Abstract
During replicative aging of primary cells morphological transformations occur, the expression pattern is altered and chromatin changes globally. Here we show that chronic damage signals, likely caused by telomere processing, impact expression of histones and lead to their depletion. Interrogation of the abundance and cell cycle expression of histones and histone chaperones revealed defects in histone biosynthesis during replicative aging. Simultaneously, epigenetic marks were redistributed across the phases of the cell cycle and the DNA damage response (DDR) machinery was activated. The age-dependent reprogramming affected telomeric chromatin itself, which was progressively destabilized, resulting in a boost of the telomere associated DDR signal with each successive cell cycle. We propose a mechanism where changes in the structural and epigenetic integrity of telomeres impact core histones and their chaperones, enforcing a self-perpetuating pathway of global epigenetic changes that ultimately leads to senescence.
Introduction
Senescence is a response to stress that limits the proliferation of damaged or aged cells. The link between telomeres, senescence and aging has become well accepted, however, it remains unclear how the localized signal at shortening telomeres is translated to a nucleus-wide response, impacting the cell on a global scale. Here we suggest a mechanism, in which telomere shortening over successive generations represents a source of chronic DNA damage within the cell that leads to a destabilization of the histone biosynthesis pathways.
Several studies indicate precedence for the sensitivity of histone biosynthesis pathways to genomic stress. Under conditions that impede DNA synthesis, histone mRNA production is post-transcriptionally curtailed 1,2. Recent studies have shown that altered expression of histone chaperones CAF1 and Asf1 confers greater sensitivity to replicative stress in S-phase in mammalian cells 3,4. In fact, histone acetylation also appears to be responsive to DNA damage and involved in the repair of lesions in DNA independently of S-phase 5,6. Thus, defects in chromatin assembly can lead to loss of epigenetic information, impair cellular function and contribute to cellular aging.
We hypothesized that chronic exposure to DNA damage signals, such as those emitting from shortening telomeres during replicative aging, affect histone biosynthesis. Here we demonstrate that synthesis of new histones is reduced upon aging in culture of human diploid fibroblasts (HDFs). This coincides with reduced levels of SLBP and histone chaperones Asf1 and CAF1, implicating intrinsic changes in histone biosynthesis and chromatin assembly. As a result, the abundance and cell cycle distribution of post-translational histone modifications is altered. These changes participate in a self-enforcing regulatory loop that ultimately affects telomeric and non-telomeric chromatin. Therefore telomere erosion over successive generations appears to trigger genome wide epigenetic adaptation. We propose that this may represent a mechanism, through which telomeric stress is amplified such that it impacts the cell on a global level and eventually leads to growth arrest.
Results
Reduction of histone levels upon DNA damage and replicative aging
When cycling early passage IMR90 fibroblasts were cultivated under conditions of chronic damage due to the presence of bleomycin, H3 and H4 levels were found lowered in a dose dependent manner (Fig. 1a, Supplementary Fig. 2b). Down regulation of histone synthesis was independent of p53 and pRB, since less H3 and H4 was expressed in HCT116 cells with and without functional p53 7 and IMR90 cells expressing HPV16 E6 and E7 oncoproteins (Supplementary Fig. 2a). We concluded that histone synthesis is sensitive to damage signaling.
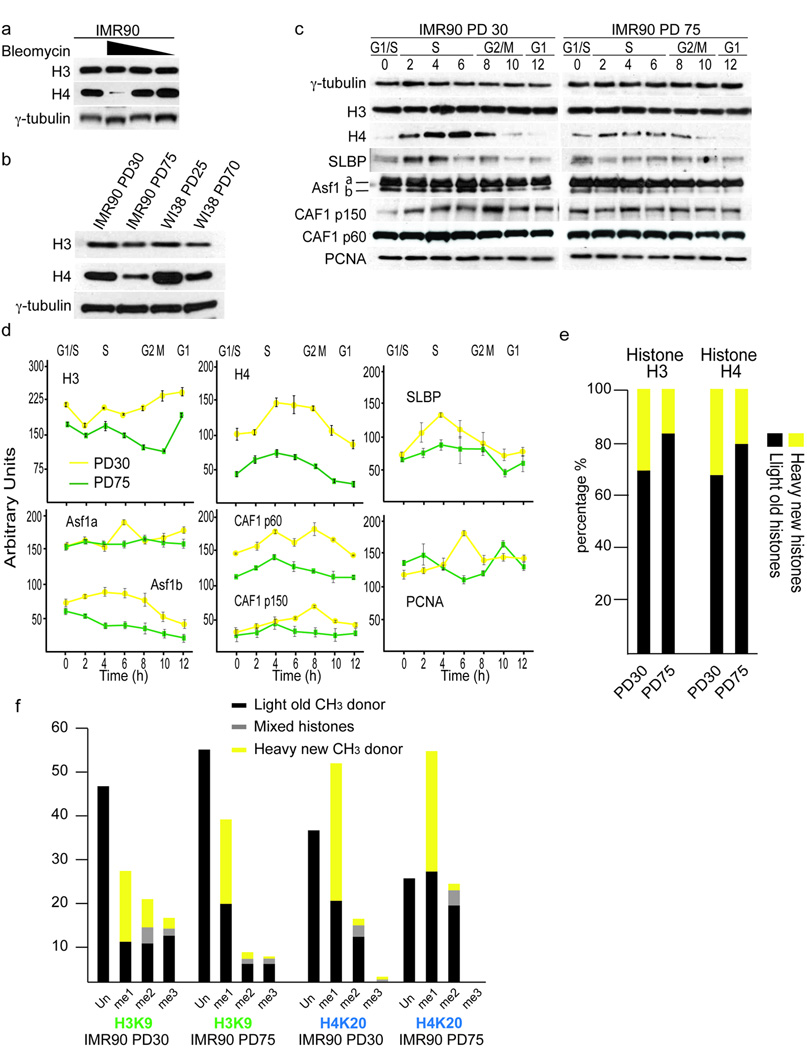
Figure 1.
Altered histone biosynthesis and redistribution of epigenetic marks upon chronic damage and cellular aging. (a) Effects of Bleomycin on histone expression. Early passage IMR90 (PD21) were subjected to 500, 50 or 5ng ml−1 of bleomycin for 6 days. Expression of H3 and H4 is shown. γ-tubulin serves as loading control. (b) Expression of H3 and H4 in early passage (PD30, PD25) and late passage (PD75, PD70) asynchronously growing IMR90 and WI38 fibroblasts. γ-tubulin serves as loading control. (c) Cell cycle expression of H3, H4, SLBP, Asf1a/b and CAF1p150, CAF1p60 and PCNA in PD30 and PD75 HDFs. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis and indicated. γ-tubulin serves as loading control. (d) Quantifications of cell cycle expression patterns of histones and histone chaperones from PD30 (yellow lines) and PD75 (green lines) HDFs. Shown are cell cycle patterns of H3, H4, Asf1a/b, Caf1p60, CAF1p150 and PCNA. Enrichments were calculated relative to γ-tubulin. Cell cycle phases are indicated on top and hours after release from G1/S on the bottom. Corresponding cell cycle phases have been determined by FACS analysis. The error bars represent the standard deviation of three independent experiments. (e) SILAC labeling of PD30 and PD75 HDFs and mass spectroscopy of histone proteins. Shown are percentages of newly synthesized histones (yellow) and histones from previous cell cycle (black). (f) SILAC labeling of PD30 and PD75 HDFs and mass spectroscopy of histone H3K9 and H4K20 methylation events. Shown are percentages of newly synthesized histone modifications (yellow), modifications from previous cell cycles (black) and those of mixed origin i.e. both old and new methyl groups co-exist forming me2 and/or me3 (grey).
To investigate the effects of telomere shortening on histone expression we compared H3 and H4 expression in early and late-passage IMR90 and WI38 fibroblasts (Fig. 1b). Consistent with the hypothesis that the shortening of telomeres induces damage signals, which affects histone synthesis, we found reduced H3 and H4 expression in the late-passage cells (Fig. 1b). To analyze the expression of histones during the cell cycle and replicative aging we investigated synchronized early and late-passage HDFs that were maintained from population doubling (PD) 20 to replicative senescence at PD85. To address the events that lead to telomere erosion associated replicative senescence, we compared early and late populations of cycling cells with similar cell cycle dynamics. To clarify the distinction between late-passage cycling (PD75) and post-mitotic senescent (PD85) cultures we performed flow cytometry of the cell cycle of PD30, PD75 and senescent cells by propidium iodide (PI), H3S10 phosphorylation and BrdU incorporation. Staining of exponentially growing asynchronous cultures showed a 4% increase in G1 cells at PD 75 (63%) when compared to PD30 (59%) (Supplementary Fig. 1a). We noted small decreases in H3S10ph (1.95% at PD30 vs. 1.75% at PD75) (Supplementary Fig. 1b) and BrdU positive cells (31% at PD30 vs. 27% at PD75) (Supplementary Fig. 1c). These differences are marginal when compared to those obtained from senescent cultures (PD85), which showed a 17% increase in cells in G1 and almost complete loss of mitotic and BrdU incorporating cells (Supplementary Fig. 1a). Further analysis of the cell cycle of synchronized early and late passage cells revealed subtle differences in S-phase between the cell cycle profiles, although the staging of the individual phases was identical (Supplementary Fig. 1d). We observed that upon aphidicolin removal 10–15% less late passage cells exit G1/S and enter S-phase (T=2hrs, PD30 at 53% vs. PD75 at 38%; T=4, PD30 at 37% vs. PD75 at 27% (Supplementary Fig. 1d). To address whether this represented differences in the competency of late passage cells to replicate we synchronized both early and late passage cells and pulsed cells with BrdU for 1hr prior to harvesting. Analysis of 2 hr intervals revealed that slightly fewer cells enter S-phase, but that there were no differences in cell cycle dynamics and the timing of S-phase (Supplementary Fig. 1e). Therefore, cells at PD75 displayed similar cell cycle dynamics as cells at PD30, even though small delays in cell cycle progression exist.
Western analysis of PD30 and PD75 cells across the cell cycle revealed that levels of histones H3 and H4 were reduced in late-passage cells and the accumulation of histones was more restricted to early S-phase (Fig. 1c, 1d). The stem loop binding protein SLBP binds tightly to the conserved hairpin at the 3’ end of histone mRNAs and is required for their translation 8. SLBP itself displayed its peak expression level in S phase 9 (Fig. 1c, 1d). Comparing expression levels of SLBP in PD30 and PD75 cells revealed that this regulation of SLBP was lost in late passage cells and less protein was expressed in S phase (Fig. 1c, 1d). SLBP was also down regulated upon chronic exposure to bleomycin (Supplementary Fig. 2b). Histone H1 was not found down regulated (Supplementary Fig. 2c), demonstrating that not all histones respond equally to replicative aging and damage signaling.
Next we examined histone chaperones Asf1a/b and CAF1-p150/p60 during the cell cycle and found the levels of these factors substantially reduced in late passage cells (Fig. 1c, 1d). It has been suggested that PCNA (proliferative cell nuclear antigen) couples DNA replication and the restoration of the parental epigenome through interactions with histone chaperones 10,11. This was reinforced here by the finding that PCNA levels were reduced in PD75 cells.
To biochemically quantify changes in histone synthesis in PD30 and PD75 cells, we applied SILAC (stable isotope labeling with amino acids in cell culture), where labeling of cells with L-methionine (methyl-13CD3) allowed us to monitor the biosynthesis of new histones during a single cell cycle by quantitative mass spectroscopy, which is especially important for H3, since western analysis cannot distinguish between H3 variants. We observed a 43% and 47% reduction in the production of H3 and H4 in late-passage cells, respectively (Fig. 1e). A concern with many in vitro studies of replicative aging is the possibility that there is an accumulation of post-mitotic senescent cells, represented as G1 in flow cytometry, thereby producing a mosaic of cells that are at different stages in their proliferative lifespan. To rule out any possible bias we labeled cells with Hoechst and sorted pure populations of cycling G1 and S/G2/M cells 12 (Supplementary Fig. 2d). Examination of H3 and H4 levels in these separate fractions again showed the most prominent loss of histones H3 and H4 in the cycling fractions of late-passage cells (Supplementary Fig. 2e). Therefore, the changes we observed do not arise from heterogeneity of cell populations.
Having confirmed the deregulation of histone production we rationalized that the establishment and maintenance of epigenetic histone marks during replicative ageing could be affected. Several of these marks, such as H2AX, H3K9ac, H3K56ac, H3K79me2, and H4K20me2 have been implicated in the response to DNA damage and replicative stress. Indeed, chronic exposure to bleomycin altered the levels of H3K56ac, H3K79me2 and H4K20me2 in a stringently dose dependent (Supplementary Fig. 2f) and checkpoint dependent manner 6 (data not shown). Therefore, it was likely that epigenetic marks would also respond to age related accumulation of DNA damage.
SILAC labeling followed by mass spectroscopy facilitates the analysis of changes in parental and newly catalyzed abundant marks such as H3K9 and H4K20 methylation. We analyzed the percentages of unmodified residues and of mono-, di- and tri-methylation of H3K9 and H4K20 in early and late-passage cells and observed a 66% reduction of both total H3K9me2 and H3K9me3 with a simultaneous increase of H3K9me1, which was largely newly catalyzed (Fig. 1f). In contrast, we found a 30% increase in H4K20me2, and detected a ~30% reduction of unmodified H4K20 and complete loss of H4K20me3 during cellular aging (Fig. 1f). Despite the depletion of newly synthesized histones by ~50%, the abundance of many histone modifications did not mirror this degree of reduction. Rather, these data potentially suggested the altered activities of chromatin modifying enzymes, such as those regulating the transition of H3K9me1 to H3K9me3 and the H4K20me2 KMTs during cellular aging.
Redistribution of histone modifications across the cell cycle
Next, we examined the cell cycle distribution and dynamics of histone acetylation and methylation events, since these marks provide an indicator of the state of epigenetic regulation 13. We used site-specific antibodies that detect acetylated and methylated states of lysine residues (Supplementary Table 1) in synchronized cell populations. H3K4 and H3K27 have been associated with activation and repression of transcription 14. In PD30 cells, H3K4me1 and H3K4me2 were present across the cell cycle, whereas H3K4me3 was enriched specifically at the end of S-phase and G2 (Fig. 2a, Left Panel). We observed a similar pattern of H3K27me1, H3K27me2 and H3K27me3, although H3K27me3 was enriched in S-phase and in G2/M. In PD75 cells, we observed a redistribution of these marks. There was less H3K4me1 and H3K4me2 appeared restricted to early S-phase (Fig. 2a, right panel). H3K4me3 exhibited a bi-phasic distribution in early S-phase and G2/M. The distribution of H3K27me1 and H3K27me2 was largely similar to that observed in early passage cells, however, H3K27me3 was enriched most prominently in G2/M. Recent biochemical studies of polycomb, which can associate with H3K27me3 via its chromodomain 15, have suggested a role in DNA replication 16. The changes in both H3K4me3 and H3K27me3 that we observed in late-passage cells potentially represents alterations in both the transcriptome and in replication dynamics 17. Quantifications are provided in Supplementary Fig. 3.
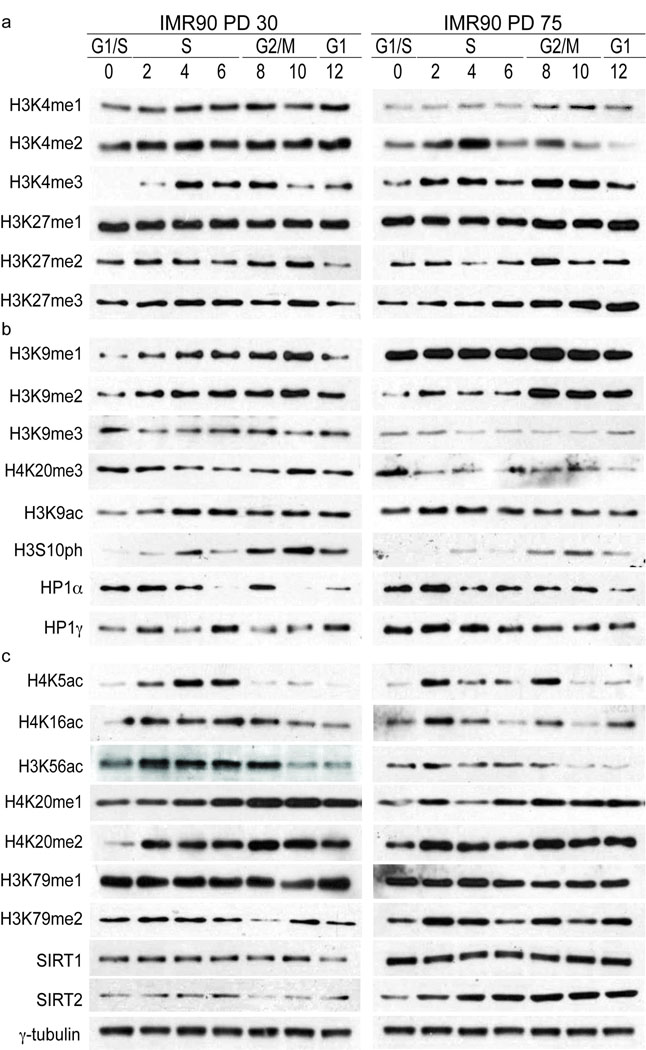
Figure 2.
Cell cycle distribution of histone modifications in early and late passage cells. (a) Cell cycle distribution of histone modifications in whole cell extracts from PD30 and PD75 HDFs. Shown are cell cycle patterns of modifications involved in transcriptional regulation (H3K4me1/2/3 and H3K27me1/2/3). 75,000 cell equivalents were loaded per lane and histones were visualized by ponceau staining prior to probing with primary antibodies. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis. Images shown represent western analysis performed, stained and exposed simultaneously to film. Quantification of western analysis can be seen in Fig. S2. Abundance of post-translational modifications was calculated relative to the total amount of that particular protein i.e. the abundance of H3K9me3 was quantified relative to total H3. (b) Shown are cell cycle patterns of modifications and events involved in heterochromatin regulation (H3K9me1/2/3, H4K20me3, H3K9Ac, H3S10ph, HP1α and HP1γ). Western analysis and quantifications were performed as in (A). (c) Cell cycle patterns of H4K5Ac, H4K16Ac, H3K56Ac, H4K20me1/2, H3K79me1/2, SIRT1 and SIRT2. Western analysis and quantifications were performed as in (A). The intensity of signals from westerns of individual modifications does not reflect the levels of each modification in the cell. However, these images reflect the levels of the same individual modification in early and late passage cells. γ-tubulin serves as loading control for SIRT1 and SIRT2.
Methylation of H3K9 and H4K20 has been implicated in heterochromatin formation and maintenance. Analysis of these marks revealed interesting features in PD30 cells: (I) They were enriched in G1, removed during S-phase and re-established in G2/M, much like the patterns of H3K9me in fission yeast 18,19. (II) The loss of H3K9me3 coincided with the increase in H3K9me1 and H3K9me2 in S-phase, likely reflecting the dynamic demethylation of H3K9me3 and/or progressive transition from H3K9me1 to H3K9me3 via H3K9me2. This is reminiscent of chromatin in Suv39h−/− mouse cells suggesting reduced Suv39h activity during replicative aging 20. (III) H3K9me3 and H3K9ac appeared to be marks, which are exclusive within the cell cycle, with H3K9ac becoming more abundant as the tri-methylation of H3K9 was removed. As previously shown by mass spectroscopy (Fig. 1f), we observed a ~30% reduction in H3K9me3 and H3K9me2 in PD75 cells, whereas H4K20me3 was almost completely lost. This loss has been confirmed in the cell cycle western analysis here in PD75 cells (Fig. 2b).
Several histone modifications have been linked with the propagation of epigenetic memory at replication forks as well as in the repair of DNA lesions. In PD30 cells we observed that acetylation of H4K5, H3K9 and H4K16 and H3K56 was elevated throughout S-phase and extending into G2 (Fig. 2c, left panel). H4K20me2 and H3K79me2 were enriched across S-phase/G2 chromatin. These marks have strong links to chromatin replication (H4K5ac), compaction (H4K16ac) and transcriptional control and DNA damage sensing (H3K9ac, H3K56ac, H4K20me2 and H3K79me2). In PD75 cells H3K5ac, H3K9ac and H4K16ac, substrates for SIRT2 21 and SIRT6 22, displayed a prominent enrichment in early S-phase with an additional H3K5ac and H4K16ac enrichment in G2 (Fig. 2c, right panel). The strong correlation between the pattern of H4K5ac and chromatin replication could be indicative of altered DNA replication dynamics in late-passage cells. H3K56ac did not exhibit this common pattern. Rather, there was substantial reduction in PD75 cells. Normally, K56ac is required for chromatin assembly during replication and marks the completion of chromatin assembly following DNA repair 23, but is lost in response to persistent DNA damage 6. The correlation between removal of this mark and the loss of proliferative capacity in replicative aging could indicate an epigenetic checkpoint. In cancer cells this mark persists by an unknown mechanism 5. H3K56 can be deacetylated by SIRT1 and SIRT2 5, enzymes found upregulated in late passage cells (Fig. 2c), pointing out the direct link between changes in the levels of modifying enzymes and their targets. Notably, several lines of study have shown that the accumulation of H3K56ac is Asf1a/b dependent. Asf1a/b expression is regulated by E2F 24 and can be also be limited by replicative stress 25. Therefore, it appears that the levels of H3K56ac, and potentially other S-phase modifications, are dictated by the availability of Asf1a/b and can be differentially affected by distinct checkpoints. This is further emphasized by the similar changes that we observed for H3K9ac, H4K20me2 and H3K79me2, marks that are linked with DDR and checkpoint control 26. As with several states of acetylation, each exhibited a single prominent enrichment in early S-phase with an additional enrichment in G2 (Fig. 2c, right panel). Therefore, it appears clear that the epigenome of late-passage cells is under considerable strain.
Biphasic activation of DDR in late-passage cells
We suggest that the changes observed in late passage cells were a response to chronic stress. By analyzing several key players in the DDR, we sought to pinpoint the cell cycle stage at which the response was activated. ATM serine 1981 phosphorylation (γ-ATM) was increased in PD75 cells, where analysis of the cell cycle expression revealed peaks in early S phase and G2/M. ATR expression in early-passage cells was confined to S and G2 phases, consistent with the kinases’ role during replication. RPA32, an important ATR target that facilitates resection and subsequent repair at breaks, exhibited a virtually identical change in its expression as that of ATR (Fig. 3, Supplementary Fig. 4).
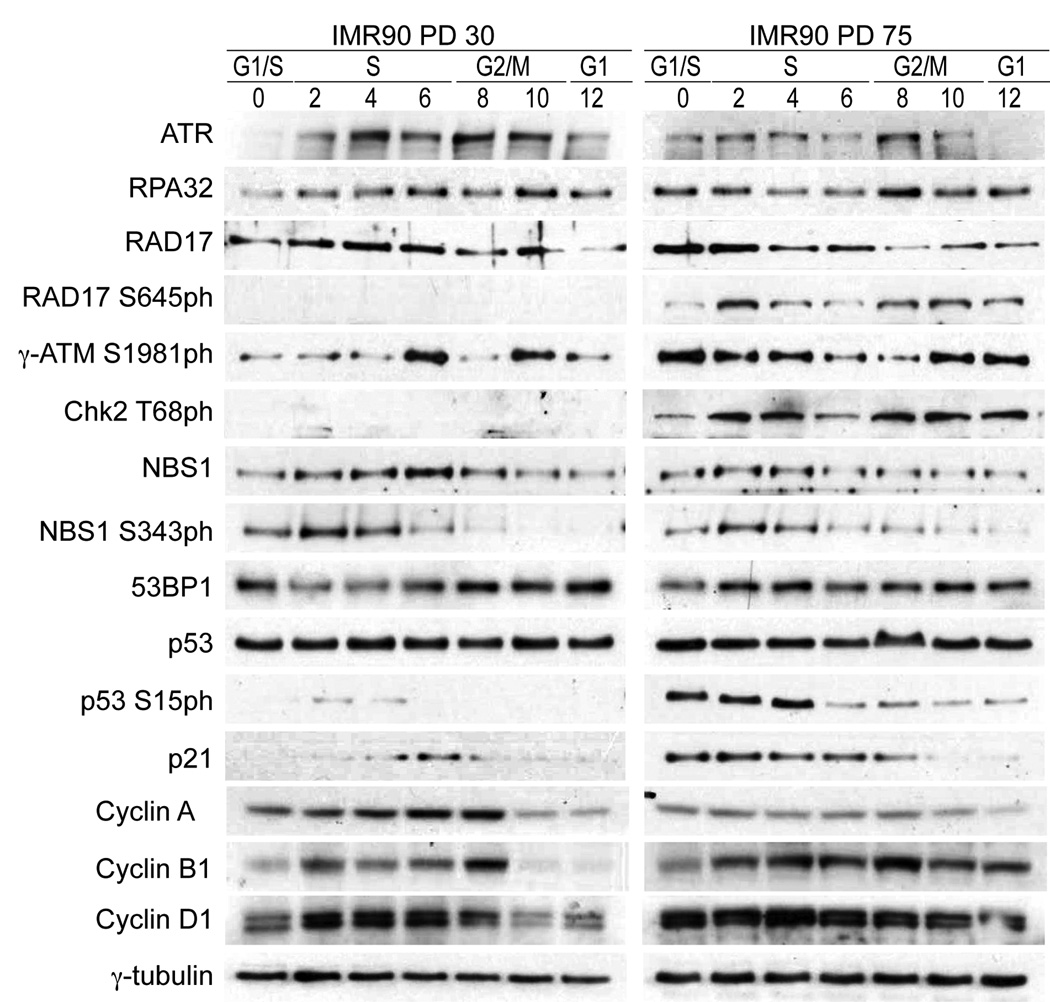
Figure 3.
DNA damage accumulation and DDR activation upon cellular aging. Expression of PCNA, Rpa32, Rad51, ATR, Rad17, Rad17-pS645, γ-ATM pS1981, Chk2-pT68, NBS1, NBS1S343ph, 53BP1, p53, p53S15ph, p21 and Cyclins A/B1/D1 in whole cell extracts of PD30 and PD75 HDFs. 75,000 cell equivalents were loaded per lane and equal loading was confirmed by ponceau staining prior to probing with primary antibodies. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis and indicated. γ-tubulin serves as loading control. Quantification of expression can be found in Fig. S5.
A key trigger of downstream DDR signaling is remobilization of the MRN complex 27, which acts as a transducer of both γ-ATM and ATR signaling to downstream targets, including Chk2 and Rad17. Phosphorylation of Rad17 has been implicated in the stabilization of proteins such as PCNA and RPA32 at stalled forks to facilitate repair 26. Chk2 plays a role in the suppression of the oncogenic potential of DNA lesions by activating the G2/M checkpoint 28. Thus, we compared the cell cycle expression of NBS1, Rad17 and Chk2 in early and late-passage cells. NBS1 expression in PD75 cells was shifted to early S-phase but there was no additional G2/M peak. NBS1 serine 343 phosphorylation was observed in both populations during early S-phase (Fig. 3, Supplementary Fig. 4). Rad17 was expressed throughout the cell cycle of both populations, but the ATR dependent phosphorylation at serine 645 was observed only in PD75 cells (Fig. 3 right panel, Supplementary Fig. 4). The γ-ATM target Chk2 threonine 68 was observed uniquely in phosphorylated state in PD75 cells, and displayed a similar biphasic pattern as γ-ATM.
While we observed p53 phosphorylation and increased p21 expression in late passage cells, this DDR was not sufficient to induce cell cycle arrest (Fig. 3, Supplementary Fig. 4). One reason might be the failure to target genes that are regulated by the activity of cyclin dependent kinases (CDKs). In early and late passage cells the expression patterns of cyclins A, B1 and D1 were altered, which is consistent with the slight delay in cell cycle dynamics observed (Supplementary Fig. 1). We detected a decline in cyclin A and an increase in cyclin D1, suggesting that cells that approach the end of their replicative life span start to display a deregulation of cyclin/cdk complexes (Fig. 3). Furthermore, the finding that late-passage cells cycle similarly to PD30 cells in the presence of an active DDR suggest that cells can tolerate a certain level of damage signaling without exiting the cell cycle.
Expression of telomerase resets the epigenome of late-passage cells
To investigate whether changes in histone metabolism play a role in human aging we compared fibroblasts from a young individual (GM00038, 9 years) with those from an old individual (AG09602, 92 years) 29 and found altered expression of H3, H4, SLBP and Asf1a/b in the old cells (Fig. 4a). Similarly to late passage IMR90 fibroblasts H3K79me2 and H4K20me2 displayed a more pronounced biphasic expression pattern in the old cells. Down regulation of H3 and H4 has been confirmed by SILAC, which demonstrated a ~50% decline in their synthesis in the old cells (Fig. 4b), suggesting that down-regulated histone synthesis and epigenetic changes are an integral part of aging.
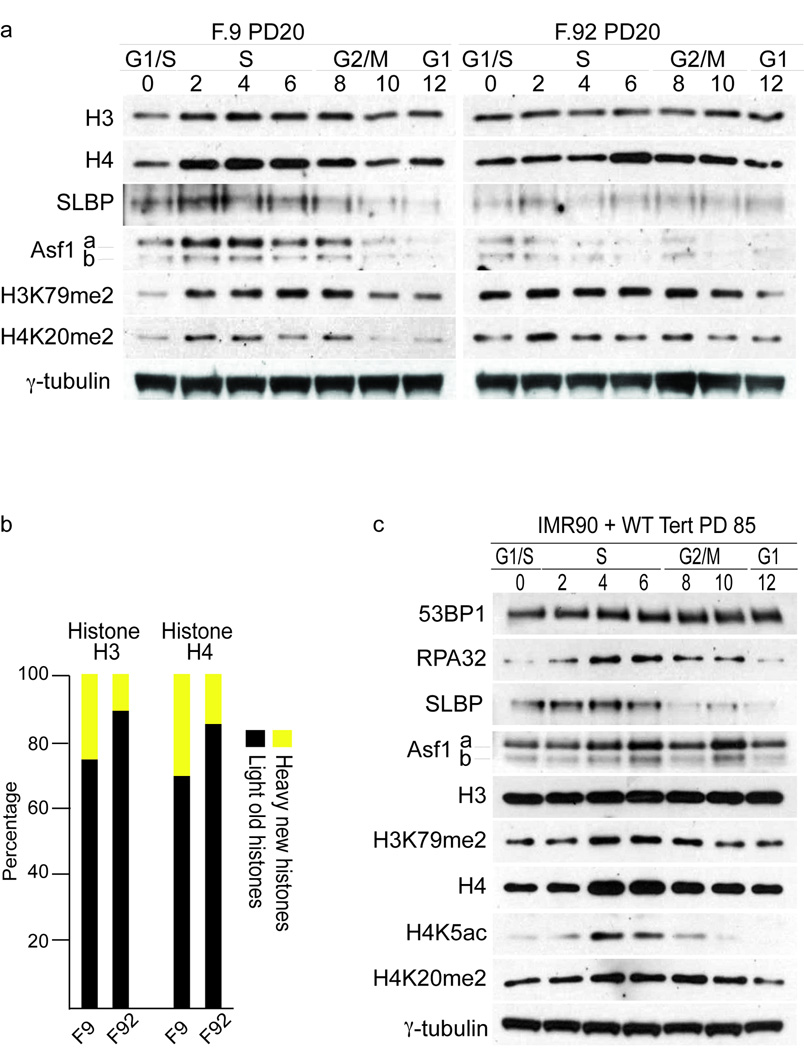
Figure 4.
Telomerase expression resets late passage cells to early passage cells. (a) Expression of H3, H4, SLBP, Asf1, H3K79mes and H4K20mes in synchronized cells from a young (left panel) and old individual (right panel). 75,000 cell equivalents were loaded per lane and equal loading was confirmed by ponceau staining prior to probing with primary antibodies. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis and indicated. γ-tubulin serves as loading control. (b) SILAC labeling of F9 and F92 HDFs and mass spectroscopy of histone proteins. Shown are percentages of newly synthesized histones (yellow) and histones from previous cell cycle (black). (c) Expression of 53BP1, Rpa32, SLBP, Asf1a/b, H3, H3K79me2, H4, H4K5ac and H4K20me2 in whole cell extracts of PD85 HDFs expressing catalytically active hTERT. 75,000 cell equivalents were loaded per lane and equal loading was confirmed by ponceau staining prior to probing with primary antibodies. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis and indicated. γ-tubulin serves as loading control.
Next we addressed whether the epigenetic changes were reversible by elongating the telomeres in PD75 cells. Only the expression of the wt hTERT allele led to detectable telomerase activity, telomere elongation and continuous cell growth beyond PD80, whereas control cells or cells expressing an inactive hTERT allele 30 senesced rapidly (Supplementary Fig. 5a,b). Analyses of the patterns of epigenetic marks and DDR proteins revealed that expression of 53BP1 and RPA32, which were substantially altered in PD75 cells (Fig. 3 right panels), were reduced to levels similarly to early passage cells (Fig. 4c, Supplementary Fig. 5d). Likewise, the expression levels of histones H3 and H4 returned to near those of early passage cells, which we confirmed by SILAC (Fig. 4c, Supplementary Fig. 5e, f). P53S15 and Chk2T68 phosphorylation declined sharply, indicating that DNA damage signaling pathways were deactivated by telomerase-dependent rescue of short telomeres (Supplementary Fig. 5c). Analysis of the cell cycle distribution of ChkT68 and RAD17S645 expression demonstrated that these marks are no longer expressed in hTERT expressing cells (Supplementary Fig. 5d). We no longer observed the biphasic distribution of H4K5ac, H4K20me2 or H3K79me2 that was seen in PD75 cells and expression was now restricted to a single peak in S/G2 phase (Fig. 4c, Supplementary Fig. 5d), as in PD30 cells (Fig. 2c, left panel). The second H4K20me2 peak in G2 was still visible, suggesting that not all marks have returned to the level of early passage cells yet. Given the association of these marks with replicative stress and DDR, the restoration of these patterns reflects a cessation of DDR activation. Furthermore, SLBP and Asf1a/b are again readily detectable upon telomere elongation. These data suggest that telomere stabilization in late-passage cells is sufficient to render them indistinguishable from early passage cells and strongly supports our view that the chronic stress associated with telomere shortening is the dominant signal that initiates the cellular senescence program. Our experiments suggest that changes in telomere integrity affect histone synthesis and genome wide chromatin structure and address the implications of expressing telomerase in pre-senescent cells, namely by reprogramming the epigenome.
A boundary distinguishes telomeric and sub-telomeric regions
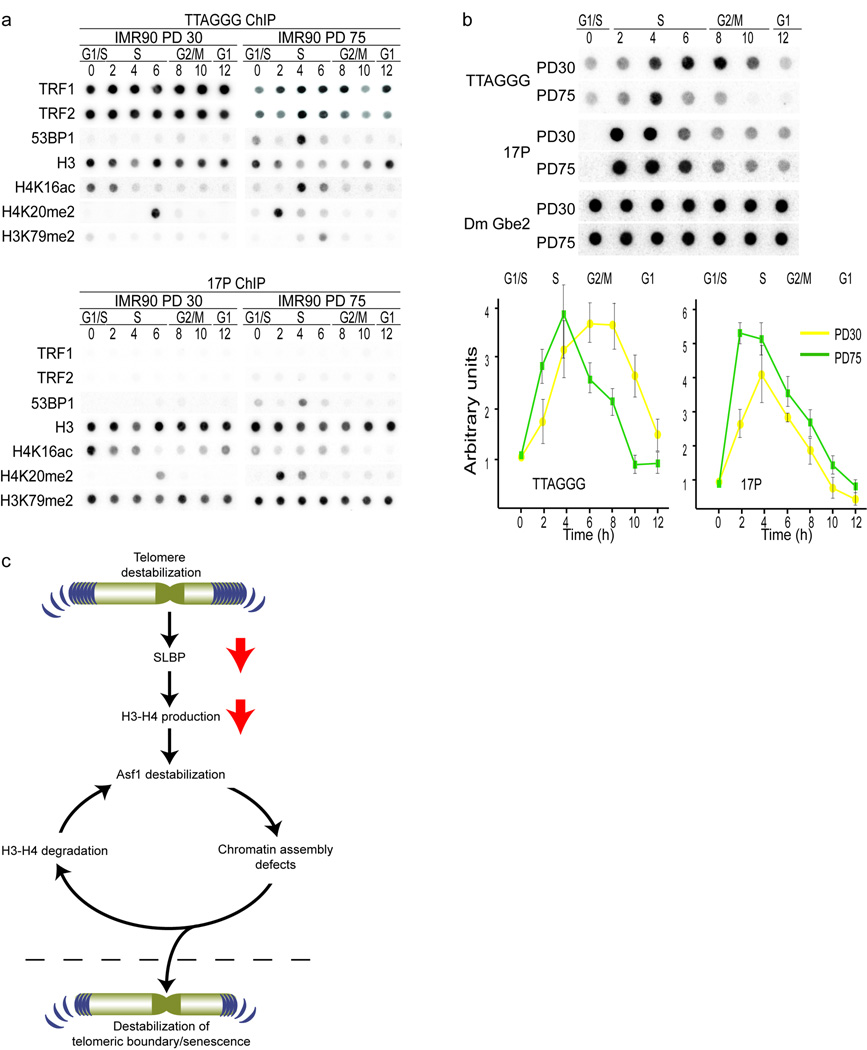
Next we investigated whether telomere erosion would impact telomeric chromatin itself. We compared the distribution of epigenetic marks at TTAGGG repeats with the subtelomeric 17P region by ChIP. Comparison of TRF1 and TRF2 occupancy of telomeres revealed reduced levels in PD75 cells (Fig. 5a, upper panel, Supplementary Fig. 6). As expected TRF1 and TRF2 were absent at subtelomeric regions (Fig. 5a, lower panel). Analysis of histone H3 in PD30 cells suggested that telomeres contained fewer nucleosomes than 17P (Fig. 5a, Supplementary Fig. 6). In PD75 cells, telomeres displayed an even more pronounced loss of nucleosomes (Fig. 5a, right panel, Supplementary Fig. 6). However, we acknowledge that due to differences in probes used we can only detect relative changes in protein occupancy and not absolute protein amounts, limiting the ability to compare histone binding between telomeres and 17P.
Figure 5.
Cellular aging results in an altered chromatin landscape of human telomeres. (a) Synchronized ChIP experiments with chromatin prepared from PD30 (left panels) and PD75 (right panels) HDFs followed by dot-blot southern hybridization with probes to telomeric TTAGGG repeats (upper panels) and sub-telomeric 17P (lower panels). Antibodies used for precipitation are indicated on the left. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis and indicated. Quantification of ChIP analysis can be seen in Fig. S5.
(b) DNA-IP with anti-BrdU antibodies followed by dot-blot southern hybridization with probes to telomeric TTAGGG repeats, sub-telomeric 17P and DmGbe2 (spiked IP control) in early (PD30) and late passage (PD75) HDFs. Hours after G1/S release and cell cycle phase are indicated above the blots. Quantifications of DNA-IPs from three independent experiments are shown in the lower panel and the error bars represent the standard deviation. The signal at T=0 was deduced from each signal as background. Hours after G1/S release are indicated and corresponding cell cycle phases have been determined by FACS analysis and indicated.
(c) DNA damage signals at short telomeres affect SLBP and in turn H3/H4 biosynthesis. Down regulation of H3/H4 destabilizes Asf1, leading to genome wide epigenetic changes, which amplify the damage signal during many cell cycles and population doublings. Eventually, epigenetic modifications spread into telomeres enabling association of DNA damage and repair proteins with chromosome ends. This self-perpetuating cycle continues until a critical damage threshold is exceeded, prompting exit from the cell cycle.
53BP1 was not detectable at telomeres or sub-telomeric chromatin in early-passage cells, but accumulated during S-phase in PD75 cells (Fig. 5a, Supplementary Fig. 6). Analyzing the major histone marks at telomeric and subtelomeric chromatin, we unexpectedly found the levels of H3K9me3, H4K20me3 and H3K27me3 to be relatively low (data not shown), indicating a clear distinction between human and mouse telomeric chromatin 31. Telomeres in laboratory-mice not only differ from human telomeres in that they are much longer, but also in that murine telomeres display extensive H3K9me3 and H4K20me3 methylation, hallmarks of constitutive heterochromatin 31. Consideration of these data and the changes in global cell cycle distribution of histone modifications between early and late-passage cells prompted us to look primarily at those marks that are redistributed during S-phase. Detailed interrogation demonstrated that telomeres in PD30 cells are hypo-acetylated at H4K16 and hypo-methylated at H3K79me2, whereas each of these were strongly enriched at the sub-telomeric region (Fig. 5a, Supplementary Fig. 6). However, in PD75 cells H4K16ac displayed S-phase enrichments at telomeres, while it was reduced at 17P. We observed a shift in H4K20me2 from late S-phase/G2 in early-passage cells to early S-phase in late-passage cells at both telomeres and 17P. Both, the S phase enrichment of H4K16ac and the shift in H4K20me2 were reversed by telomerase expression (data not shown). Based on this evidence we suggest that the interplay of epigenetic marks at human telomeres mirrors that of the telomeric heterochromatin boundary of S. cerevisiae 32.
In yeast, shortened or damaged telomeres replicate earlier in the cell cycle 33. The S-phase associated changes in DDR protein expression and redistribution of histone modifications that we observed raised the possibility that replication dynamics of telomeres and sub-telomeric regions are altered during the cellular aging process. Assaying for BrdU incorporation at different regions in the genome during the cell cycle revealed that telomeres in early passage cells replicated throughout S-phase into G2, whereas telomeres of PD75 cells replicated much earlier, with BrdU incorporation largely complete by the end of S-phase (Fig. 5b). The sub-telomeric 17P completed replication hours earlier than telomeres in early passage cells, but in late passage cells the replication timing of 17P was relatively unchanged.
Discussion
We propose a model in which the chronic stress signal generated by shortening telomeres leads to down regulation of H3 and H4 synthesis. This alters Asf1 expression and function and as a result, the capacity to faithfully restore the chromatin landscape following replication and DNA damage chromatin is progressively limited as cells age. This serves to amplify the DDR signal of telomere shortening by further abrogating the histone-chaperone equilibrium, and illustrates how localized damage signals at telomeres can be translated genome-wide. The loop continues until changes in the chromatin environment at telomeres facilitate the invasion of damage markers into the TTAGGG repeats beyond a threshold that the cell can tolerate, which prompts irreversible cell cycle exit and senesce (Fig. 5c). Since all these effects can simply be suppressed by elongation of telomeres points towards short telomeres as the main inducer of cellular senescence.
Histone synthesis during replicative aging
Even slight changes in the histone-DNA equilibrium disrupt DNA synthesis, compromise chromatin architecture and challenge cell viability 34,35. Transient replicative stress activates mechanisms that curtail the production of histones 36. SLBP is required for histone pre-mRNA processing 37, export of histone mRNA and stimulates histone translation. RNAi of SLBP causes proliferation defects, presumably by compromising the reconstitution of adequate levels of S-phase histones. Asf1 regulates chromatin structure by promoting chromatin disassembly, accepting histones from parental DNA strands, and promoting their deposition to daughter strands via other histone chaperones 38. Under conditions of replicative stress there is a notable increase in the association of histones with Asf1, thought to reflect a “histone buffer” to be utilized as soon as chromatin assembly proceeds 3. In human cells, even a marginal increase of histones impeded Asf1 function and prevented chromatin disassembly, indicating that changes in the level of histones in the cell can directly influence the properties of proteins involved in their biogenesis 38. The depletion of Asf1 resulted in a similar failure to complete S-phase following release from a replicative block. We observed that the production of new histones was reduced in late-passage cells. This decline might correlate with the destabilization of Asf1 activity, prompting degradation of histones, and also with reduced expression of SLBP, leading to less efficient translation of histone messages.
Genome-wide chromatin reorganization during replicative aging
We observed that acetylation of H3K9, H3K56, H4K5 and H4K16 is distributed throughout S-phase and G2/M and that distribution of these marks was highly responsive to the epigenetic changes during cellular aging, as has been observed for H4K16 during aging in yeast 39. We noted increases in H3K9ac and H4K16ac and a bi-phasic cell cycle distribution for those marks, as well as for H4K5ac. In contrast H3K56ac was substantially reduced, potentially due to targeting by multiple SIRT histone deacetylases 5,40, or more likely, due to lower levels of Asf1 and Caf1 23,41,42. The correlation between H3K56ac and the loss of proliferative capacity in replicative aging might indicate that the removal of this mark represents an epigenetic checkpoint, as has been suggested in yeast 43.
In contrast to histone acetylation that can exist on unassembled histones, the establishment of several methyl-marks requires a nucleosomal substrate 44–46. Therefore, it is likely that changes in histone acetylation are likely to influence other chromatin modifications. Genetic studies have shown a requirement for H4K16ac for H3K79me2 to be set, whereas H4K20me2 is inhibited by this mark 47,48. We showed that the cell cycle distribution of acetylation, H4K20me2 and H3K79me2 largely overlapped and their redistribution correlated with that of 53BP1 and DDR proteins. Thus it is possible that changes in histone acetylation, H4K20me2 and H3K79me2 on replicating chromatin serves as a genome surveillance mechanism that provides a chromatin environment conducive to repair of lesions and reactivation of stalled forks.
A new link for telomeres in replicative senescence
Critically short telomeres are often invoked as the principal determinant of the replicative potential of cells, however, TTAGGG repeats in senescent cells have by no means been exhausted. The accumulation of 53BP1 (or γH2AX) at dysfunctional telomeres has been observed in senescent cells 49, as well as at telomeres devoid of TRF2 and Pot1 50,51. In the latter case binding of 53BP1 appears to be at least in part dependent on the accumulation of H4K20me2 and has been shown to alter the local chromatin environment to promote NHEJ 52. Therefore, similar changes are likely to occur, albeit at reduced kinetics and levels, during replicative senescence and acute telomere uncapping.
Here we have identified a chromatin boundary as a novel feature of human telomeres. This boundary is defined by four factors: (I) the start of the telomeric sequence, (II) the potential decline in nucleosome occupancy between sub-telomeric regions and telomeres, (III) the TRF proteins and (IV) the differential distribution of histone modifications across sub-telomeric and telomeric regions.
The model implies that the association of DDR proteins with telomeres is proportional to the stability of the boundary. Therefore, as a result of the chronic DDR at shortening telomeres and due to reduced histone biosynthesis over the course of several generations this boundary is destabilized. Notably, this boundary is strongly reminiscent of the heterochromatic boundary that exists at telomeres in S. cerevisiae, where differential accumulation of H4K16ac and H3K79me between telomeres and adjacent regions are proposed to generate a barrier for the spreading of telomeric heterochromatin 32,48. Much like our observations made in human cells, telomeres in budding yeast are normally hypoacetylated and hypomethylated. The general similarities between telomeres in aging yeast and human cells suggest that the loss of histones during aging could be an evolutionary conserved mechanism that regulates replicative lifespan, employing changes in telomere structure or metabolism, particularly during S-phase of the cell cycle.
Methods
Cell lines, culture and treatments
Early passage (PD25) IMR90 HDFs (ATCC) were grown in Glutamax-DMEM (Invitrogen) with 15% (v/v) fetal bovine serum, at 7.5% (v/v) CO2 and 3% (v/v) O2. Cells were passaged for an additional 60 population doublings where the new PD was calculated as PD= PD at plating + [ln (#harvested/#seeded)/ln 2]. Cells from young (GM00038) and old (AG09602) individuals (Coriell Cell Repository) were cultured as above. Cells were counted with the Z1 Coulter Counter (Beckman Coulter). Retroviruses were produced and cells were infected as described 53. Cells to be used for flow cytometry were fixed in 70% (v/v) EtOH and stained according to standard protocols. For cell sorting, asynchronous logarithmically growing cultures of IMR90 were labeled for 1 hour prior to harvest with 5ug ml−1 lipophilic Hoechst 33342. Cells were sorted into pure G1 and S-G2/M populations as described 12. Bleomycin treatment was done for six days at concentrations that did not inhibit cell growth (500, 50 and 5 ng ml−1).
SILAC labeling
IMR90 cells were plated at 1×106 cells in 15cm dishes and grown overnight. Cells were washed twice in DPBS and new SILAC medium containing 30mg l−1 L-methionine-13CD3 (Sigma Isotec) and 63mg l−1 L-Cystine DiHydrochloride (Sigma) added. Cells were incubated for 12hrs and harvested by trypsin treatment, washed in DPBS and lysed in NuPage LDS sample buffer (Invitrogen) at 104 cells per µ.
Mass Spectroscopy 54
Histones were separated by 8–12% (v/v) SDS-PAGE and stained with Coomassie. The histone region was excised and digested by a modified in-gel digestion protocol. Briefly, gel bands were destained overnight with 50% (w/v) acetonitrile/50mM NH4HCO3. Then free primary and secondary amines were propionylated: To the gel pieces 20µl 50mM NH4HCO3 were added followed by 50µl 30% (v/v) propionic anhydride in methanol. This procedure was repeated twice. Then gel bands were washed with 100mM NH4HCO3, 50mM NH4HCO3 and 50% (w/v) acetonitrile/50mM NH4HCO3. Cysteines were reduced with 100µl 10mM dithiothreitol (DTT) at 56 ° C for 20min and then carbamidomethylated by addition of 200µl 5mM iodoacetamide in 100mM NH4HCO3 for 20min at RT, protected from light. Samples were washed twice with 100mM NH4HCO3 and twice with 50mM NH4HCO3. Then 200µl acetonitrile were added and the gel pieces shrunk on a speedvac. Enough trypsin (12.5ng µl−1 in 50mM NH4HCO3) was added so that the gel pieces were covered and incubated for 5min. Then the supernatant was removed, 20µl 50mM NH4HCO3 were added and the samples incubated over night at 37°C. Peptides were extracted by removal of the supernatant from the gel pieces followed by two extractions with 20µl 5% (v/v) formic acid on an ultrasonic bath for 10min. The three supernatants were combined and analyzed by mass spectrometry.
HPLC–MS
Peptide samples were analyzed on a ThermoFisher Scientific Orbitrap mass spectrometer coupled to an Agilent 1100 nano-LC pump fitted with a 12.5cm × 75µm column pulled and packed in house with Reprosil C18-AQ 3µm packing material. For quantification, mass chromatograms corresponding to expected masses of 1+ and 2+ ions [2+ and 3+ ions for H3(27–40) peptides] were analyzed using Xcalibur software (Thermo Electron Corp., Waltham, MA, USA). Mass accuracy was set to 20ppm. Peak areas were calculated and corrected for different mass spectrometric responses as determined by synthetic peptide standards. The total abundance of any modification was calculated from the sum of all corrected peak areas of differentially modified fragments covering a particular amino acid sequence.
Cell cycle synchronization
Cells were synchronized by growing IMR90 HDFs to 50% confluence in 15cm plates and incubated overnight with 2mM thymidine (Sigma). The cells were washed three times with PBS at 37°C, and fresh medium at 37°C was added. After 9–10 hr in the incubator, 1 µg ml−1 aphidicolin (AG Scientific) was added for 12hrs. Finally, the cells were washed three times with PBS at 37°C, and fresh media were added.
Cell Cycle Immunoblotting
Following release from aphidicolin block cells were harvested every 2 hr by trypsin treatment, washed in DPBS and lysed in NuPage LDS sample buffer (Invitrogen) at 104 cells per µl. Proteins were homogenized using benzonase (Novagen), denatured for 10 minutes at 68°C and resolved by SDS-PAGE, transferred to nitrocellulose, blocked in 2–5% (w/v) BSA and 0.1% (v/v) Tween20 for 20 minutes and probed. As secondary antibodies, HPR-linked anti-rabbit or anti-mouse (Amersham) was used, and the HPR signal was visualized with Supersignal West Pico Sensitivity substrate (Pierce). Abundance of post-translational modifications was calculated relative to the total amount of that particular protein. For other proteins the enrichment was calculated relative to γ-tubulin. All comparative western images shown were processed in parallel and membranes were exposed on the same film for equal time.
Synchronized Chromatin Immunoprecipitation
After release, cells were harvested every 2 hrs, washed with PBS, and fixed with 1% (v/v) formaldehyde in PBS for 30 min at RT. Chromatin was sonicated to an average fragment size of 500bp and precipitations were performed as described 55. DNA was then used in dot-blot southern hybridization with TTAGGG and 17P (sub-telomeric) probes. For the quantifications, the enrichment of histone modifications was calculated as a percentage of total H3 signal. For other proteins, the enrichment was calculated as a percentage of input signal.
BrdU DNA Immunoprecipitation
IMR90 cells were synchronized as described. Cells were incubated with 50 µM Br-dUTP (Sigma) for 1 hr prior to harvesting by trypsin treatment. Immunoprecipitation of BrdU labeled DNA was performed according to protocols outlined in 56. DNA was purified by Phenol-Chloroform extraction and used for dot-blot southern hybridization with TTAGGG and 17P probes.
Antibodies
See supplemental Table 1.
Figures
Figures were generated using Adobe Photoshop and Adobe Illustrator.
Supplementary Material
Acknowledgements
We thank Genevieve Almouzni (Institute Curie, Paris) for sharing Asf1a/b antibodies and helpful discussions. We are grateful to T. Jenuwein (Max Planck Institute, Freiburg, Germany)for the gift of the H3K9, H3K27 and H4K20 series of antibodies, Tony Hunter (The Salk Institute, USA) for Cyclin antibodies, Titia de Lange (The Rockefeller University, USA) for advice on chronic damage protocols, Jake Jaffe (Broad Institute, USA) and Steve Carr (Broad Institute, USA) for access to the Broad Proteomics Platform, expert advice and helpful discussion. We also thank David Chambers (The Salk Institute, USA) and Jonna Barrie (The Salk Institute, USA) for technical assistance with flow cytometry. We are grateful to members of the Karlseder Lab for critical discussion of the manuscript. ROS is supported by the George E. Hewitt Foundation for Medical Research, SK is supported by a postdoctoral fellowship of the Ernst Schering Research Foundation and the European Union, and JK acknowledges support by the NIH (RO1 GM06525 and RO1 AG025837)
ROS designed and carried out experiments and wrote the manuscript, SK did the MS analysis, SLS provided advice and access to the MS facilities and JK designed experiments and wrote the manuscript.
References
- 1.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su C, et al. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. Embo J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groth A, et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3' end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 9.Whitfield ML, et al. Stem-loop binding protein, the protein that binds the 3' end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 11.Gerard A, et al. The replication kinase Cdc7-Dbf4 promotes the interaction of the p150 subunit of chromatin assembly factor 1 with proliferating cell nuclear antigen. EMBO Rep. 2006;7:817–823. doi: 10.1038/sj.embor.7400750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan G, Hernando E, Cordon-Cardo C. Separation of live cells in different phases of the cell cycle for gene expression analysis. Cytometry. 2002;49:170–175. doi: 10.1002/cyto.10173. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein S. Replicative senescence: the human fibroblast comes of age. Science. 1990;249:1129–1133. doi: 10.1126/science.2204114. [DOI] [PubMed] [Google Scholar]
- 18.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 19.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 21.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 22.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi R, et al. Transcriptional regulation of human chromatin assembly factor ASF1. DNA Cell Biol. 2007;26:91–99. doi: 10.1089/dna.2006.0515. [DOI] [PubMed] [Google Scholar]
- 25.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 26.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 28.Stracker TH, Couto SS, Cordon-Cardo C, Matos T, Petrini JH. Chk2 suppresses the oncogenic potential of DNA replication-associated DNA damage. Mol Cell. 2008;31:21–32. doi: 10.1016/j.molcel.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart SA, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci U S A. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 32.Grunstein M. Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 33.Bianchi A, Shore D. Early replication of short telomeres in budding yeast. Cell. 2007;128:1051–1062. doi: 10.1016/j.cell.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 34.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 35.Steger DJ, Workman JL. Transcriptional analysis of purified histone acetyltransferase complexes. Methods. 1999;19:410–416. doi: 10.1006/meth.1999.0877. [DOI] [PubMed] [Google Scholar]
- 36.Harris ME, et al. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominski Z, Zheng LX, Sanchez R, Marzluff WF. Stem-loop binding protein facilitates 3'-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 39.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 46.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 47.Nishioka K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 48.Altaf M, et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.d'Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 50.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 51.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 52.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 54.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 56.Azuara V. Profiling of DNA replication timing in unsynchronized cell populations. Nat Protoc. 2006;1:2171–2177. doi: 10.1038/nprot.2006.353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.