Abstract
Down syndrome (DS) is associated with many neural defects, including reduced brain size and impaired neuronal proliferation, highly contributing to the mental retardation. Those typical characteristics of DS are closely associated with a specific gene group “Down syndrome critical region” (DSCR) on human chromosome 21. Here we investigated the molecular mechanisms underlying impaired neuronal proliferation in DS and, more specifically, a regulatory role for dual-specificity tyrosine-(Y) phosphorylation-regulated kinase 1A (Dyrk1A), a DSCR gene product, in embryonic neuronal cell proliferation. We found that Dyrk1A phosphorylates p53 at Ser-15 in vitro and in immortalized rat embryonic hippocampal progenitor H19-7 cells. In addition, Dyrk1A-induced p53 phosphorylation at Ser-15 led to a robust induction of p53 target genes (e.g. p21CIP1) and impaired G1/G0-S phase transition, resulting in attenuated proliferation of H19-7 cells and human embryonic stem cell-derived neural precursor cells. Moreover, the point mutation of p53-Ser-15 to alanine rescued the inhibitory effect of Dyrk1A on neuronal proliferation. Accordingly, brains from embryonic DYRK1A transgenic mice exhibited elevated levels of Dyrk1A, Ser-15 (mouse Ser-18)-phosphorylated p53, and p21CIP1 as well as impaired neuronal proliferation. These findings suggest that up-regulation of Dyrk1A contributes to altered neuronal proliferation in DS through specific phosphorylation of p53 at Ser-15 and subsequent p21CIP1 induction.
Keywords: Neurodevelopment, p53, Phosphorylation Enzymes, Protein Phosphorylation, Transgenic, Down Syndrome, Dyrk1A, Proliferation
Introduction
Down syndrome (DS)2 is the most common genetic disorder and is caused by the presence of all or part of an extra human chromosome 21 (1). The patients have many abnormalities such as mental retardation, deficits in learning and memory, and early onset Alzheimer disease (AD) (2, 3). The brains of DS patients exhibit an arrest of neurogenesis in many CNS regions, including the hippocampus at all ages, even the fetal stage (4–6). Cell proliferation has been shown to be impaired in human fetal DS brains and Ts65Dn mouse brains (7, 8). Ts65Dn mouse possesses an extra copy of a part of chromosome 16, which corresponds to human chromosome 21, and also shows learning and memory impairments and altered neuronal proliferation in the hippocampus (8–10). However, the molecular mechanisms underlying impaired neuronal proliferation in DS are unknown.
The typical characteristics of DS are thought to be closely associated with a gene group mapped to a specific region of human chromosome 21q22 “Down syndrome critical region” (DSCR) (3). Dual-specificity tyrosine-(Y) phosphorylation-regulated kinase 1A (Dyrk1A), one of the DSCR genes, encodes a proline-directed serine/threonine kinase, which phosphorylates several transcription factors, including NFAT, CREB, and FKHR (11, 12). DYRK1A transgenic (Tg) mice, which express human DYRK1A present on a bacterial artificial chromosome, exhibit significant impairment in hippocampal-dependent memory tasks and altered synaptic plasticity, features that are similar to those seen in DS patients (13). Several other studies also suggest that Dyrk1A appears to contribute to AD-like neuropathological features in DS by modulating the formation of intracellular Tau inclusions and the production of β-amyloid (14–16).
Dyrk1A likely contributes not only to AD-like neuropathology, but also to impaired neurogenesis. Mice bearing the 152F7 fragment of the yeast artificial chromosome containing DYRK1A gene show learning and memory deficits as well as reduced neuronal density in the cerebral cortex (17). During embryonic development, Dyrk1A mRNA is co-expressed with mRNA of an anti-proliferative gene Tis21 in chick (18). Moreover, Dyrk1A expression precedes the onset of neurogenesis and occurs in the presence of very few S phase cells in mouse (19). Although a few sparse clues exist, the correlation between Dyrk1A and neuronal proliferation and the underlying molecular mechanism remain elusive.
The present study was conducted to investigate the mechanism by which Dyrk1A impairs neuronal proliferation. Using immortalized rat embryonic hippocampal progenitor H19-7 cells, human embryonic stem cells-derived neural precursor cells, and DYRK1A Tg mice, we provide evidence that Dyrk1A attenuates neuronal proliferation by direct phosphorylation of p53, an effect that may underlie reduced brain size and neuronal number as well as impaired neuronal proliferation in DS.
EXPERIMENTAL PROCEDURES
Immunoprecipitation and Immunoblot Analysis
Cells were harvested by trypsinization, pelleted, lysed with 1% Nonidet P40 lysis buffer (50 mm Tris, pH 7.5, 137 mm NaCl, 1% Nonidet P40, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 0.2 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mm sodium orthovanadate, and 10 mm sodium fluoride), and briefly sonicated. Lysates were clarified by centrifugation at 13,000 × g for 15 min at 4 °C. For immunoprecipitation, 1 μg of suitable antibody was incubated with 1 mg of cell lysates overnight at 4 °C. The mixture was then incubated with 30 μl of a 1:1 Protein A-Sepharose bead suspension for 2 h. Beads were pelleted by centrifugation at 13,000 × g for 30 s and washed three times with 1% Nonidet P40 lysis buffer. In some cases, Exactacruz B and E (Santa Cruz Biotechnology) were used to eliminate IgG signals. Immunocomplexes were dissociated by boiling in SDS-PAGE sample buffer, separated on SDS-PAGE gels, and transferred to nitrocellulose membranes. Membranes were blocked in TBST buffer (25 mm Tris, pH 7.5, 137 mm NaCl, 2.7 mm KCl, and 0.1% Tween 20) plus 5% nonfat dry milk for 1 h at room temperature. They were then probed overnight at 4 °C with TBST buffer containing 3% nonfat dry milk and primary antibodies. Membranes were washed three times in TBST buffer and incubated for 2 h at room temperature with TBST buffer containing 3% nonfat dry milk and horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG antibodies. The membranes were washed three times with TBST buffer, and signals were visualized with an enhanced chemiluminescence reagent.
Cell Proliferation Analysis
H19-7/Dyrk1A and H19-7/pTK cells (2.0 × 104 cells) were seeded onto poly-l-lysine-coated dishes and cultured for the indicated times. The number of viable cells was then estimated using the Cell Counting kit-8 (Dojindo Molecular Technology), according to the manufacturer's protocol. Cellular proliferation was also analyzed using the in situ cell proliferation kit FLUOS (Roche Applied Science), according to the manufacturer's protocol. Samples were visualized using an LSM 510 META confocal microscope (Carl Zeiss) or an Olympus BX61 microscope with a DP70 camera (12.5 megapixels, Olympus).
Flow Cytometry
H19-7 cells were trypsinized and washed three times with ice-cold PBS. To count the number of cells at each phase, cells were fixed with 70% ethanol/PBS overnight at 4 °C and then analyzed as described previously (14). If necessary, cells were treated with 2 mm thymidine for 24 h to synchronize cells in G1/G0-S phase transition. After release by washing twice with PBS and adding fresh media, cells were cultured for the indicated times, and analyzed.
In Vitro Kinase Assay
H1299 cells were transfected with plasmid encoding Xpress-tagged wild-type or K188R Dyrk1A for 24 h. Cells were then lysed in 1% Nonidet P40 lysis buffer, and lysates were immunoprecipitated with an anti-Xpress antibody overnight at 4 °C. The lysate-antibody mixture was then incubated with 30 μl of a 1:1 Protein A-Sepharose bead suspension for 2 h at 4 °C with gentle inverting. Beads were centrifuged at 13,000 × g for 30 s at 4 °C, washed twice with 1% Nonidet P40 lysis buffer, and washed twice with 1× Dyrk1A buffer (20 mm HEPES, pH 7.4, 20 mm MgCl2, 5 mm MnCl2, and 1 mm dithiothreitol). Dyrk1A immunocomplexes (or bacterial recombinant Dyrk1A proteins for supplemental Fig. S1A) were mixed with 1 μg of recombinant p53 proteins (substrate) and then mixed with 1× Dyrk1A buffer containing 0.2 mm sodium orthovanadate and 10 μm ATP. The in vitro kinase reaction was initiated by the addition of 10 μCi of [γ-32p]ATP. The reaction was allowed to proceed for 15 min at 30 °C and was terminated by the addition of SDS-PAGE sample buffer. The protein samples were resolved by SDS-PAGE, and incorporated γ-32P radioisotope was detected by autoradiography.
Luciferase Reporter Assay
H19-7 cells (5.0 × 104 cells) were seeded onto poly-l-lysine-coated dishes. The following day, H19-7 cells were co-transfected with firefly luciferase reporter plasmids containing a synthetic p53 responsive element, the p21CIP1 promoter, or the Mdm2 promoter. Co-transfection of pRL plasmids, which constitutively express Renilla luciferase, was used to normalize transfection efficiency. A subset of cells was co-transfected with plasmid encoding Xpress-tagged wild-type or K188R Dyrk1A. Twenty-four hours after transfection, cells were lysed and analyzed using the Dual-luciferase reporter assay system (Promega, Madison, WI).
Preparation of DYRK1A Tg Mouse Brain and Human DS Frontal Cortex Samples
All mice experiments were performed in accordance with the guidelines set forth by Inje University Council Directive for the proper care and use of laboratory animals. DYRK1A Tg mice expressing the human DYRK1A gene on a bacterial artificial chromosome were generated and maintained as previously described (13). The expression of DYRK1A in each Tg mouse was confirmed with PCR. Mice were sacrificed by cervical dislocation. Whole or specific regions of mouse brains were isolated with a scalpel blade. For immunoblot analyses, brain samples were disrupted in 1% Nonidet P-40 lysis buffer using a Dounce homogenizer with a tight fitting pestle (25 strokes). Lipid layers were then removed with a pipette, and homogenates were clarified by centrifugation at 13,000 × g for 15 min at 4 °C.
To analyze neuronal proliferation of embryonic DYRK1A Tg mice, BrdU administration and immunohistochemical analysis of BrdU-labeled tissues were performed as described elsewhere (7) with minor modifications. In brief, pregnant mice at E13.5 were intraperitoneally injected once with BrdU (100 μg per gram of body weight) and sacrificed 24 h after injection. The E14.5 embryos were fixed on 4% paraformaldehyde/PBS and cryoprotected in 30% sucrose for 2 days at 4 °C. After freezing in OCT compound (Sakura Finetek), coronal sections (10 μm) of frozen embryos were mounted on ProbeOn Plus slides (Fisher Scientific), and immunostained using primary anti-BrdU (1:100 dilution, BD Biosciences) and secondary Alexa Fluor 488-conjugated anti-mouse IgG antibodies (1:100 dilution, Invitrogen).
Primary cortical neural stem cell precursors from DYRK1A Tg and littermate control mice were prepared, as described elsewhere (20), with some modifications. In brief, mouse neural precursors were isolated from E12.5 embryo cortices carefully and incubated in 0.05% trypsin with 10 μg/ml DNase I in Hanks' balanced salt solution for 5 min at 37 °C. After stopping trypsinization by adding five times volume of DMEM/F-12 (1:1) media, the tissue digests were dissociated using several rounds of trituration. The cells were washed once with Hanks' balanced salt solution and plated on poly-l-lysine and laminin-coated coverslips in DMEM/F-12 media with 20 ng/ml EGF and 20 ng/ml basic FGF.
Postmortem brain lysates from human DS patients (n = 3; 23 weeks gestational age, 339 and 427 days old; postmortem interval: 18.67 ± 12.10 h) and age-matched controls (n = 3; 22 weeks gestational age, 332 and 587 days old; postmortem interval: 11.33 ± 2.31 h) were prepared as described elsewhere (21). 25 μg of lysates were analyzed by immunoblot. Densitometric quantification was performed using Multi Gauge v3.1 software (Fujifilm).
Data Deposition
The sequences reported herein have been deposited in the GenBankTM data base (accession numbers NP_036923 (rat Dyrk1A), NP_112251 (rat p53), and NP_000537 (human p53)).
Statistical Analysis
Group means were compared using a Student's t test. Values of p less than 0.05 were accepted as significant. All data are expressed as mean ± S.D. unless otherwise indicated.
Note
The detailed information and protocols for DNA constructs, transfection and lentiviral transduction, purification of recombinant proteins, and others are provided as supplemental data.
RESULTS
Dyrk1A Interacts and Co-localizes with p53
We previously observed that ∼2-fold overexpression of Dyrk1A leads to AD-like neuropathological features, increased susceptibility to serum deprivation-induced apoptosis (14), and growth suppression in H19-7 cells.3 To gain insight into the mechanisms by which Dyrk1A affects cell proliferation of H19-7 cells, we performed microarray analysis with DNA chips containing oligonucleotide probes from 20,000 rat genes (Agilent Technologies). Genes were screened with respect to expression profiles, gene ontology classes, and transcription factor target genes. A group of pro-apoptotic genes and p53 target genes were significantly up-regulated in Dyrk1A-overexpressing H19-7 cells (H19-7/Dyrk1A) compared with parental control cells.3 These included genes encoding cyclin-dependent kinase (CDK) inhibitor 1A (p21CIP1/Cdkn1A), proliferating cell nuclear antigen (PCNA), growth arrest and DNA damage-inducible gene 45 (Gadd45), Fas, and Mdm2. Based on the numerous reports that p53 plays a pivotal role in controlling cell proliferation by transactivating a variety of downstream target genes, we further investigated whether and how Dyrk1A and p53 may affect each other.
Because the modulation of p53 activity primarily occurs through phosphorylation of serine and threonine residues within the N-terminal transactivation domain (22) and Dyrk1A is a proline-directed serine/threonine kinase (23), we firstly determined whether Dyrk1A interacts with p53. HEK293 cells were co-transfected with plasmids encoding Xpress-tagged rat Dyrk1A and V5-tagged rat p53, and total cell lysates were immunoprecipitated with an anti-V5 antibody. Immunoblot analysis of V5 immunoprecipitates with anti-Xpress antibody revealed an interaction between ectopically expressed Dyrk1A and p53 (Fig. 1A). Endogenous Dyrk1A also bound to endogenous p53 in H19-7 cells, as seen by immunoblot analysis of anti-p53 immunoprecipitates with anti-Dyrk1A antibody (Fig. 1B).
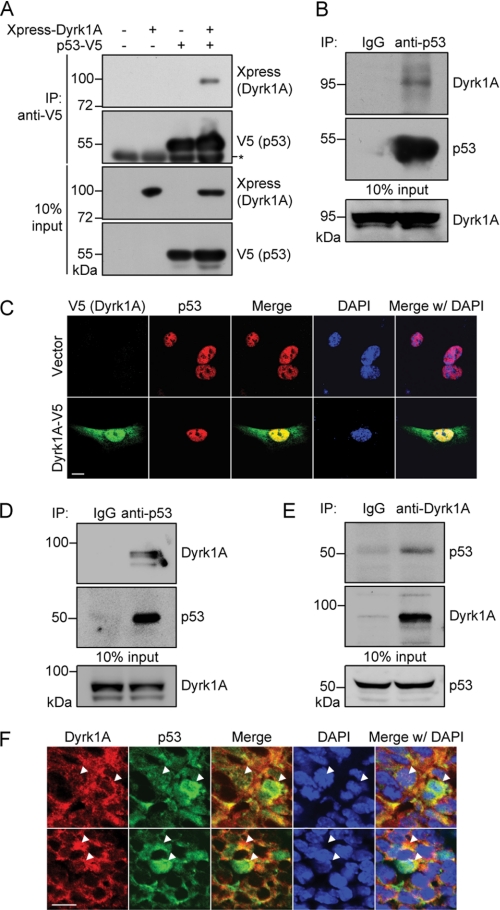
FIGURE 1.
Dyrk1A interacts and co-localizes with p53. A, co-immunoprecipitation/immunoblot analysis of interaction between Xpress-tagged Dyrk1A and V5-tagged p53 in HEK293 cells. Proper expression of the transiently expressed proteins was verified by immunoblot analysis with anti-Xpress or anti-V5 antibodies. The asterisk indicates immunoglobulin heavy chains. B, co-immunoprecipitation/immunoblot analysis of interaction between endogenous Dyrk1A and p53 in H19-7 cells. Immunoprecipitation was performed with rabbit polyclonal anti-p53 antibody or normal rabbit immunoglobulin G (as a negative control). C, representative confocal images of immunostaining for V5-tagged Dyrk1A (green) and p53 (red) in H19-7 cells. Nuclei were counterstained with DAPI (blue). Scale bar = 10 μm. D and E, reciprocal co-immunoprecipitation/immunoblot analyses of Dyrk1A-p53 interactions in E18.5 rat whole brain lysates. F, representative confocal images of immunohistochemistry for Dyrk1A (red) and p53 (green) in the hippocampus from E14.5 mice. Nuclei were counterstained with DAPI (blue). Scale bar = 10 μm. Arrowheads indicate the co-localization of Dyrk1A and p53 in the nuclear, perinuclear, and cytoplasmic regions.
To determine whether Dyrk1A co-localizes with p53, we transfected H19-7 cells with Dyrk1A-V5 or mock transfected these cells (control). Immunostaining with anti-V5 and anti-p53 antibodies revealed that labels for endogenous p53 and ectopically expressed Dyrk1A overlapped within the cell nucleus (Fig. 1C). To determine whether the Dyrk1A-p53 interactions observed in cultured cells could be extended to whole tissues, we performed co-immunoprecipitation analyses of whole brain lysates from embryonic day (E) 18.5 Sprague-Dawley rats. As shown in Fig. 1D, immunoblot analysis of anti-p53 immunoprecipitates using anti-Dyrk1A antibody clearly showed that Dyrk1A and p53 interact in the embryonic rat brain. This interaction was also observed in reciprocal co-immunoprecipitation experiments (Fig. 1E), confirming that Dyrk1A-p53 interactions are not an artifact arising from DNA transfection in cultured cell lines. In addition, immunohistochemical analysis of the hippocampus from E14.5 mice revealed that Dyrk1A co-localizes with p53 mainly in the perinuclear regions as well as less frequently in the cytoplasmic and nuclear regions in embryonic hippocampal cells (Fig. 1F).
Dyrk1A Specifically Phosphorylates p53 at Ser-15
To address whether Dyrk1A directly phosphorylates p53, we performed in vitro kinase assays using p53-null human lung carcinoma H1299 cells expressing Xpress-tagged wild-type or a kinase-inactive mutant (K188R) Dyrk1A. Anti-Xpress immunoprecipitates were incubated with recombinant His6-tagged human wild-type p53 and [γ-32p]ATP. Autoradiographic analysis of the reaction products clearly showed that Dyrk1A directly phosphorylates p53 in vitro (Fig. 2A). This finding was further supported by in vitro kinase assay using bacterial recombinant Dyrk1A proteins (supplemental Fig. S1A). The ability of p53 to serve as a substrate of Dyrk1A in human breast carcinoma MCF-7 cells and which type of amino acids are phosphorylated by Dyrk1A were also investigated. Cells were co-transfected with Xpress-Dyrk1A and FLAG-p53, and anti-FLAG immunoprecipitates were probed with anti-phospho-serine and anti-phospho-threonine antibodies. As shown in Fig. 2B, transient Dyrk1A overexpression increased p53 phosphorylation at serine residues. However, no significant phosphorylation was observed at threonine residues (Fig. 2B) or at tyrosine residues (data not shown).
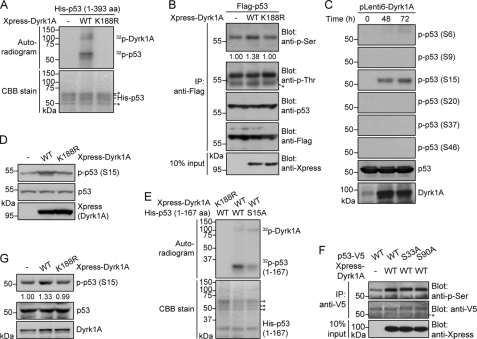
FIGURE 2.
Dyrk1A directly phosphorylates human p53 at Ser-15. A, in vitro kinase assay of Dyrk1A-mediated phosphorylation of human p53. H1299 cells were transfected with plasmids encoding Xpress-tagged WT or kinase-inactive (K188R) Dyrk1A. Anti-Xpress immunoprecipitates were incubated with recombinant His6-tagged human p53 and [γ-32p]ATP. The reaction products were separated by SDS-PAGE, and autoradiograms were obtained. Asterisks indicate immunoglobulin heavy chains and nonspecific protein bands. B, Dyrk1A phosphorylates human p53 at serine residues. MCF-7 cells were transfected with plasmids encoding FLAG-tagged human p53 and Xpress-tagged WT or K188R Dyrk1A. Anti-FLAG immunoprecipitates were then probed with anti-phospho-serine, anti-phospho-threonine, anti-p53, and anti-FLAG antibodies. The asterisk indicates immunoglobulin heavy chains. C, immunoblot analysis of serine-phosphorylated p53 in U2OS cells transiently expressing Dyrk1A for varying times (0–72 h). Immunoblots were performed with six separate anti-phospho-p53 antibodies against serines 6, 9, 15, 20, 37, and 46. Levels of p53 and Dyrk1A protein were analyzed by re-probing membranes with anti-p53 and anti-Dyrk1A antibodies. D, immunoblot analysis of Ser-15-phosphorylated p53 in MCF-7 cells expressing Xpress-tagged WT or K188R Dyrk1A. E, in vitro kinase assays showing phosphorylation of human p53 at Ser-15 by Dyrk1A. Assays were performed with anti-Xpress immunoprecipitates (Xpress-tagged WT or K188R Dyrk1A) and recombinant His6-tagged WT or S15A mutant p53. Asterisks indicate immunoglobulin heavy chains and nonspecific protein bands. F, substitution of p53-S33A and p53-S90A does not affect Dyrk1A-mediated phosphorylation of p53. U2OS cells were transfected with V5-tagged human p53-WT alone or together with Xpress-tagged WT Dyrk1A. Where specified, Xpress-tagged WT Dyrk1A were co-transfected with p53-S33A or p53-S90A. Anti-V5 immunoprecipitates were then probed with anti-phospho-serine and anti-V5 antibodies. Proper expression of Dyrk1A was verified using immunoblot analysis with anti-Xpress antibody. The asterisk indicates immunoglobulin heavy chains. G, immunoblot analysis of Ser-15-phosphorylated p53 in primarily cultured rat cortical neurons expressing Xpress-tagged WT or K188R Dyrk1A.
To identify the one or more p53 domains that serve as binding and phosphorylation sites of Dyrk1A, we analyzed in vitro interactions between Dyrk1A and His6-tagged p53 deletion mutants. Lysates from Dyrk1A-V5-expressing H1299 cells were incubated with bacterially expressed p53 deletion mutants (supplementary Fig. S1B). Immunoblot analysis of anti-V5 immunoprecipitates using anti-p53 antibody showed that wild-type Dyrk1A interacts with a region of p53 encompassing amino acids 94–167 (supplemental Fig. S1C), which contains part of the DNA-binding domain. To identify the one or more regions of p53 that are phosphorylated by Dyrk1A, we performed an in vitro kinase assay by mixing anti-Dyrk1A immunocomplexes with various recombinant p53 truncation mutants containing the Dyrk1A-binding domain. As shown in supplemental Fig. S1D, Dyrk1A phosphorylated the N-terminal region of human p53 spanning amino acids 1–93.
The fragment of human p53 spanning amino acids 1–93 contains eight serine residues: serines 6, 9, 15, 20, 33, 37, 46, and 90. To narrow down which amino acid(s) are targeted by Dyrk1A, human osteosarcoma U2OS cells were transduced with lentiviral particles containing wild-type Dyrk1A. Immunoblot analyses of total cell lysates were then performed with six separate phospho-specific antibodies against serines 6, 9, 15, 20, 37, and 46. Among these residues, Ser-15 was markedly phosphorylated by Dyrk1A (Fig. 2C).
The selective and remarkable increase in p53 phosphorylation at Ser-15 was also observed in MCF-7 cells (Fig. 2D). Consistent with this finding, in vitro kinase assays revealed that substitution of Ser-15 with alanine (S15A) blocked p53 phosphorylation by Dyrk1A (i.e. anti-Dyrk1A immunoprecipitates) by >50%, indicating that Dyrk1A mainly phosphorylates Ser-15 of p53 (Fig. 2E). In contrast to Ser-15, Ser-33 and Ser-90 did not appear to be major targets of Dyrk1A, because substitution of these residues with alanine did not significantly affect p53 phosphorylation by Dyrk1A (Fig. 2F). Finally, the level of p53-Ser-15 was also increased by >30% in rat cortical neurons expressing wild-type Dyrk1A, but not its kinase-defective mutant (Fig. 2G), confirming the actual occurrence of Dyrk1A-mediated p53 phosphorylation in neurons. These results suggest that Dyrk1A specifically phosphorylates p53 at Ser-15.
Dyrk1A-induced Phosphorylation of p53 Increases Its Transcriptional Activity and the Expression of Downstream Target Genes in H19-7 Cells
The phosphorylation of p53 enhances its transcriptional activity, leading to the induction of target genes such as p21CIP1, PCNA, Gadd45, Fas, and Mdm2 (22). Specifically, phosphorylation of Ser-15 induces the transcriptional activity of p53, whereas phosphorylation within the C-terminal domain regulates the oligomerization and nuclear translocation of p53 (22). Thus, we investigated the effect of Dyrk1A overexpression on the transcriptional activity of p53 and the expression of p53 target genes in H19-7 cells. Firstly, both immunoblot analysis and in vitro kinase assay confirmed that Dyrk1A-induced p53 phosphorylation occurs at Ser-15 in H19-7 cells (Fig. 3A and supplemental Fig. S2A). Analysis of a luciferase reporter driven by a synthetic p53-responsive element showed that the overexpression of Xpress-tagged wild-type Dyrk1A, but not its kinase-dead mutant (K188R), increased p53 activity by ∼2.1-fold (p < 0.01, Fig. 3B). The induction of p53 target genes was also analyzed using luciferase reporters. Dyrk1A overexpression augmented the activity of a luciferase reporter driven by the p21CIP1 promoter by ∼1.7-fold (p < 0.001, Fig. 3C). The validity of the p21CIP1 promoter reporter assay was confirmed by p53-dependent induction in H19-7 cells (p < 0.001, supplemental Fig. S2B). Transient Dyrk1A overexpression also induced luciferase reporter activity driven by the Mdm2 promoter by ∼1.6-fold (p < 0.01, supplemental Fig. S2C). In addition, immunoblot analysis of cell lysates revealed a remarkable increase of p21CIP1 protein levels under this condition (Fig. 3D).
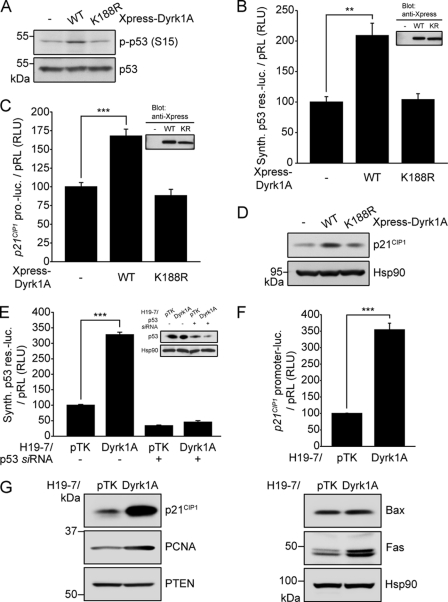
FIGURE 3.
Dyrk1A induces the expression of p53 target genes in H19-7 cells. A, immunoblot analysis of p53 and p53-phosphoSer-15 in H19-7 cells expressing Xpress-tagged WT or K188R Dyrk1A. B and C, luciferase activity of reporter plasmids containing a synthetic p53 responsive element (B) or the p21CIP1 promoter (C) in H19-7 cells expressing Xpress-tagged WT or K188R Dyrk1A (n = 3; **, p < 0.01 and ***, p < 0.001). Immunoblotting was used to verify proper expression of transiently expressed Dyrk1A proteins (anti-Xpress antibody). D, immunoblot analysis of p21CIP1 in H19-7 cells expressing Xpress-tagged WT or K188R Dyrk1A. Hsp90 served as a loading control. E and F, luciferase activity of reporter plasmids containing a synthetic p53 responsive element (E) or the p21CIP1 promoter (F) in H19-7/Dyrk1A cells (n = 3; ***, p < 0.001). Knockdown of endogenous p53 was verified with anti-p53 antibody. G, immunoblot analysis of p53 target gene products in H19-7/Dyrk1A cells. Hsp90 served as a loading control.
We also investigated the expression of p53 target genes in H19-7/Dyrk1A cells. Consistent with previous findings, the luciferase reporter activity driven by the synthetic p53-responsive element (3.3-fold, p < 0.001, Fig. 3E), p21CIP1 promoter (3.5-fold, p < 0.001, Fig. 3F), or Mdm2 promoter (3.0-fold, p < 0.001, supplemental Fig. S2D) was higher in H19-7/Dyrk1A cells than in parental control cells (H19-7/pTK). Moreover, knockdown of endogenous p53 with p53-specific small interfering RNA (siRNA) markedly reduced luciferase activity driven by the synthetic p53-responsive element, indicating that the augmented luciferase activity in H19-7/Dyrk1A cells results from Dyrk1A-induced p53 activation (Fig. 3E). The protein levels of p53 target gene products were also higher in H19-7/Dyrk1A cells than H19-7/pTK cells. Especially, p21CIP1 level was more remarkably increased than the other p53 target gene products such as PCNA and Fas, whereas Bax and phosphatase and tensin homolog (PTEN) did not show any increase (Fig. 3G). These results suggest that up-regulation of Dyrk1A in H19-7 cells leads to the induction of p53 target genes, especially p21CIP1, which is closely associated with cell cycle regulation.
Up-regulation of Dyrk1A Attenuates Proliferation of H19-7 Cells via Impairing G1/G0-S Phase Transition
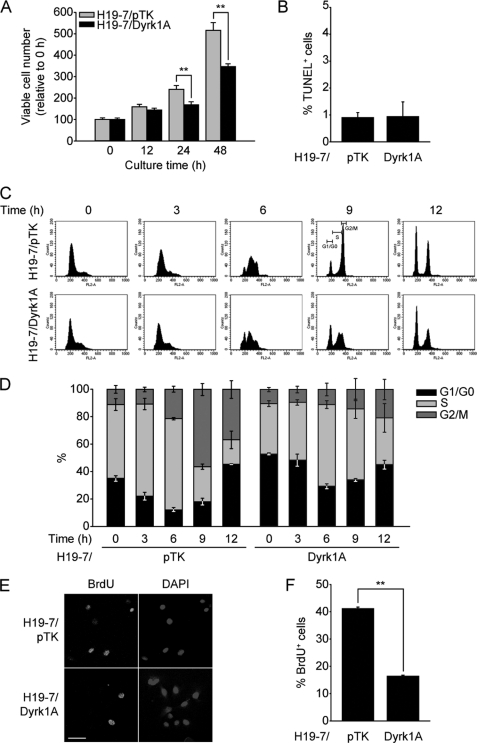
Based on the finding that p21CIP1 inhibits CDKs and regulates G1/G0-S phase transition (24), the ability of Dyrk1A to induce p21CIP1 implicates that it may also influence G1/G0-S phase transition in H19-7 cells. To test this possibility, we firstly analyzed cell growth of H19-7/Dyrk1A cells comparing to H19-7/pTK cells. As shown in Fig. 4A, the number of viable H19-7/Dyrk1A cells was ∼70% (24 h) and 67% (48 h) of that in H19-7/pTK cells (p < 0.001). To examine whether it may result from an increased cell death, the percentage of apoptotic cells was compared in H19-7/Dyrk1A and H19-7/pTK cells. The terminal dUTP nick end labeling (TUNEL) assay revealed that both cells exhibit no significant difference (p > 0.9) in apoptosis (Fig. 4B), which was consistent with the previous report (14). To further analyze the pattern of cell cycle, we performed thymidine synchronization and release, and subsequently FACS analysis using propidium iodide. As shown in Fig. 4, C and D, H19-7/Dyrk1A cells showed slower G1/G0-S phase transition (H19-7/Dyrk1A G1/G0 phase: 48.38% versus H19-7/pTK: 22.01% at 3 h; p < 0.001) than parental control cells as well as more G1/G0 accumulation at 0 h (synchronized for 24 h) (H19-7/Dyrk1A G1/G0 phase: 52.68% versus H19-7/pTK: 35.04%; p < 0.001). This result was further supported by the finding that, after 6 h of culture, 5-bromo-2-deoxyuridine (BrdU) labeling was lower in H19-7/Dyrk1A cells (16.33 ± 0.47%) than in H19-7/pTK cells (41.16 ± 0.53%; p < 0.001) (Fig. 4, E and F). Moreover, this negative effect of Dyrk1A on H19-7 cell proliferation was dependent on its kinase activity (supplemental Fig. S3, A–C). These data suggest that an approximate 2-fold up-regulation of Dyrk1A attenuates H19-7 cell proliferation by impairing G1/G0-S phase transition.
FIGURE 4.
Dyrk1A overexpression delays cell proliferation and induces G1/G0 phase accumulation in H19-7 cells. A, proliferation of H19-7/Dyrk1A cells and H19-7/pTK cells, as measured using a tetrazolium-based assay. Data are expressed in arbitrary units (n = 12; **, p < 0.001). B, cell death of H19-7/Dyrk1A and H19-7/pTK cells under the same condition, as measured using TUNEL assay. Data are expressed as the percentage of TUNEL-positive cells (846 cells were counted for H19-7/pTK, 1130 cells for H19-7/Dyrk1A) (p > 0.9). C and D, cell cycle phase analysis of H19-7/Dyrk1A and H19-7/pTK cells after release from thymidine synchronization. Representative histogram images at each time point after release from thymidine synchronization (2 mm for 24 h) are shown (C). Data are expressed as the percentage of cell population in each phase (D) (n = 3; p < 0.001 for each comparison of G1/G0 phase population at 0–9 h). E and F, BrdU labeling in H19-7/Dyrk1A and H19-7/pTK cells. Cells were labeled with BrdU for 6 h. Scale bar = 50 μm. Data are expressed as the percentage of BrdU-positive cells (141 cells were counted for H19-7/pTK, 159 cells for H19-7/Dyrk1A) (**, p < 0.001).
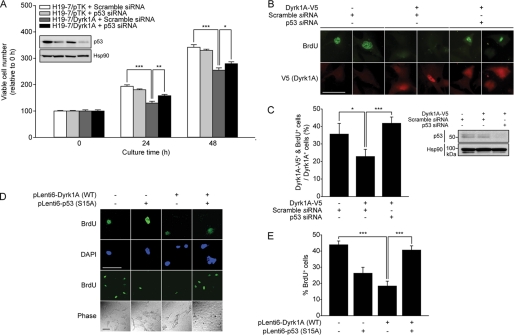
Dyrk1A Overexpression Impairs G1/G0-S Phase Transition in a p53-Ser-15 Phosphorylation-dependent Manner
We next examined whether the increase of p53 activity contributes to the attenuation of cell proliferation in H19-7/Dyrk1A cells. As shown in Fig. 5A, the addition of siRNA targeting to endogenous p53, but not of scrambled siRNA, resulted in the increase of cell growth in H19-7/Dyrk1A cells (p < 0.01 at 24 h and p < 0.05 at 48 h). This result indicates that the attenuation of cell proliferation in H19-7/Dyrk1A cells may occur through the p53-dependent pathway. Next we assessed the effect of Dyrk1A overexpression on p53-dependent reduction of cell proliferation. We transfected H19-7 cells with Dyrk1A-V5 and then performed BrdU labeling assay for 6 h. Transient Dyrk1A overexpression reduced the number of BrdU-positive cells to ∼64% of that seen in mock transfected control cultures (p < 0.01) (Fig. 5, B and C). Moreover, specific siRNA targeting p53, but not scrambled siRNA, restored the Dyrk1A-induced reduction in BrdU labeling to control levels (p < 0.0001). These data demonstrate that Dyrk1A-mediated attenuation of cell proliferation occurs through p53 activation. In contrast to much smaller recovery of cell growth in Fig. 5A, which may result from low transfection efficiency of RNA duplexes into H19-7 cells as well as the incomplete depletion of endogenous p53, p53 dependence on Dyrk1A-induced reduction of cell proliferation was obviously shown in Fig. 5, B and C, which exclusively counted V5 staining-positive cells transfected with Dyrk1A.
FIGURE 5.
Overexpression of Dyrk1A attenuates proliferation of H19-7 cells through p53 phosphorylation at Ser-15. A, effect of p53-specific or scramble siRNAs (30 nm) on cell proliferation in H19-7/Dyrk1A cells and H19-7/pTK cells, as measured using a tetrazolium-based assay. Data are expressed in arbitrary units (n = 4; *, p < 0.05, **, p < 0.01, and ***, p < 0.001). Knockdown of endogenous p53 was verified using anti-p53 antibody. Hsp90 served as a loading control. B and C, effect of p53 siRNA (30 nm) or scramble siRNA (30 nm) on BrdU labeling in H19-7 cells expressing V5-tagged Dyrk1A. Immunostaining for V5 was performed to identify Dyrk1A-V5-positive cells. Scale bar = 50 μm. Data are expressed as the percentage of BrdU-positive cells. A total of 435 cells was counted for mock transfection and scrambled siRNA, 107 cells for Dyrk1A-V5 and scrambled siRNA, and 130 cells for Dyrk1A-V5 and p53 siRNA (*, p < 0.01 and ***, p < 0.0001). Knockdown of endogenous p53 was verified using anti-p53 antibody. Hsp90 served as a loading control. D and E, BrdU labeling in H19-7 cells expressing WT Dyrk1A and/or p53-S15A mutant. Scale bar = 50 μm. Data are expressed as the percentage of BrdU-positive cells and are reported as the mean ± S.E. A total of 593 cells was counted for mock transfection, 428 cells for p53-S15A, 625 cells for Dyrk1A WT, and 551 cells for Dyrk1A WT and p53-S15A (***, p < 0.0001).
Similar to transient Dyrk1A-V5 overexpression, lentiviral Dyrk1A expression reduced BrdU-positive H19-7 cells by ∼41% (p < 0.0001), and co-expression of the p53-S15A mutant rescued the reduced BrdU labeling to control levels (p < 0.0001) (Fig. 5, D and E). In addition, transduction of the p53-S15A mutant alone considerably reduced BrdU labeling, suggesting that the S15A mutation is not sufficient to inactivate the anti-growth effect of human p53 on H19-7 cells. Taken together, these results suggest that up-regulation of Dyrk1A impairs G1/G0-S phase transition in H19-7 cells through p53 phosphorylation at Ser-15.
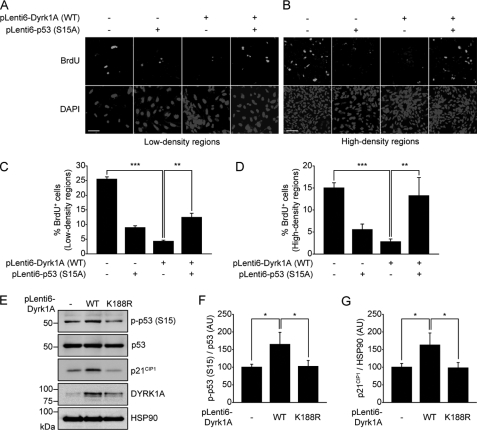
Dyrk1A Overexpression Reduces Proliferation of Human Embryonic Stem Cell-derived Neural Precursor Cells through p53 Phosphorylation at Ser-15
We have shown that Dyrk1A phosphorylates p53 at Ser-15, leading to the induction of p53 target genes and delaying proliferation of H19-7 cells. To validate the physiological relevance of our findings, we investigated whether Dyrk1A also has an anti-growth role in human neural precursor cells. Human neural precursor cells derived from embryonic stem cells (hES-NP) were prepared (25), transduced with lentiviral particles containing wild-type Dyrk1A and/or p53-S15A mutant, and then labeled with BrdU for 6 h. BrdU incorporation was analyzed separately for low density (Fig. 6, A and C) and high density cell population regions (Fig. 6, B and D). Dyrk1A overexpression diminished the number of BrdU-positive hES-NP cells to ∼17 and 19% of the number of BrdU-positive control cells in low and high density regions, respectively (p < 0.0001 for both) (Fig. 6, C and D). Expression of the p53-S15A mutant partially blocked the Dyrk1A-induced decrease in BrdU labeling (p < 0.001), resulting in a number of BrdU-positive cells that was ∼49 and 88% of the number of BrdU control cells in low and high density regions, respectively (Fig. 6, C and D). As in H19-7 cells, expression of the p53-S15A mutant alone significantly suppressed BrdU labeling. Finally, overexpression of wild-type Dyrk1A increased levels of Ser-15-phosphorylated p53 and p21CIP1 (p < 0.05), whereas the kinase-inactive Dyrk1A-K188R mutant had no effect (Fig. 6, E–G). These results indicate that overexpression of Dyrk1A in hES-NP cells leads to p53 phosphorylation at Ser-15, subsequent transcriptional activation, and reduced cell growth.
FIGURE 6.
Overexpression of Dyrk1A attenuates proliferation of hES-NP cells through phosphorylation of p53 at Ser15. A–D, BrdU labeling in hES-NP cells overexpressing WT Dyrk1A and/or p53-S15A. Representative images of labeling in low density regions (A) and high density regions (B) are shown. Scale bar = 50 μm. In low density regions, a total of 656 cells was counted for mock transfection, 677 cells for p53-S15A, 906 cells for Dyrk1A WT, and 1,278 cells for Dyrk1A WT and p53-S15A (C). In high density regions, a total of 3,419 cells was counted for mock transfection, 1,657 cells for p53-S15A, 4,243 cells for Dyrk1A WT, and 2,743 cells for Dyrk1A WT and p53-S15A (D) (**, p < 0.001 and ***, p < 0.0001). E–G, immunoblot analysis of Ser-15-phosphorylated p53, p53, and p21CIP1 in hES-NP cells transiently expressing WT or K188R mutant Dyrk1A. Proper expression of Dyrk1A was verified with the anti-Dyrk1A antibody. Band intensity of p53-phosphoSer-15 and p21CIP1 from three independent immunoblots was quantified using Multi Gauge v3.1 software and normalized to that of p53 and HSP90, respectively. Data are expressed in arbitrary units (*, p < 0.05).
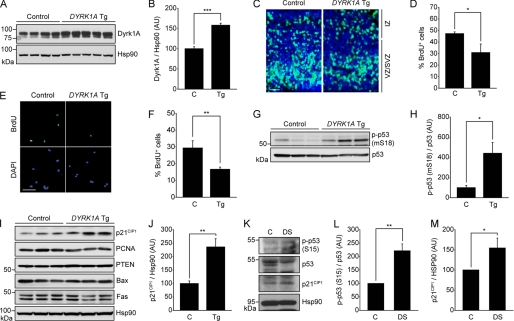
DYRK1A Tg Mouse Brains Exhibit Impaired Neuronal Proliferation as Well as Elevated Levels of Phosphorylated p53 and p21CIP1
Next, we investigated whether Dyrk1A also has an anti-growth role in brains of embryonic DYRK1A Tg mice, which exhibit an approximate 1.5-fold increase in total Dyrk1A (Fig. 7, A and B). This level of Dyrk1A is similar to the levels observed in DS patients (26). These mice have significant impairments in hippocampal-dependent memory tasks, altered synaptic plasticity, and AD-like neuropathological features, including Tau hyperphosphorylation and β-amyloid production (13, 15, 16). To examine whether and how Dyrk1A overexpression affects cell proliferation, we conducted BrdU pulse labeling assay in the neocortex and primary cultured neural precursor cells from Dyrk1A Tg mice. Immunohistochemical analysis of E14.5 Dyrk1A Tg mice embryos revealed that less neocortical cells were BrdU-labeled in DYRK1A Tg mice (30.84 ± 7.45%, p < 0.05) than in littermate controls (47.27 ± 1.56%), indicating impaired proliferation in embryonic neocortical neuronal cells of DYRK1A Tg brains (Fig. 7, C and D). Primary cortical neural stem cell precursors from E12.5 DYRK1A Tg embryos were also less BrdU-labeled than those from littermate controls (p < 0.01) (Fig. 7, E and F). Alteration in cell proliferation was accompanied by an increase in mouse p53 phosphorylation at Ser-18, which corresponds to Ser-15 in human p53 (p < 0.05) (Fig. 7, G and H). Furthermore, p21CIP1 level was specifically and remarkably increased while the other p53 target gene products were not (p < 0.01) (Fig. 7, I and J). Taken together, these data suggest that the 1.5-fold up-regulation of Dyrk1A enhances p53 phosphorylation and p21CIP1 levels, in turn impairing neuronal proliferation. Furthermore, these findings implicate that Dyrk1A plays a role in the embryonic stages of mouse brain development.
FIGURE 7.
Impaired neuronal proliferation, elevated p53 phosphorylation, and p21CIP1 levels in embryonic DYRK1A Tg mice. A and B, immunoblot analysis of Dyrk1A expression in whole brain lysates from E14.5 DYRK1A Tg mice and littermate controls. Band intensity of Dyrk1A was quantified and normalized to the intensity of Hsp90. Data are expressed in arbitrary units (n = 5 for Tg, n = 4 for C; ***, p < 0.001). C and D, BrdU labeling assay in the neocortex of E14.5 DYRK1A Tg mice (n = 4) and littermate controls (n = 2). E13.5-timed pregnant mice were intraperitoneally injected with BrdU for 24 h, and the embryos were surgically removed. Scale bar = 20 μm. Data are expressed as the percentage of BrdU-positive cells (green) against DAPI-stained total cells (blue) (*, p < 0.05). VZ/SVZ, ventricular/subventricular zone; IZ, intermediate zone. E and F, analysis of BrdU labeling in primarily cultured cortical neural stem cell precursors from E12.5 DYRK1A Tg embryos (n = 5) and in littermate controls (n = 4). Cells were labeled with BrdU for 12 h. Scale bar = 50 μm. Data are expressed as the percentage of BrdU-positive cells and are reported as the mean ± S.E. (**, p < 0.01). G and H, immunoblot analysis of Ser-18-phosphorylated p53 and p53 in whole brain lysates from E14.5 DYRK1A Tg mice and littermate controls. Band intensity of Ser-18-phosphorylated p53 was quantified and normalized to the intensity of p53 (n = 3 for each). Data are shown as the mean ± S.E. (*, p < 0.05). I and J, immunoblot analysis of p53 target gene products in whole brain lysates from E14.5 DYRK1A Tg mice and littermate controls. Band intensity of p21CIP1 was quantified and normalized to the intensity of Hsp90 (n = 3 for each). Data are shown as the mean ± S.E. (**, p < 0.01). K–M, immunoblot analysis of p53-phosphoSer-15 (L), and p21CIP1 (M) in the frontal cortices prepared from DS patient brains and age-matched controls. Band intensity was quantified and normalized to the intensity of p53 (L) or HSP90 (M) (*, p < 0.05 and **, p < 0.01).
Human Embryonic and Infant DS Patients Show Elevated Levels of Ser-15-phosphorylated p53 and p21CIP1 in the Frontal Cortex
Finally, we analyzed the extent of p53 phosphorylation at Ser-15 and p21CIP1 protein levels in frontal cortex prepared from embryonic and infant human DS patients. Autopsy brains were obtained from three DS individuals at the embryonic and infant stages (23 weeks gestational age, 339 and 427 days old) and three age-matched normal individuals (22 weeks gestational age, 332 and 587 days old). Immunoblot analysis of tissue lysates demonstrated that DS frontal cortices had ∼1.8-fold greater DYRK1A expression than the age-matched controls (p < 0.05) (supplemental Fig. S4). This is consistent with a previous study showing that DS patient brains exhibit an approximate 1.5-fold increase (1.3- to 1.8-fold) in DYRK1A protein levels (26). In addition, levels of Ser-15-phosphorylated p53 (p < 0.01) and p21CIP1 (p < 0.05) were significantly greater in DS frontal cortices than in controls (Fig. 7, K–M). These results suggest that the accumulation of Dyrk1A in DS patients increases Ser-15-phosphoylation of p53, leading to up-regulation of p21CIP1 levels during late embryonic and early infant stages.
DISCUSSION
The present study demonstrated that one of the DSCR gene products, Dyrk1A, directly phosphorylates p53 at Ser-15 in vitro and in mammalian neuronal cells. In addition, Dyrk1A up-regulation by ∼1.5- to 2-fold, an extent of up-regulation similar to that seen in DS patients, leads to the induction of p53 target genes such as p21CIP1. Moreover, these events attenuated proliferation of rat and human neural progenitor cells through impairing G1/G0-S phase transition. This stimulatory effect of Dyrk1A on p53 and p21CIP1 activity and its functional role during neuronal proliferation were further supported by evidence obtained from brains of embryonic DYRK1A Tg mice and human DS frontal cortices.
The transcription factor, p53, is known to be phosphorylated at several serine and threonine residues in the N-terminal transactivation domain in response to various stimuli (22). The events elicited by p53 phosphorylation depend on the kinase phosphorylating each individual site. For example, casein kinases 1δ and ϵ phosphorylate Ser-6 and Ser-9 of p53, and Ser-20 is phosphorylated by checkpoint kinases 1 and 2 (27–29). Regarding Ser-15, a number of kinases such as ATM, ATR, DNA-PK, p38, and hSMG-1 are known to phosphorylate Ser-15 of p53, which subsequently stimulates p53 transactivation activity (30–35). Among Dyrk family members, homeodomain-interacting protein kinase 2 has been reported to phosphorylate p53 at Ser-46 and regulate p53 activity (36, 37). In addition, Dyrk2 was recently reported to phosphorylate p53 at Ser-46 in response to genotoxic stress (38). In contrast to those two kinases, Dyrk1A directly phosphorylates p53 at Ser-15, but not at Ser-46 (Fig. 2C). Moreover, our data reveal that Dyrk1A-mediated phosphorylation of p53 at Ser-15 activates transcription to suppress growth of embryonic neuroprogenitor cells.
Although the involvement of Dyrk family members (e.g. Dyrk1A, Dyrk2, and homeodomain-interacting protein kinase 2) in apoptotic cell death has been well documented (14, 37, 38), their association with cell proliferation has not been intensively studied. Nevertheless, a few reports suggest that Dyrk family is closely involved in cell cycle regulation. For example, Dyrk1B becomes maximally active in the G0 phase of fibroblasts and stabilizes CDK inhibitor p27KIP1 by phosphorylating at Ser-10 (39). Dyrk1B also phosphorylates cyclin D1 at Thr-288 and eventually causes G1/G0 arrest through the destabilization of cyclin D1 (40). Although Dyrk1A and Dyrk1B similarly altered G1/G0-S phase transition, their downstream signals appear to transmit through quite different pathways. Dyrk1A overexpression caused a specific increase of p21CIP1 in E14.5 mouse brains (Fig. 7I), but not other CDK inhibitors, including p27KIP1 and p15INK4B (data not shown). Similar to our finding, Yabut et al. also reported very recently that transiently overexpressed Dyrk1A by in utero electroporation inhibits neural cell proliferation in embryonic mouse neocortex (41). Although more studies should be performed, the current finding indicates that Dyrk1A overexpression leads to impaired neuronal proliferation through the signaling pathway containing p53 and p21CIP1 during late embryonic brain development.
The attenuation of H19-7 cell proliferation caused by stable Dyrk1A overexpression was shown to be largely dependent on the phosphorylation and consequent activation of p53. Correspondingly, specific knockdown of p53 restored the inhibition of cell proliferation induced by Dyrk1A to the control level (Fig. 5, B and C). Expression of p53-S15A mutant also recovered the reduced BrdU labeling by Dyrk1A, verifying a link between Dyrk1A-mediated p53-Ser-15 phosphorylation and the attenuated proliferation (Figs. 5B, 5C, and 6, A–D). Of course, it is debatable how to interpret the finding that cell proliferation was much greater in cells co-transfected with Dyrk1A and p53-S15A than in cells with p53-S15A alone. In addition, one should be very cautious to interpret the effect of p53-S15A on Dyrk1A-induced suppression of cell proliferation, because Dyrk1A-involving downstream signaling pathways and consequent cell phenotypes could be altered depending on the Dyrk1A levels. Moreover, we cannot rule out the possibility that one or more other factors may interact with p53-S15A and have an additional effect on cell growth. Nevertheless, the finding that p53-S15A expression considerably rescued the negative effect of Dyrk1A on neuronal proliferation obviously indicates that Dyrk1A-induced attenuation of embryonic neuronal proliferation occurs through p53-Ser-15 phosphorylation.
The interaction between Dyrk1A and p53 was observed in both rat embryonic neuronal cells (Fig. 1B) and E18.5 rat whole brain (Fig. 1, D and E) and in 7-week-old rat whole brain (supplemental Figs. S5A and S5B). Dyrk1A phosphorylation of p53 was also observed in whole brains of E14.5 mouse embryos (Fig. 7, G and H) as well as in whole brains of infant (postnatal 3 day) and in the hippocampus, frontal cortex, and cerebellum of adult (2-month-old) mouse brains (supplemental Figs. S5C and S5D). These results suggest that the interaction between Dyrk1A and p53 and subsequent p53 phosphorylation seem to occur in both proliferating and post-mitotic neurons. Given the identification of their interaction and p53 phosphorylation at postnatal and adult stages, they may have additional function(s) in neural systems other than embryonic neuronal proliferation, which should be further investigated. However, because Dyrk1A is known to be strongly expressed at embryonic stages and at birth and its expression gradually decreases thereafter until lower expression levels stabilize at around 3 weeks (42), we currently focused on the biological significance of their interaction and phosphorylation in embryonic development.
With regard to the functional role, transcription factor p53 induces a lot of downstream target genes, which are mostly associated with the regulation of cell cycle and cell death (22). Among the tested target gene products, the levels of p21CIP1, PCNA, and Fas were up-regulated in H19-7/Dyrk1A cells compared with parental control cells (Fig. 3G). Although Fas mediates extrinsic apoptotic signaling pathway, p21CIP1 and PCNA are closely involved in cell cycle regulation. Especially, the increase of p21CIP1 levels was more remarkable than that of PCNA and Fas, suggesting that there is a preferential activation of p53- and p21CIP1-mediated downstream pathways (Fig. 3G). Also, this hypothesis was further supported by additional immunoblot data, which showed that caspase-3 as a notable marker of apoptotic cell death was not activated under the condition of cell proliferation (at the zero time point in supplemental Fig. S6), even though Fas levels were increased. Moreover, the protein levels of even PCNA and Fas were not changed in E14.5 DYRK1A Tg brains compared with littermate controls (Fig. 7I). In contrast, the p21CIP1 level was consistently higher in DYRK1A Tg brains, suggesting that the subsequent activation of p53 and p21CIP1 comprises a downstream signaling pathway of Dyrk1A in hippocampal H19-7 cells and in embryonic mouse brains.
The differential expression pattern of p53 target genes between H19-7 cells (Fig. 3G) and E14.5 mouse brains (Fig. 7I) and the preferential activation of p21CIP1 among these may result from the complexities of transcriptional regulatory modes. Like many other genes, the promoters of p53 targets also consist of various regulatory elements and multiple transcription factor-binding sites, respectively. For example, p21CIP1 promoter is regulated by AP2, BRCA1, STAT family, and C/EBP family as well as p53 (43–48). Differential transcription factors and/or their combinatorial effects would tightly regulate the temporal and spatial modes of a specific gene expression. Thus, differential expression of p53 target genes in H19-7 cells and E14.5 mouse brains could be generated, because these two neural systems may have quite different cellular contexts, including different intracellular signaling cascades and various effectors. Moreover, they showed different levels of Dyrk1A expression (∼1.5-fold in DYRK1A Tg and ∼2.0-fold in H19-7/Dyrk1A cells). Nevertheless, the specific induction of p21CIP1 by Dyrk1A-mediated p53 phosphorylation was consistently observed in three distinct systems such as H19-7 cells, hES-NP cells, and E14.5 mouse brains. As an in vivo model, DYRK1A Tg mice are more suitable to validate the biological significance of actual Dyrk1A overexpression rather than cultured neuronal cells. Furthermore, embryonic DYRK1A Tg mouse brains closely reflect the condition of DS patient brains, which exhibit ∼1.5-fold increase of Dyrk1A protein levels (26).
Although we hypothesized that Dyrk1A-mediated phosphorylation of p53 preferentially induces cell cycle-regulatory genes such as p21CIP1, Dyrk1A is also possibly linked to apoptotic cell death. Previously, we reported that Dyrk1A overexpression makes H19-7 cells vulnerable to serum deprivation (14), which was confirmed by an immunoblot data that H19-7/Dyrk1A cells showed a rapid activation of caspase-3 under the condition of serum deprivation, but not in cell proliferation (supplemental Fig. S6). In this context, the sensitization of H19-7 cells to apoptosis might be possibly involved in Dyrk1A-induced p53 activation and the subsequent up-regulation of Fas (Fig. 3G). Interestingly, the increases of p53 and Fas protein were observed in the frontal cortex, temporal cortex, and cerebellum of DS patients (49). Meanwhile, Dyrk1A negatively regulates intrinsic apoptotic pathway by phosphorylating caspase-9 at Thr-125 during retina development (50). In this system, Dyrk1A had no effect on the proliferation of retina progenitor cells (50), implying that we should further investigate the diverse putative roles of Dyrk1A and its possible link to many different systems.
Regarding DS, the overexpression of Dyrk1A alone would not explain all the symptoms seen in DS patients. Furthermore, Dyrk1A-mediated phosphorylation of p53 would not be the only one cause of impaired neuronal proliferation in DS, because Dyrk1A has a lot of substrates involved in various cellular processes (42). For example, nuclear factor of activated T cells (NFAT) inhibition by Dyrk1A phosphorylation could contribute to the altered proliferation (51, 52). In addition, Dyrk1A dosage imbalance reduces neuron-restrictive silencer factor (NRSF/REST) levels and deregulates chromosomal clusters of genes located near NRSF/REST-binding sites, suggesting that Dyrk1A overexpression may perturb overall gene expression (53, 54). Dyrk1A is also known to phosphorylate proteins associated with mRNA splicing (Cyclin L2, SF2, and SF3) and translation (eIF2Bϵ) (42). Interestingly, Yabut et al. recently reported that transiently overexpressed Dyrk1A by in utero electroporation inhibits neural cell proliferation in embryonic mouse neocortex, in which the nuclear export and degradation of cyclin D1 could act as a key cell cycle modulator (41). This study also supports our finding in several aspects. First, our finding also demonstrated that Dyrk1A overexpression inhibits neuronal proliferation during embryonic development. Second, as shown in supplemental Fig. S7A, H19-7/Dyrk1A cells also showed a reduction in cyclin D1 protein levels compared with parental control cells. Nonetheless, the goal of our study was to determine the role of the p53-p21CIP1 activation pathway as a possible cell cycle modulator, because p21CIP1 up-regulation was constantly observed in H19-7 cells and DYRK1A Tg brains (Figs. 3G and 7I), while there was no change of cyclin D1 levels in E14.5 mouse brains (supplemental Figs. S7B and S7C). This discrepancy might result from the difference of sample preparation and experimental protocols, because the outcome of stable overexpression by human DYRK1A gene on a bacterial artificial chromosome in Tg mice could be different from that of the transient overexpression of GFP-fused Dyrk1A by in utero electroporation.
Furthermore, we observed the elevation of Ser-15-phosphorylated p53 and p21CIP1 levels in the frontal cortex of autopsy brains obtained from three human DS patients at the embryonic and infant stages (Fig. 7, K–M). These data suggest a role for Dyrk1A in DS pathogenesis, because we demonstrated that up-regulation of Dyrk1A in DS brains may impair neuronal proliferation through p53 phosphorylation and p21CIP1 induction. These findings help us to expand the current understanding of cell cycle alteration and impaired neuronal proliferation seen in DS patients.
Supplementary Material
Acknowledgments
We are deeply grateful to W. Becker, J. W. Cho, and C. O. Joe for generously providing plasmids.
This study was supported by a grant of the Korea Healthcare Technology R&D Project, Ministry of Health, Welfare & Family Affairs, Republic of Korea (A092004 to K. C. C. and W.-J. S.). This work was also supported by the Brain Research Center of the 21st Century Frontier Research Program Technology (2009K-001251 to K. C. C.) funded by the Ministry of Education, Science and Technology (MEST), Republic of Korea. This work was also partially supported by the National Research Foundation of Korea (NRF) grant funded by MEST (2010-0018916 to K. C. C.), by Basic Science Research Program through NRF (2010-0001668 to K. C. C.), and by a grant from Korea Science and Engineering Foundation (R01-2007-000-11910-0 to W.-J. S.).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBIDataBankwith accession number(s) NP_036923, NP_112251, and NP_000537.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S7.
J. Park and K. C. Chung, unpublished observation.
- DS
- Down syndrome
- AD
- Alzheimer disease
- CDK
- cyclin-dependent kinase
- DSCR
- Down syndrome critical region
- Dyrk1A
- dual-specificity tyrosine-(Y) phosphorylation-regulated kinase 1A
- Gadd45
- growth arrest and DNA damage-inducible gene 45
- p21CIP1/Cdkn1A
- CDK inhibitor 1A
- E
- embryonic day
- Tg
- transgenic
- PTEN
- phosphatase and tensin homolog
- NFAT
- nuclear factor of activated T cells
- NRSF/REST
- neuron-restrictive silencer factor/RE1 silencing transcription factor.
REFERENCES
- 1.Lejeune J., Gautier M., Turpin R. (1959) C. R. Hebd. Seances Acad. Sci. 248, 1721–1722 [PubMed] [Google Scholar]
- 2.Wisniewski K. E., Wisniewski H. M., Wen G. Y. (1985) Ann. Neurol. 17, 278–282 [DOI] [PubMed] [Google Scholar]
- 3.Antonarakis S. E., Lyle R., Dermitzakis E. T., Reymond A., Deutsch S. (2004) Nat. Rev. Genet. 5, 725–738 [DOI] [PubMed] [Google Scholar]
- 4.Wisniewski K. E., Laure-Kamionowska M., Wisniewski H. M. (1984) New Engl. J. Med. 311, 1187–1188 [DOI] [PubMed] [Google Scholar]
- 5.Tanzi R. E. (1996) Nat. Med. 2, 31–32 [DOI] [PubMed] [Google Scholar]
- 6.Larsen K. B., Laursen H., Graem N., Samuelsen G. B., Bogdanovic N., Pakkenberg B. (2008) Ann. Anat. 190, 421–427 [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti L., Galdzicki Z., Haydar T. F. (2007) J. Neurosci. 27, 11483–11495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contestabile A., Fila T., Ceccarelli C., Bonasoni P., Bonapace L., Santini D., Bartesaghi R., Ciani E. (2007) Hippocampus 17, 665–678 [DOI] [PubMed] [Google Scholar]
- 9.Escorihuela R. M., Vallina I. F., Martínez-Cué C., Baamonde C., Dierssen M., Tobeña A., Flórez J., Fernández-Teruel A. (1998) Neurosci. Lett. 247, 171–174 [DOI] [PubMed] [Google Scholar]
- 10.Lorenzi H. A., Reeves R. H. (2006) Brain Res. 1104, 153–159 [DOI] [PubMed] [Google Scholar]
- 11.Yang E. J., Ahn Y. S., Chung K. C. (2001) J. Biol. Chem. 276, 39819–39824 [DOI] [PubMed] [Google Scholar]
- 12.Park J., Oh Y., Chung K. C. (2009) BMB Reports 42, 6–15 [DOI] [PubMed] [Google Scholar]
- 13.Ahn K. J., Jeong H. K., Choi H. S., Ryoo S. R., Kim Y. J., Goo J. S., Choi S. Y., Han J. S., Ha I., Song W. J. (2006) Neurobiol. Dis. 22, 463–472 [DOI] [PubMed] [Google Scholar]
- 14.Park J., Yang E. J., Yoon J. H., Chung K. C. (2007) Mol. Cell Neurosci. 36, 270–279 [DOI] [PubMed] [Google Scholar]
- 15.Ryoo S. R., Cho H. J., Lee H. W., Jeong H. K., Radnaabazar C., Kim Y. S., Kim M. J., Son M. Y., Seo H., Chung S. H., Song W. J. (2008) J. Neurochem. 104, 1333–1344 [DOI] [PubMed] [Google Scholar]
- 16.Ryoo S. R., Jeong H. K., Radnaabazar C., Yoo J. J., Cho H. J., Lee H. W., Kim I. S., Cheon Y. H., Ahn Y. S., Chung S. H., Song W. J. (2007) J. Biol. Chem. 282, 34850–34857 [DOI] [PubMed] [Google Scholar]
- 17.Smith D. J., Stevens M. E., Sudanagunta S. P., Bronson R. T., Makhinson M., Watabe A. M., O'Dell T. J., Fung J., Weier H. U., Cheng J. F., Rubin E. M. (1997) Nat. Genet. 16, 28–36 [DOI] [PubMed] [Google Scholar]
- 18.Hämmerle B., Vera-Samper E., Speicher S., Arencibia R., Martínez S., Tejedor F. J. (2002) Dev. Biol. 246, 259–273 [DOI] [PubMed] [Google Scholar]
- 19.Hämmerle B., Elizalde C., Tejedor F. J. (2008) Eur. J. Neurosci. 27, 1061–1074 [DOI] [PubMed] [Google Scholar]
- 20.Currle D. S., Hu J. S., Kolski-Andreaco A., Monuki E. S. (2007) J. Vis. Exp. 2, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo H., Sonntag K. C., Isacson O. (2004) Ann. Neurol. 56, 319–328 [DOI] [PubMed] [Google Scholar]
- 22.Appella E., Anderson C. W. (2001) Eur. J. Biochem. 268, 2764–2772 [DOI] [PubMed] [Google Scholar]
- 23.Himpel S., Tegge W., Frank R., Leder S., Joost H. G., Becker W. (2000) J. Biol. Chem. 275, 2431–2438 [DOI] [PubMed] [Google Scholar]
- 24.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 25.Park C. H., Minn Y. K., Lee J. Y., Choi D. H., Chang M. Y., Shim J. W., Ko J. Y., Koh H. C., Kang M. J., Kang J. S., Rhie D. J., Lee Y. S., Son H., Moon S. Y., Kim K. S., Lee S. H. (2005) J. Neurochem. 92, 1265–1276 [DOI] [PubMed] [Google Scholar]
- 26.Dowjat W. K., Adayev T., Kuchna I., Nowicki K., Palminiello S., Hwang Y. W., Wegiel J. (2007) Neurosci. Lett. 413, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knippschild U., Milne D. M., Campbell L. E., DeMaggio A. J., Christenson E., Hoekstra M. F., Meek D. W. (1997) Oncogene 15, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 28.Hirao A., Kong Y. Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S. J., Mak T. W. (2000) Science 287, 1824–1827 [DOI] [PubMed] [Google Scholar]
- 29.Shieh S. Y., Ahn J., Tamai K., Taya Y., Prives C. (2000) Genes Dev. 14, 289–300 [PMC free article] [PubMed] [Google Scholar]
- 30.Shieh S. Y., Ikeda M., Taya Y., Prives C. (1997) Cell 91, 325–334 [DOI] [PubMed] [Google Scholar]
- 31.Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 32.Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. (1998) Science 281, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 33.Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., Abraham R. T. (1999) Genes Dev. 13, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S. J., Hwang S. G., Shin D. Y., Kang S. S., Chun J. S. (2002) J. Biol. Chem. 277, 33501–33508 [DOI] [PubMed] [Google Scholar]
- 35.Brumbaugh K. M., Otterness D. M., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R. S., Maquat L. E., Abraham R. T. (2004) Mol. Cell 14, 585–598 [DOI] [PubMed] [Google Scholar]
- 36.Hofmann T. G., Mincheva A., Lichter P., Droge W., Schmitz M. L. (2000) Biochimie (Paris) 82, 1123–1127 [DOI] [PubMed] [Google Scholar]
- 37.Hofmann T. G., Möller A., Sirma H., Zentgraf H., Taya Y., Dröge W., Will H., Schmitz M. L. (2002) Nat. Cell Biol. 4, 1–10 [DOI] [PubMed] [Google Scholar]
- 38.Taira N., Nihira K., Yamaguchi T., Miki Y., Yoshida K. (2007) Mol. Cell 25, 725–738 [DOI] [PubMed] [Google Scholar]
- 39.Deng X., Mercer S. E., Shah S., Ewton D. Z., Friedman E. (2004) J. Biol. Chem. 279, 22498–22504 [DOI] [PubMed] [Google Scholar]
- 40.Zou Y., Ewton D. Z., Deng X., Mercer S. E., Friedman E. (2004) J. Biol. Chem. 279, 27790–27798 [DOI] [PubMed] [Google Scholar]
- 41.Yabut O., Domogauer J., D'Arcangelo G. (2010) J. Neurosci. 30, 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park J., Song W. J., Chung K. C. (2009) Cell Mol. Life Sci. 66, 3235–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng Y. X., Somasundaram K., El-Deiry W. S. (1997) Nat. Genet. 15, 78–82 [DOI] [PubMed] [Google Scholar]
- 44.Somasundaram K., Zhang H., Zeng Y. X., Houvras Y., Peng Y., Zhang H., Wu G. S., Licht J. D., Weber B. L., El-Deiry W. S. (1997) Nature 389, 187–190 [DOI] [PubMed] [Google Scholar]
- 45.Chin Y. E., Kitagawa M., Su W. C., You Z. H., Iwamoto Y., Fu X. Y. (1996) Science 272, 719–722 [DOI] [PubMed] [Google Scholar]
- 46.Matsumura I., Ishikawa J., Nakajima K., Oritani K., Tomiyama Y., Miyagawa J., Kato T., Miyazaki H., Matsuzawa Y., Kanakura Y. (1997) Mol. Cell Biol. 17, 2933–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timchenko N. A., Wilde M., Nakanishi M., Smith J. R., Darlington G. J. (1996) Genes Dev. 10, 804–815 [DOI] [PubMed] [Google Scholar]
- 48.Chinery R., Brockman J. A., Peeler M. O., Shyr Y., Beauchamp R. D., Coffey R. (1997) Nat. Med. 3, 1233–1241 [DOI] [PubMed] [Google Scholar]
- 49.Seidl R., Fang-Kircher S., Bidmon B., Cairns N., Lubec G. (1999) Neurosci. Lett. 260, 9–12 [DOI] [PubMed] [Google Scholar]
- 50.Laguna A., Aranda S., Barallobre M. J., Barhoum R., Fernández E., Fotaki V., Delabar J. M., de la Luna S., de la Villa P., Arbonés M. L. (2008) Dev. Cell 15, 841–853 [DOI] [PubMed] [Google Scholar]
- 51.Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., Yamasaki N., Miyakawa T., Francke U., Graef I. A., Crabtree G. R. (2006) Nature 441, 595–600 [DOI] [PubMed] [Google Scholar]
- 52.Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P. G., Rao A. (2006) Nature 441, 646–650 [DOI] [PubMed] [Google Scholar]
- 53.Canzonetta C., Mulligan C., Deutsch S., Ruf S., O'Doherty A., Lyle R., Borel C., Lin-Marq N., Delom F., Groet J., Schnappauf F., De Vita S., Averill S., Priestley J. V., Martin J. E., Shipley J., Denyer G., Epstein C. J., Fillat C., Estivill X., Tybulewicz V. L., Fisher E. M., Antonarakis S. E., Nizetic D. (2008) Am. J. Hum. Genet. 83, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lepagnol-Bestel A. M., Zvara A., Maussion G., Quignon F., Ngimbous B., Ramoz N., Imbeaud S., Loe-Mie Y., Benihoud K., Agier N., Salin P. A., Cardona A., Khung-Savatovsky S., Kallunki P., Delabar J. M., Puskas L. G., Delacroix H., Aggerbeck L., Delezoide A. L., Delattre O., Gorwood P., Moalic J. M., Simonneau M. (2009) Hum. Mol. Genet. 18, 1405–1414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.