structural alignment of the active sites of the SET7/9 Y305F and SET8 Y334F mutants bound to cognate dimethylated peptides illustrates that the coordinates of the dimethyl-lysyl side chains are virtually superimposable, with one methyl group oriented toward the methyltransfer pore and the second positioned within the vacant solvent pocket (Fig. 2F). Taken together, the structures of the SET7/9 Y305F complexes and the similarities in the dimethyl-lysine conformations in the SET7/9 Y305F and SET8 Y334F mutants imply that the Phe/Tyr switch governs product specificity through a conserved mechanism whereby it indirectly influences the binding modes of the methyl-lysine side chain by modulating the affinity of the water molecule (water 1) bound in the solvent pocket.
Biochemical Characterization of the SET7/9 Y245A Mutant
Previous studies by Xiao et al. (11) reported that the Y245A mutation yields an unusual change in the product specificity of SET7/9, converting the enzyme to a trimethyltransferase with weak monomethyltransferase activity. We determined that the SET7/9 Y245A could mono-, di-, and trimethylate the TAF10-K189 peptide by mass spectrometry (data not shown) and TLC (Table 3), confirming the earlier studies of Xiao et al. (11). ITC analysis revealed that the Y245A mutant displayed comparable KD values for the unmodified and methylated TAF10-K189 peptides (Fig. 1), although its affinity for the trimethylated peptide was modestly diminished in comparison with the other peptides (Table 1). The ITC data demonstrate that the Y245A mutant bound the unmodified, mono-, and dimethylated substrates with equivalent affinities, suggesting that a kinetic effect or a structural alteration in the active site may be responsible for its diminished activity toward unmodified substrates.
To gain further insight into its peculiar product specificity, we characterized the kinetic properties of the SET7/9 Y245A mutant. Steady state analysis demonstrated that this mutant displayed similar Km values for the unmodified, mono- and dimethylated TAF10 peptides (Table 2). However, the turnover number for the TAF10-K189 peptide was diminished over 10-fold versus the methylated peptides and was reduced ∼30-fold versus the WT enzyme, in agreement with the weak monomethyltransferase activity reported by Xiao et al. (11). In addition, we investigated whether this mutant catalyzes lysine trimethylation via a processive or distributive mechanism as described for SET7/9 Y305F. The TLC data illustrate that the mono- and dimethyl-lysine intermediates accumulated at quantities greater than that of the enzyme used in the assay, indicating that SET7/9 Y245A obeys a distributive mechanism, analogous to the Y305F mutant (Table 3).
Structures of SET7/9 Y245A Bound to Unmodified and Methylated TAF10 Peptides
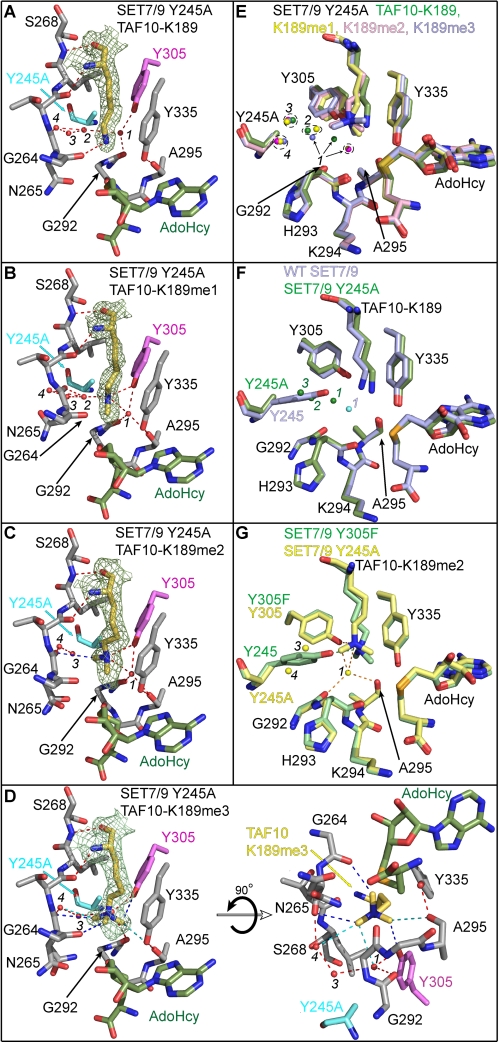
To elucidate the mechanism underlying its unusual product specificity, we determined the crystal structures of SET7/9 Y245A in complex with AdoHcy and unmodified, mono-, di-, and trimethylated TAF10 peptides (supplemental Table 1). These complexes superimpose with the structure of the WT SET7/9·AdoHcy·TAF10-K189 complex with root mean squared differences of less than 0.4 Å for all aligned atoms, indicating that the Y245A mutant does not perturb the overall structure of the enzyme. Simulated annealing omit maps illustrate that K189 side chains are bound within the lysine binding channel through hydrogen bonds and van der Waals contacts (Fig. 3, A–D), although the interactions and binding modes are distinct from those in the complexes of WT SET7/9 and the Y305F mutant (Fig. 2, A–D). In the unmodified TAF10 peptide complex, the K189 ϵ-amino group forms a weak hydrogen bond with the carbonyl oxygen of Gly-264 (Fig. 3A), whereas the ϵ-amino groups of K189me1 and K189me2 hydrogen bond to the hydroxyl group of Tyr-305 in the mono- and dimethylated peptide complexes (Fig. 3, B and C). The conformations of the K189me1 and K189me2 side chains are further stabilized by water-mediated hydrogen bonding and through CH–O hydrogen bonding to their methyl groups. In the TAF10-K189me3 peptide complex, the trimethyl-lysine side chain is coordinated exclusively through direct and water-mediated CH–O hydrogen bonds to its methyl groups because the quaternary ϵ-ammonium cation cannot engage in hydrogen bonding (Fig. 3D).
FIGURE 3.
Crystal structures of the SET7/9 Y245A mutant in complex with AdoHcy and unmodified and methylated TAF10 peptides. Active site of SET7/9 Y245A bound to AdoHcy and TAF10-K189 (A), TAF10-K189me1 (B), TAF10-K189me2 (C), and TAF10-K189me3 peptides (D). A–D, Fo – Fc simulated-annealing omit maps (contoured at 2.5 σ) for the unmodified and methylated K189 side chains are illustrated. The residues and hydrogen bonds in each complex are colored as in Fig. 2. The water molecules in the lysine binding channels of the Y245A complexes are numbered 1–4, as described in the text. E, superimposition of the active sites of the Y245A complexes bound to the four methylated states of TAF10-K189 (unmodified, mono-, di-, and trimethylated shown in green, yellow, pink, and blue, respectively). F, overlay of WT SET7/9 (blue carbons) and SET7/9 Y245A (green carbons) in complex with TAF10-K189. Waters corresponding to the WT and Y245A structures are colored cyan and green, respectively. G, alignment of the active sites of the Y245A (yellow carbons) and Y305F (green carbons) mutants in complex with the TAF10-K189me2 peptide. Hydrogen bonds from the Y305F structure are shown as green dashed lines, and waters and hydrogen bonds in the Y245A structure are shown in yellow and orange, respectively.
A structural alignment of the four SET7/9 Y245A complexes illustrates distinct binding modes for the unmodified versus the methylated K189 side chains, highlighting the selectivity of this mutant for methylated substrates. The side chains of K189me1, K189me2, and K189me3 roughly overlay with their respective ϵ-amino groups superimposed and adopt slightly kinked conformations (Fig. 3E), as opposed to the extended trans geometry of the unmodified and methylated lysines in the complexes of the WT enzyme and the Y305F mutant (Fig. 2, A–D). Conversely, the unmodified K189 side chain does not superimpose with its methylated counterparts and is oriented in an alternative configuration due to its hydrogen bonding to Gly-264 (Fig. 3, A and E). An overlay of the structures of the WT enzyme and Y245A mutant bound to the unmodified TAF10 peptide illustrates that the side chains of K189 do not superimpose and that the K189 ϵ-amino group appears to be misaligned with AdoHcy in the Y245A complex (Fig. 3F). This suboptimal alignment may explain the diminished kcat value of SET7/9 Y245A mutant toward substrates with unmodified lysines (Table 2).
A comparison of the structures of the SET7/9 Y245A and Y305F complexes yields a molecular explanation for the different product specificities of these two mutants. In the SET7/9 Y305F complexes, Tyr-245 aligns the K189 ϵ-amino group for methyl transfer through hydrogen bonding to its hydroxyl group (Fig. 2, B–D). Conversely, in the Y245A mutant, the K189me1 and K189me2 ϵ-amino groups are oriented through hydrogen bonding to Tyr-305 (Fig. 3, B and C). These distinct hydrogen bond patterns impart differences in the conformations of the lysyl side chains due to the relative orientations of Tyr-245 and Tyr-305 in the lysine binding channel. Specifically, the kinked conformation adopted by the K189me1 and K189me2 side chains in the Y245A complexes (Fig. 3, B and C) may contribute to the differences in the turnover numbers of this mutant versus those of the WT enzyme and the Y305F mutant (Table 2). In addition, the dimethyl ϵ-amino group of the K189me2 side chain binds in distinct orientations in the Y245A and Y305F mutants due to their hydrogen bonding to Tyr-305 and Tyr-245, respectively (Fig. 3G). In the Y305F mutant, hydrogen bonds to the dimethyl ϵ-amino group coupled with steric constraints in the lysine binding channel prevent the K189me2 side chain from undergoing a conformational change that is conducive to trimethylation (Fig. 2D), consistent with its dimethyltransferase activity. However, in the Y245A mutant, the alanine substitution enlarges the diameter of the lysine binding channel, accommodating trimethyl-lysine (Fig. 3D). In addition, the larger diameter would permit the dimethyl-lysine substrate to undergo the conformational reorganization necessary to align the ϵ-amino group in a productive geometry for trimethylation.
A major difference in the active site of the Y245A mutant versus the other SET7/9 structures is the presence of several water molecules bound in the cavity generated by the Y245A mutation. In the structure of the Y245A mutant bound to TAF10-K189, three water molecules (waters 2–4) occupy this cavity and are arranged in a triangular geometry (Fig. 3A). In addition, water 1 shifts ∼1.6 Å from its position in the solvent pocket toward water 2 to which it forms a hydrogen bond (Fig. 3, A and E). The shift in water 1 was unexpected given its conserved orientation in the solvent pocket of the SET7/9 WT and Y305F complexes (Fig. 2, A–C) as well as in the structures of other SET domain KMTs (8). This displacement is presumably related to the alternative conformation of the K189 side chain whose ϵ-amino group is too distant (4.3 Å) to form a productive hydrogen bond to water 1. Conversely, in the Y245A complexes bound to TAF10-K189me1 and TAF10-K189me2, water 1 remains tightly bound in the solvent pocket through hydrogen bonds to Tyr-305 hydroxyl group, the carbonyl oxygens of Gly-292 and Ala-295, and the K189 ϵ-amino group (Fig. 3, B, C, and E), analogous to its binding in the WT enzyme (Fig. 2A). However, in the TAF10-K189me3 complex, one of the methyl groups of the trimethyl ϵ-ammonium cation is oriented into the solvent pocket (Fig. 3D), similar to the dimethyl-lysine binding mode observed in the Y305F mutant (Fig. 2D). The binding of the methyl group in the solvent pocket displaces water 1 by 3.2 Å relative to its position in the TAF10-K189me1 complex (Fig. 3E), thereby avoiding a steric clash with the trimethylated ϵ-ammonium group. Variations in the occupancy of water 2 are also seen in the different Y245A structures. Water 2 is bound in similar orientations in the active site of the unmodified and monomethylated peptide complexes but is absent in the di- and trimethylated peptide complexes due to the binding of a methyl group in this position (Fig. 3, A–E). In summary, the changes in the positions or occupancies of waters 1 and 2 correlate with the binding modes of the unmodified and methylated K189 within the active site of the Y245A mutant.
Catalytic Models of Lysine Multiple Methylation by SET7/9 Y245A, and Y305F
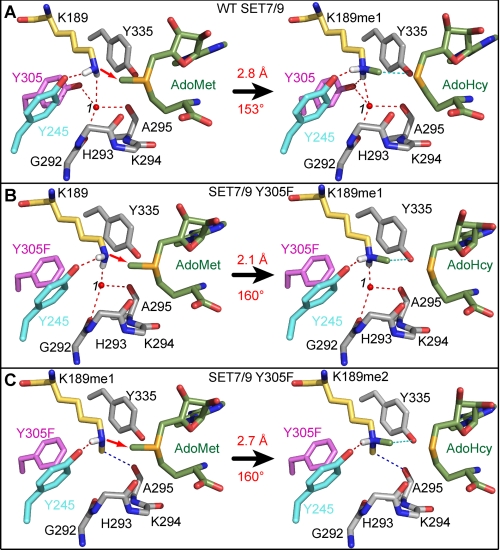
The structures of the SET7/9 complexes reported here offer a prime opportunity to generate stepwise models for lysine mono-, di-, and trimethylation by a SET domain KMT. We modeled the AdoMet-bound Michaelis complexes by superimposing the SET7/9 product complexes with the previously reported structure of the SET7/9-AdoMet binary complex (Fig. 4) (39). The conformations of the mono- and dimethyl ϵ-amino groups in the Michaelis complexes were inferred from the coordinates of the corresponding dimethyl- and trimethyl-lysine products, respectively. In addition, we modeled the ϵ-amino group in a deprotonated state with its hydrogen atoms oriented toward the hydrogen bond acceptors that align the lysyl side chain for methylation. As a basis for this comparison, we first modeled the monomethylation reaction catalyzed by WT SET7/9 (Fig. 4A). In the substrate ternary complex, the lysine ϵ-amine is aligned with the methyl group and sulfonium cation of AdoMet through a hydrogen bond to the Tyr-245 hydroxyl group and water 1 in the solvent pocket. The values of the reaction distance and angle are 2.8 Å and 153°, respectively, in approximate agreement with the linear geometry of a SN2 methyl transfer reaction calculated in other modeled substrate complexes (8, 10). In the product complex, the monomethyl-lysine side chain is bound in an extended conformation with its methyl group oriented within the methyl transfer pore, thereby obstructing AdoMet binding. Furthermore, water 1 remains tightly coordinated in the solvent pocket through four hydrogen bonds to Gly-292, Ala-295, Tyr-305, and the monomethyl ϵ-amino group. These interactions hinder the dissociation of water 1 and the related rearrangement of the monomethyl-lysine side chain required for a second methyl transfer reaction, explaining why the WT enzyme cannot catalyze di- and trimethylation. These findings concur with the model for SET7/9 product specificity reported in previous structural and functional studies (6, 11).
FIGURE 4.
Catalytic models of the methyl transfer reactions catalyzed by WT SET7/9 and the Y305F mutant. A, monomethylation of TAF10-K189 by the WT enzyme. The reaction scheme depicts the modeled substrate ternary complex (left) and the product complex (right) for the transfer of the methyl group from AdoMet (green carbon atoms) to K189 in TAF10 (yellow carbons), yielding AdoHcy and K189me1. The red arrow indicates the direction of the nucleophilic attack of the deprotonated ϵ-amino group on the AdoMet methyl group. The transferred methyl group is colored green, and the white atoms represent the hydrogens of the ϵ-amino group. Hydrogen bonds and residues in the enzyme active site are illustrated as in Fig. 2. The reaction distance and angle are labeled in red. B and C, models of the Y305F mutant for the first methyl transfer reaction with TAF10-K189 (B) and second methyl transfer reaction with TAF10-K189me1 (C). Color schemes are the same as in A.
Similar reaction geometry is observed in the model for the monomethyl transfer reaction catalyzed by SET7/9 Y305F. Hydrogen bonds from the Tyr-245 hydroxyl group and water 1 align the lysine ϵ-amino group with the AdoMet methyl group at a distance of 2.1 Å and an angle of 160°, equivalent to those measured in the Michaelis complex of the WT enzyme (Fig. 4, A and B). In the product complex, monomethyl-lysine adopts an extended trans configuration analogous to that in the WT enzyme. For dimethylation to occur, the monomethyl-lysine must undergo a conformational change in which its methyl group is rotated out of the methyl transfer path with AdoMet. The structure of the Y305F mutant bound to the dimethylated TAF10 peptide (Fig. 2D) implies that this rearrangement occurs through the dissociation of water 1 due to the loss of the Tyr-305 hydrogen bond in the solvent pocket. The dissociation of water 1 would enable the monomethyl-lysine side chain to adopt an alternative conformation through a rotation about its Cϵ–Nζ bond, projecting the methyl group into the solvent pocket (Fig. 4C). This rotation reorients the methyl group out of the methyl transfer path while realigning the monomethyl ϵ-amino group for a second methylation reaction through a direct hydrogen bond to the Tyr-245 hydroxyl group and a CH–O hydrogen bond between its methyl group and Ala-295. The modeled reaction geometry for monomethyl-lysine substrate complex of the Y305F mutant (2.7 Å, 160°) is equivalent to that of the first methyl transfer reaction in SET7/9 Y305F. These geometries concur with our previous models for mono- and dimethylation catalyzed by SET8 Y334F (8), illustrating that the orientation of a methyl group into the solvent pocket is a conserved feature of SET domain KMTs that catalyze multiple methylation.
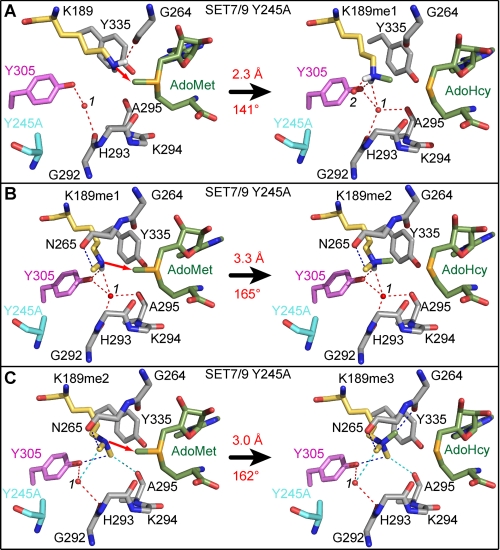
In addition, we modeled the methyl transfer reactions catalyzed by SET7/9 Y245A (Fig. 5). In the model of the lysine substrate complex, the ϵ-amino group is aligned for methyl transfer by a hydrogen bond to the carbonyl oxygen of Gly-264, resulting in a short reaction distance (2.3 Å) and a suboptimal reaction angle (141°) with the methyl group of AdoMet (Fig. 5A). This misalignment appears to be a direct consequence of the Y245A mutation that abolishes hydrogen bonding to the ϵ-amino group, illustrating that the suboptimal orientation of the ϵ-amine likely contributes to the diminished activity of this mutant toward unmodified substrates (11). Conversely, in the modeled monomethyl-lysine substrate complex for SET7/9 Y245A, Tyr-305 hydrogen bonds to the ϵ-amino group, aligning it for methyl transfer (Fig. 5B). In addition, CH–O hydrogen bonds to the monomethyl-lysine methyl group and the dissociation of water 2 from the active site also contribute to repositioning the ϵ-amino group for dimethylation. Collectively, these interactions orient the ϵ-amine in a reaction angle of 165° that is more conducive to methyl transfer. However, the reaction distance for dimethylation is ∼0.6 Å longer than that in the corresponding Y305F model because Tyr-305 is positioned further from AdoMet than Tyr-245 (Figs. 4C and 5B).
FIGURE 5.
Models for the methyl transfer reactions catalyzed by the SET7/9 Y245A mutant. Models of the Y245A mutant for the first methyl transfer reaction with TAF10-K189 (A), the second methyl transfer reaction with TAF10-K189me1 (B), and the third methyl transfer reaction with TAF10-K189me2 (C). Residues and hydrogen bonds are depicted as in Fig. 3.
In the third methyl transfer reaction catalyzed by SET7/9 Y245A, the lone pair of electrons of the dimethyl-lysine ϵ-amino group acts as the nucleophile and thus cannot engage in hydrogen bonding. The structure of the trimethyl-lysine product complex (Fig. 3D) implies that the dimethyl ϵ-amine is aligned via CH–O hydrogen bonds to its methyl groups, as shown in the model of the Michaelis complex for this reaction (Fig. 5C). These CH–O hydrogen bonds restrain the orientation of the ϵ-amino group and position one of the methyl groups into the solvent pocket, displacing water 1 as discussed earlier (Fig. 3, D and E). These interactions cumulatively align the ϵ-amino group and AdoMet methyl group with a reaction distance of 3.0 Å and angle of 162° (Fig. 5C). Taken together, the models of the substrate complexes for SET7/9 Y245A suggest that CH–O hydrogen bonds play an increasingly important role in aligning the methylated ϵ-amino group in successive rounds of methyl transfer.
DISCUSSION
The structural and functional characterization of the SET7/9 Y245A and Y305F mutants presented here yields new insights into the mechanism underlying the product specificity of SET domain KMTs. Importantly, it resolves a general paradox concerning this specificity. How does the active site constrain the motion of the lysine ϵ-amino group to align it for methyl transfer with AdoMet, while providing adequate volume to accommodate the mono-, di-, and trimethylated lysine side chain generated during multiple methyl transfer reactions? The structures of the Y305F and Y245A mutants resolve this paradox, illustrating that alterations in the positions or occupancies of water molecules within their active sites generate the space required to accommodate the multiply methylated ϵ-amine produced during successive catalytic cycles. Minor perturbations in the side chains of certain active site residues, such as Tyr-305, are also observed in alignments of the WT enzyme and the Y245A and Y305F complexes, although these changes are modest compared with the displacement or dissociation of the water molecules in the active site. These findings suggest that the waters function as transient place holders that facilitate the SN2 methyl transfer reaction. During monomethylation, they function to constrain the movement of the lysine ϵ-amino group by mediating hydrogen bonds between the substrate and enzyme, thereby promoting the linear alignment with the methyl group and sulfonium cation of AdoMet (Fig. 4, A and B). During di- and trimethylation, the water molecules either relocate within the lysine binding channel or dissociate from the enzyme, yielding the space required to rotate the methyl group away from the methyl transfer pore and to realign the ϵ-amine in productive geometry for the next methyl transfer reaction (Figs. 4C and 5, B and C). These findings agree with our prior analysis of the SET8 Phe/Tyr switch mutant in which we demonstrated that the Y334F substitution attenuates hydrogen bonding to the water molecule bound in the solvent pocket, promoting its dissociation and the conformational changes necessary for lysine dimethylation (8). Indeed, there is a nearly identical alignment of the dimethyl-lysine side chains in the structures of SET7/9 Y305F and SET8 Y334F complexes, despite the differences in the orientations of the Phe-305 and Phe-334 side chains in each structure (Fig. 2F). Finally, both the SET8 Y334F (8) and SET7/9 Y305F mutants (Table 2) displayed diminished catalytic efficiencies for lysine dimethylation versus monomethylation. These differences may reflect the kinetics of the reorganization within the active site, including the dissociation of the water molecule from the solvent pocket and the concomitant realignment of the monomethyl-lysine into a productive geometry for dimethylation.
In addition to their place-holding role, the active site waters may also facilitate the deprotonation of the lysine ϵ-amino group between methyl transfer reactions. For methylation to occur, the ϵ-amino group must be deprotonated to function as the nucleophile in the SN2 methyl transfer reaction with AdoMet (Figs. 4 and 5). Although the pKa value of the lysine ϵ-amine in solution is ∼10.5, molecular dynamics simulations by Zhang and Bruice (25, 26) indicate that this value diminishes to 8.2 upon formation of the SET7/9 Michaelis complex due to the proximity of the AdoMet sulfonium cation and the low dielectric constant of the active site. Furthermore, their simulations show that a chain of water molecules facilitates the deprotonation of the ϵ-amino group prior to methyl transfer, transferring the proton to bulk solvent. Although these water molecule chains are not evident in our crystal structures, the Y305F and Y245A complexes suggest another potential mechanism for deprotonation. In the dimethyl-lysine complexes of the Y305F and Y245A mutants, the dissociation of water 1 and 2, respectively, from the lysine binding channel requires that the solvent-mediated hydrogen bond to the ϵ-amino group is broken (Figs. 2D and 3C). It is conceivable that these waters dissociate from the active site as hydronium ions, promoting the realignment and deprotonation of the methyl ϵ-amino group for the next methyl transfer reaction.
A comparison of the SET7/9 Y305F and SET8 Y334F complexes yields insights into the mechanism by which the Phe/Tyr switch influences water binding within the solvent pocket. The phenylalanine substitution in the Phe/Tyr switch results in the loss of a single hydrogen bond to the water molecule (water 1) in the solvent pocket compared with the four hydrogen bonds that coordinate the solvent molecule in WT SET7/9 (Fig. 2, A and B) and SET8 (7, 8). Although this attenuation in hydrogen bonding may appear insignificant, this difference is nonetheless important for at least two reasons. First, theoretical calculations indicate that, on average, water molecules form ∼3.5 hydrogen bonds in solutions (40, 41). This value is greater than the number of hydrogen bonds coordinating water 1 in the solvent pocket in SET7/9 Y305F (Fig. 2, B and C) as well as in SET8 Y334F and other di- and trimethyltransferases that possess a hydrophobic residue in the Phe/Tyr switch site (8). From the perspective of the water molecule, the greater hydrogen bonding potential in solution would tend to thermodynamically favor its dissociation from the solvent pocket in SET domain KMTs that lack a tyrosine in the Phe/Tyr switch position. Second, the ordered binding of water molecules observed in the active sites of SET domain ternary complexes represents an unfavorable entropy compared with their diffusion in bulk solvent. In WT SET7/9 (Fig. 2A) and SET8 (7, 8), this entropic penalty can be partially offset through the favorable enthalpy of binding associated with the four hydrogen bonds that coordinate the water within the solvent pocket. It is conceivable that the loss of the tyrosine-mediated hydrogen bond in the Phe/Tyr switch shifts the equilibrium in favor of dissociation of the water molecule from the solvent pocket, thereby facilitating dimethylation in SET7/9 Y305F, SET8 Y334F, and other di- and trimethyltransferases.
The structures of the SET7/9 Y245A and Y305F complexes illustrate the interactions that align the lysine ϵ-amino group during the methyl transfer reactions in each enzyme. In the WT enzyme and the Y305F mutant, hydrogen bonding to the hydroxyl group of Tyr-245 appears to be critical in properly aligning the ϵ-amine for methyl transfer (Fig. 4). Tyr-245 is conserved in the sequences of many SET domain KMTs (8, 42), and substitutions of this residue generally impair or abolish activity, indicating its importance in catalysis (8, 43). However, SET7/9 appears to be an exception to this rule, as the Y245A mutant is not only active but is capable of catalyzing lysine trimethylation. In this mutant, Tyr-305 appears to assume the role of Tyr-245 by hydrogen bonding to the monomethylated ϵ-amino group to align it for methyl transfer with AdoMet, as illustrated in the modeled substrate complex for the dimethylation reaction (Fig. 5B). Conversely, in the model for trimethylation, the Tyr-305 hydroxyl group does not hydrogen bond to the ϵ-amine but instead participates in a CH–O hydrogen bond with one of the methyl groups to assist in aligning the dimethylated ϵ-amine for the methyl transfer reaction (Fig. 5C). Additional structural and functional studies of the SET domain trimethyltransferases will aid in further illuminating the roles of CH–O hydrogen bonds in facilitating lysine multiple methylation.
Supplementary Material
Acknowledgments
We acknowledge S. Schiebold for assistance in protein expression, purification, and crystallization and S. Anderson and R. Sanishvili for their assistance with x-ray data collection. We also thank S. Bulfer and S. Horowitz for reading the manuscript and providing useful comments. This work utilized the Protein Structure Facility of the Michigan Diabetes Research and Training Center, University of Michigan, supported by National Institutes of Health Grant DK020572, NIDDK. Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. GM/CA CAT has been funded in whole or in part by National Institutes of Health NCI Grant Y1-CO-1020 and NIGMS Grant Y1-GM-1104. Use of the LS-CAT Sector 21 was supported by Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor Grant 085P1000817 for the support of this research program.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM073839 (to R. C. T.) and National Institutes of Health Administrative Supplement GM073839-04S1 (to R. C. T.) funded through the American Recovery and Reinvestment Act. This work was also supported by Department of Energy Grant DE-FG02-92ER20075 (to R. L. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- AdoMet
- S-adenosylmethionine
- AdoHcy
- S-adenosylhomocysteine
- KMT
- lysine methyltransferase
- ITC
- isothermal titration calorimetry
- CH–O
- carbon-oxygen hydrogen bond
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Huang J., Berger S. L. (2008) Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 2.Morgunkova A., Barlev N. A. (2006) Cell Cycle 5, 1308–1312 [DOI] [PubMed] [Google Scholar]
- 3.Yang X. D., Lamb A., Chen L. F. (2009) Epigenetics 4, 429–433 [DOI] [PubMed] [Google Scholar]
- 4.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins R. E., Tachibana M., Tamaru H., Smith K. M., Jia D., Zhang X., Selker E. U., Shinkai Y., Cheng X. (2005) J. Biol. Chem. 280, 5563–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X., Yang Z., Khan S. I., Horton J. R., Tamaru H., Selker E. U., Cheng X. (2003) Mol. Cell 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couture J. F., Collazo E., Brunzelle J. S., Trievel R. C. (2005) Genes Dev. 19, 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couture J. F., Dirk L. M., Brunzelle J. S., Houtz R. L., Trievel R. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20659–20664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian C., Wang X., Manzur K., Sachchidanand, Farooq A., Zeng L., Wang R., Zhou M. M. (2006) J. Mol. Biol. 359, 86–96 [DOI] [PubMed] [Google Scholar]
- 10.Trievel R. C., Flynn E. M., Houtz R. L., Hurley J. H. (2003) Nat. Struct. Biol. 10, 545–552 [DOI] [PubMed] [Google Scholar]
- 11.Xiao B., Jing C., Wilson J. R., Walker P. A., Vasisht N., Kelly G., Howell S., Taylor I. A., Blackburn G. M., Gamblin S. J. (2003) Nature 421, 652–656 [DOI] [PubMed] [Google Scholar]
- 12.Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 13.Ea C. K., Baltimore D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18972–18977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estève P. O., Chin H. G., Benner J., Feehery G. R., Samaranayake M., Horwitz G. A., Jacobsen S. E., Pradhan S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. (2004) Mol. Cell 14, 175–182 [DOI] [PubMed] [Google Scholar]
- 16.Masatsugu T., Yamamoto K. (2009) Biochem. Biophys. Res. Commun. 381, 22–26 [DOI] [PubMed] [Google Scholar]
- 17.Munro S., Khaire N., Inche A., Carr S., La Thangue N. B. (2010) Oncogene 29, 2357–2367 [DOI] [PubMed] [Google Scholar]
- 18.Pagans S., Kauder S. E., Kaehlcke K., Sakane N., Schroeder S., Dormeyer W., Trievel R. C., Verdin E., Schnolzer M., Ott M. (2010) Cell Host Microbe 7, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian K., Jia D., Kapoor-Vazirani P., Powell D. R., Collins R. E., Sharma D., Peng J., Cheng X., Vertino P. M. (2008) Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., Gaudet F., Li E., Chen T. (2009) Nat. Genet. 41, 125–129 [DOI] [PubMed] [Google Scholar]
- 21.Yang X. D., Huang B., Li M., Lamb A., Kelleher N. L., Chen L. F. (2009) EMBO J. 28, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H. B., Guo H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8797–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu P., Wang S., Zhang Y. (2008) J. Am. Chem. Soc. 130, 3806–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu P., Zhang Y. (2006) J. Am. Chem. Soc. 128, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Bruice T. C. (2007) Biochemistry 46, 14838–14844 [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Bruice T. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5728–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couture J. F., Collazo E., Hauk G., Trievel R. C. (2006) Nat. Struct. Mol. Biol. 13, 140–146 [DOI] [PubMed] [Google Scholar]
- 28.Trievel R. C., Beach B. M., Dirk L. M., Houtz R. L., Hurley J. H. (2002) Cell 111, 91–103 [DOI] [PubMed] [Google Scholar]
- 29.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Vagin A., Teplyakov A. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 36.Collazo E., Couture J. F., Bulfer S., Trievel R. C. (2005) Anal. Biochem. 342, 86–92 [DOI] [PubMed] [Google Scholar]
- 37.Chirpich T. P., Zappia V., Costilow R. N., Barker H. A. (1970) J. Biol. Chem. 245, 1778–1789 [PubMed] [Google Scholar]
- 38.Couture J. F., Hauk G., Thompson M. J., Blackburn G. M., Trievel R. C. (2006) J. Biol. Chem. 281, 19280–19287 [DOI] [PubMed] [Google Scholar]
- 39.Kwon T., Chang J. H., Kwak E., Lee C. W., Joachimiak A., Kim Y. C., Lee J., Cho Y. (2003) EMBO J. 22, 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandra A., Chowdhuri S. (2002) J. Phys. Chem. B 106, 6779–6783 [Google Scholar]
- 41.Guardia E., Marti J., Garcia-Tarres L., Laria D. (2005) J. Mol. Liq. 117, 63–67 [Google Scholar]
- 42.Dillon S. C., Zhang X., Trievel R. C., Cheng X. (2005) Genome Biol. 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Tamaru H., Khan S. I., Horton J. R., Keefe L. J., Selker E. U., Cheng X. (2002) Cell 111, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.