Abstract
Recent efforts in systematically profiling embryonic stem (ES) cells have yielded a wealth of high-throughput data. Complementarily, emerging databases and computational tools facilitate ES cell studies and further pave the way toward the in silico reconstruction of regulatory networks encompassing multiple molecular layers. Here, we briefly survey databases, algorithms, and software tools used to organize and analyze high-throughput experimental data collected to study mammalian cellular systems with a focus on ES cells. The vision of using heterogeneous data to reconstruct a complete multilayered ES cell regulatory network is discussed. This review also provides an accompanying manually extracted dataset of different types of regulatory interactions from low-throughput experimental ES cell studies available at http://amp.pharm.mssm.edu/iscmid/literature.
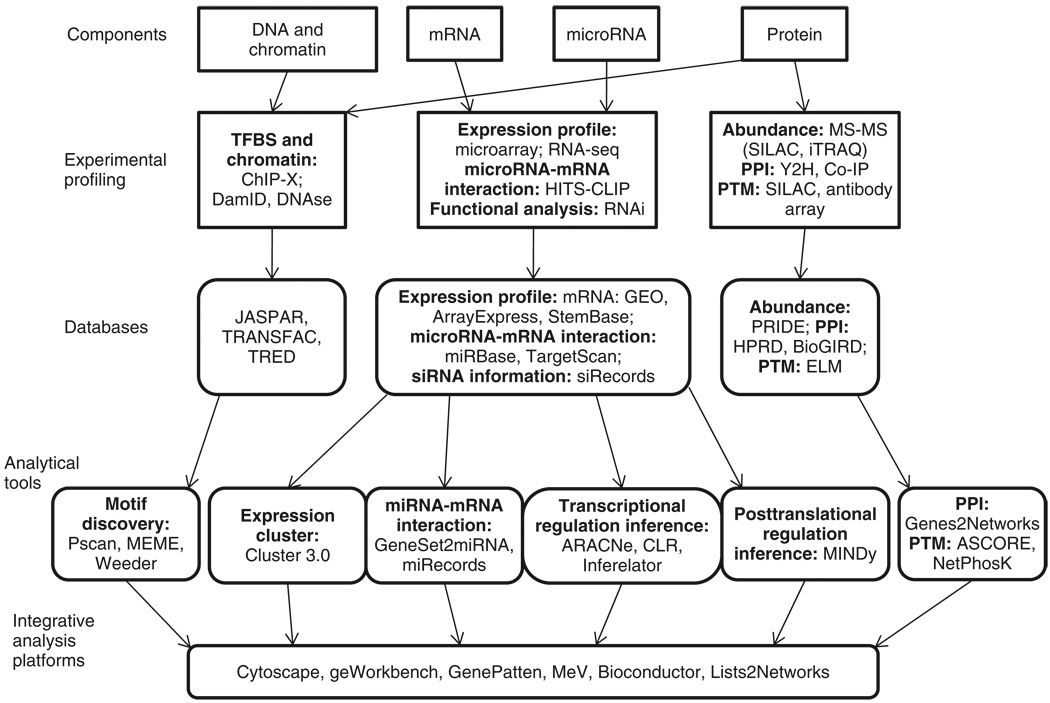
Pluripotent embryonic stem (ES) cells are derived from the inner cell mass of a developing embryo and can be cultured indefinitely in vitro. In vivo, mouse ES cells can contribute to all adult cell populations, including the germ line. Under defined in vitro conditions both mouse and human ES cells can differentiate into numerous mammalian cell types providing great promise for regenerative medicine. Recent studies have shown that adult mouse and human cells can be ‘reprogrammed’ into an induced pluripotent stem (iPS) cell state using simple combinations of transcription factors. In order to harness the exciting biomedical potential of ES/iPS cells, the molecular regulatory networks responsible for controlling pluripotency/self-renewal as well as, commitment and differentiation into different lineages, need to be characterized. Stem cell research is increasingly employing high-throughput systems biology approaches to define molecular ‘parts lists’ and regulatory interactions between the parts in ES cells and in their more differentiated progeny. How these parts are interconnected into gene and cell signaling regulatory networks ultimately responsible for self-renewal and differentiation is unclear. Approaches aimed to bridge the gap among molecules, network architectures, and dynamics in order to ultimately ‘explain’ phenotypic behavior are in their infancy. To enable these efforts, a pipeline process that couples experimental and computational approaches has emerged. An example of such a pipeline is outlined in Figure 1. First, data are collected from different molecular regulatory layers [for example: epigenomic, messenger RNA (mRNA), and proteomic data] using emerging high-throughput technologies. Second, in order to extract biological knowledge out of such rich, complex but often, noisy experimental datasets, advanced computational tools and databases are being developed. Moreover, computational methods capable of synthesizing data from numerous experimental platforms with user-friendly interactive interfaces are gradually emerging. The computational methods include tools that convert raw data into standardized database formats/records. Such data records are organized into databases where experiments from different sources can be merged. Algorithms are then used to query such databases and integrate the high-throughput data with annotated data collated from low-throughput studies and other high-throughput studies in order to obtain new biological insights. Here, the organization of experimental data into sets of biochemically related gene products and ultimately interacting gene-product networks is extremely useful. The abstraction/simplification of data into gene-sets and networks is qualitative and as such typically ignores quantitative detail. However, it provides a birds-eye-view of the system as a whole when advanced algorithms are applied to dissect the complexity and rank components. Taken together, computational tools and the algorithms embedded within them are used to make predictions that are translated into rational hypotheses that can be validated using low-throughput functional experiments. Although results from high-throughput experiments provide a global view of the many variables involved and their relationships, current technologies lack accuracy and direct functional perspective. In contrast, low-throughput techniques, while providing functional understanding of specific components and interactions, do not have the scope needed to understand the multi-factorial behavioral complexity of the system’s behavior as a whole.
FIGURE 1.
Pipeline process for systematic studies of ES cells starting with experimental methods to characterize the state of the cell at different regulatory layers. Then, data from such experiments are stored in public repositories for data consolidation and reuse. Such databases are analyzed by algorithms implemented in software tools that can make predictions and build networks from the data. Different types of networks from the different regulatory layers can be combined, whereas lists of genes/proteins can be probed for functional enrichment using integrated computational platforms.
ES cell research is an area that fits well with the systems biology pipeline process because these cells are relatively easy to handle experimentally, have defined sets of phenotypes that can be experimentally evaluated, and are relatively homogenous in gene expression and morphology (this latter point is discussed in more detail in a subsequent section). The fact that we can differentiate stem cells to different cell types also makes these cells ideal. Stem cells can be considered the ‘ideal model organism’. In this review, we first discuss the systems biology pipeline process by surveying different types of high-throughput experiments with an emphasis on the computational tools and databases associated with each specific type of experimental approach. We then describe how these methodologies have been applied so far to study ES cells at different molecular regulatory layers, whether these layers are epigenetic, transcriptional, mRNA, microRNA, proteomic, and others. We focus on data-mining approaches which include algorithms, software tools, and databases. Such methods are used to reconstruct in silico regulatory networks and develop hypotheses for further experimentation. Finally, we present an initial ES cell regulatory network constructed from low-throughput studies.
mRNA EXPRESSION
Experimental approaches
Systematic genomic approaches for profiling the state of the cell at the mRNA expression level are the most established high-throughput approach for studying ES cells. At early stages of technology development, expressed sequence tags (EST),1–3 serial analysis of gene expression (SAGE),4,5 and massively parallel signature sequencing (MPSS)6,7 have been utilized for probing mRNA expression patterns in ES cells. For example, through global comparisons of mouse versus human ES cells, Wei et al.7 showed similarities as well as discrepancies between the transcriptomes of human and mouse ES cells. Currently, the most established method to globally profile mRNA levels on a genome-wide scale is via microarray technology. Although microarray technology has matured and is now widely used, it is expected to be gradually replaced by next generation or deep, sequencing techniques8 which can be used to profile the transcriptome with greater accuracy and at lower cost. Sequencing-based profiling is capable of providing more accurate absolute gene expression values, at least at the cell population level. Simply ‘counting’ the numbers of sequence tags derived from individual genomic loci is also more statistically robust. Although powerful, the ability of deep sequencing to detect extremely low levels of gene-specific transcripts raises its own set of potential complications. For example, running a single lane using the Illumina Solexa or similar technologies can yield up to tens of gigabases of sequence information in segments of nucleotides. Collecting such large volumes of data can run into an ‘embarrassment of riches’ situation, where transcripts present at less than a single copy per cell are identified. Whether such low abundance transcripts are biologically relevant and reflective of functionally important heterogeneity of the analyzed cell population, or whether they are a product of transcriptional (or other) ‘noise’, needs to be carefully considered.
Databases
In recent years, public mRNA expression repositories have been developed for the purpose of sharing and exchanging results from mRNA microarray studies. Gene Expression Omnibus (GEO)9 (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress10 (http://www.ebi.ac.uk/microarray-as/ae/) are two leading resources. Although these are general repositories for any type of organism and cell type, more focused repositories exist. For example, Gene Expression Database (GXD) (http://www.informatics.jax.org/)11 is a community specific resource for mRNA expression in mouse; BloodExpress (http://hscl.cimr.cam.ac.uk/bloodexpress/) is a repository for the mouse hematopoesis system,12 Stem Cell Database (SCDB) (http://stemcell.mssm.edu/v2/) and StemBase (http://www.stembase.ca/?path=/) are mRNA expression databases collecting data from different types of stem cells.13 In addition, Stromal Cell Data Base (StroCDB)14 (http://stromalcell.mssm.edu/) contains gene expression data from stromal cell lines with differing hematopoietic stem cell supporting activities. Also, the FunGenES (Functional Genomics in Embryonic Stem Cells) database (http://www.fungenes.org/index.html) includes expression profiles in mouse ES cells with several analytical tools. Data exchange is typically handled through XML and OWL exchange standards such as MIAME.15
Analysis
Analysis of gene expression data remains an active area of research because the extraction of meaningful biological knowledge out of large volumes of noisy data is challenging. Different algorithms have been employed for identifying expression patterns. Unsupervised clustering methods such as hierarchical clustering,16 principle component analysis,2 and self-organizing maps17 are some of the most popular approaches. The rationale behind these different algorithms is that groups of genes that behave similarly at their mRNA expression levels are co-regulated and are therefore likely to encode components of the same pathway or biological process. Once identified, these clusters or patterns can be linked to prior biological knowledge. Specific tools to perform clustering analyses include TIGR MeV,18 Genes@Work,19 Cluster 3.0,20 BioDiscovery,21 EXPANDER 2.0,22 and the BioConductor project which implements modules using the freely available R statistical package.23
Applications to study ES cells
Gene expression microarray technology has been widely used for profiling the expression level of the mRNA of genes that control self-renewal and differentiation in ES cells. For example, Bhattacharya et al.24 analyzed expression patterns of six human ES cell lines by oligonucleotide arrays and identified an overrepresented subset of over 90 genes as ES cell signature genes, which include Nanog, Sox2, and Oct4. Similarly, Ivanova et al.25 performed gene expression analysis in mouse embryonic and neural stem cells and defined ESC- and NSC-enriched gene-sets; Ramalho-Santos et al.26 defined from transcriptional profiles 216 genes enriched in mouse embryonic, neural, and hematopoietic stem cells. Consistent with such molecular signatures of ES cell genes, Masui et al.27 used mRNA expression microarrays to screen genes that are up- or down-regulated in a Sox2-null ES cell line. They found multiple genes in the down-regulated set that are related to Oct4, pointing to the fact that Sox2 and Oct4 may function in a coordinated manner. In another study, Pritsker et al.25 conducted gain-of-function screens combined with expression microarrays to identify genes whose over-expression causes the maintenance of the undifferentiated status of mES cells cultured without leukemia inhibitory factor (LIF), an extracellular ligand sufficient for maintenance of mES cells. They found a role for genes encoding a wide scope of regulatory proteins including the gene that encodes the kinase Akt1 and several novel transcription factors. Sperger et al.28 compared expression profiles of human ES cells and several carcinoma cells. They identified sets of mRNAs that are specific for human ES cells. These mRNAs showed relative similarity of expression patterns among different ES cell lines. Westfall et al.29 conducted microarray analysis for mRNA expression in human ES cells under either 4% or 20% oxygen cell culture conditions. They identified several oxygen-sensitive genes required for the maintenance of self-renewal. Their global gene expression profiling experiments characterized the mRNA signature of ES cells during specific developmental stages and environmental conditions. One of the best characterizations of human ES cells can be found in Adewumi et al.30 In their study, they combined FACS analysis of protein surface expression with mRNA profiling.
Various other studies have reported gene expression profiling of undifferentiated human and mouse ES cells as well as their derivatives (see Refs 31–37 and one of many reviews38). These combined efforts ultimately define specific molecular signatures of ES cells and bring us closer to unraveling the architecture of ES cell regulatory networks at the transcriptional regulatory layer.
HIGH-THROUGHPUT KNOCK-DOWN STUDIES
Experiments
Complementing mRNA gene expression profiling, genome-wide knock-down screens are increasingly applied for identifying novel functional genes linked to specific phenotypes or a specific biological process. The primary methodology utilizes short interfering RNA (siRNA) screen to target specific mRNAs for degradation. Commercially available chemically synthesized siRNAs are 21 nucleotides long with symmetric 3′-overhangs of two nucleotides. Alternatively, short hairpin RNAs (shRNA) can be transcribed from expression cassettes inserted into plasmids or viral vectors.39 Another option is to use endoribonuclease-prepared short interfering esiRNAs.40 These are produced in vitro from cDNA templates transcribed into double-stranded RNA and subsequently digested by endoribonucleases to a pool of overlapping effectors. It has been reported that this approach results in less off-target effects. Robotics is often used to streamline the knock-down process and to measure changes in phenotypic outcomes using microscopy.
Databases and resources for siRNA selection
siRNA sequences validated by experiments are available through several public databases. For example, the siRecords41 is a database collecting over 17,000 validated siRNAs. Another useful resource is siRNAdb,42 which provides both siRNA experimental data as well as computationally predicted siRNAs sequences. siRNAdb provides users with the ability to evaluate siRNAs for potential specific and non-specific targets. In addition, commercially available repositories include validated siRNAs from Qiagen, and Silencer-validated siRNAs from Ambion. Several public or commercial bioinformatics prediction tools have been developed to assist in selecting siRNAs with enhanced hit likelihood and reduced potential for off-target effects. Representative computational tools include siDESIGN43 and siSearch.44 The algorithms used in these tools are based on empirical rules such as sequence asymmetry, stability, and predicted secondary structure, often implementing machine-learning techniques.
Applications to study ES cells
Several mid-scale to large-scale siRNA screens allowed stem cell researchers to systematically identify novel genes essential for ES cell pluripotency/self-renewal and differentiation. Such knockdown approaches are generally complimented with mRNA expression changes to link the knocked-down-gene with the genes it regulates. For example, Ivanova et al.36 explored the effect of shRNA-mediated silencing of seven different transcription factors on mRNA expression profiles in mouse ES cells. They identified four novel genes (Esrrb, Tbx3, Tcl1, and Dppa4) previously unreported as being essential for maintenance of ES cell self-renewal and pluripotency. Based on the expression data and further clustering analysis, Ivanova et al. defined the similarities and differences among various effects of these factors. Mid-level screens used an esiRNA approach to screen 1008 chromatin regulators. This study identified Tip60-p400 as a key regulator of ES cell identity.45 In a separate study, Schaniel et al.46 performed a synthetic RNA-based shRNA screen on 312 genes identifying several SWI/SNF chromatin remodeling complex components such as Smarcc1/Baf155 to be important for early commitment/differentiation events. Recently, two groups performed larger-scale RNAi screens in mouse ES cells to identify genes critical for stem cell self-renewal. Hu et al.47 used synthesized siRNA pools and identified more than 100 novel functional genes implicated in mouse ES cell maintenance. Another study by Ding et al.48 employed esiRNA library and found over 200 candidate genes important for ES cell self-renewal. These studies illustrate the power of genome-wide RNA interference to functionally and systematically identify the part-list components important for ES cell self-renewal.
microRNA REGULATION OF mRNA TRANSLATION AND EXPRESSION
Experiments
MicroRNAs (miRNAs) are small, approximately 22 nucleotide long non-coding endogenous RNAs which play an important role in many biological systems including the regulation of ES cells.49 The binding of the approximately 22 nucleotide long mature miRNAs to mRNAs in the RNA-induced silencing complex (RISC) triggers either degradation of mRNAs or inhibition of translation. miRNA expression and function has been observed to be critical in ES cell regulation.50 The experimental approaches for detecting miRNA expression are by miRNA array profiling, northern blots, and quantitative real-time PCR. Such empirical methods are complimented by computational algorithms that can discover miRNAs as well as predict their targets.51
Databases
Some of the primary online resources for miRNA sequences, targets, and other annotations are miRBase,52 Miranda (microRNA.org), and TargetScan.53 CoGemiR54 and TarBase55 are examples of two other emerging microRNA databases. RNAdb56 is a database for all non-protein-coding RNAs including microRNAs and small nucleolar RNAs (snoRNAs), the latter of which are participants in the process of ribosomal RNA modification and maturation.57
Tools to predict microRNA targets
Several stand-alone miRNA–target prediction tools, such as TargetScanS,53 PicTar,58 and miRanda,59 have been developed. These tools implement algorithms that employ observed base-pair rules summarized into principles extracted from known miRNA–target interactions. In addition, cross-species conservation of miRNA–target interactions is used for miRNA–target interaction prediction. Different methods use slightly different scoring schemas, detection criteria, and conservation requirements. For example, TargetScanS requires perfect complement with a miRNA seed, whereas DIANA-MicroT60 allows for targets with imperfect seed matching. Recently published tools implemented machine-learning techniques to make predictions directly based on validated miRNA targets, i.e., MirTarget261 and NBmirTar.62 Apart from sequence matching, algorithms are developed to consider secondary structure, one example is PITA63; whereas EIMMo predicts miRNA targets using evolutionary sequence conservation across different organisms combined with information about molecular and biochemical pathways.64 Such tools can be evaluated using mRNA expression data. Complementarily, several resources integrate various stand-alone databases and tools for better prediction and comprehensiveness. For example, miRecords65 comprise both manually retrieved experimentally validated miRNA–target interactions and miRNA–target interaction predictions integrated from 11 stand-alone prediction tools. Also, lists of miRNA targets or clusters of miRNAs working as a group can be predicted using enrichment statistics. For example, GeneSet2microRNA66 implements such an approach. In addition to the miRNA–target prediction tools and databases, analyses of functional annotation of predicted targets for specific miRNAs are emerging. Such analyses link miRNA–targets to gene ontology (GO) terms or to cell signaling pathways.67 These analyses are best utilized when miRNA data are integrated with mRNA expression data.68 A list of miRNA prediction tools and databases is provided in Table 1.
TABLE 1.
microRNA-Related Tools and Databases
Applications to study ES cells
miRNAs are regulators of ES cell pluripotency/self-renewal and differentiation.80–82 It was shown, for example, that Dicer-deficient ES cells are defective in proliferation and differentiation.83 Expression profiles of miRNAs in various ES and derived cell lines already revealed unique signatures in these cells. For example, Thomson et al.84 performed custom microarray-based analysis of miRNAs expression in mouse ES cells, embryoid bodies, and adult tissues. Based on their results, the expression profiles of miRNAs in ES cells are much different than in embryoid bodies or adult cells. Babiarz et al.82 identified novel Dicer-dependent non-canonical microRNAs in mouse ES cells by deep sequencing of knockout cell lines; Wu et al.85 applied genomic analysis of miRNA profiling and revealed differences between two human ES cell lines, which in turn, explains subtype-specific differentiation bias; whereas a miRNA expression microarray by Cao et al.86 compared miRNA expression between human and mouse ES cells identifying conserved expression from chromosomes 19 and X. In several other seminal studies, miRNA regulation was linked to key pluripotency transcription factors. For example, Boyer et al.87 showed that Nanog, Oct4, and Sox2 occupy the promoters of a combined 14 miRNAs, only 2 of which are bound by all 3 in hESC, suggesting that regulation of miRNAs by these core pluripotency factors is crucial to maintain the pluripotent state. Card et al.88 showed that Oct4/Sox2 regulate miR-302 which targets the mRNA encoding cyclin D in human ES cells, whereas a study reported by Barroso-delJesus et al.89 suggests that the miR-302–367 cluster is regulated by Nanog, Oct4, Sox2, and Rex1 (all self-renewal transcription factors). Xu et al.90 showed that Mir-145 targets and represses pluripotency transcription factors, whereas Tay et al.91 demonstrated that miRNA-134, miR-296, and miR-470, which are induced upon retinoic acid mediated differentiation, target the pluripotency transcription factors Nanog, Oct4, and Sox2.
One application of in silico prediction of miRNA–target interactions specific for ES cells attempted to identify miRNA–mRNA interactions essential for pluripotency and differentiation.92 The authors combined mRNA expression with predicted miRNAs to suggest a list of miRNAs that are important for maintaining pluripotency. Although computational approaches can be used to predict novel miRNA candidates important for ES cell regulation, experimental approaches are necessary to confirm such predictions. For example, Ciaudo et al.93 performed both computational prediction for miR-302 targets using two target-predicting tools: PicTar58 and EIMMo, coupled with experimental validation. They confirmed that Arid4a and Arid4b are targeted by the miR-302 family, which is enriched in male-specific differentiating ES cells. In a study that utilized a miRNA over-expression strategy in Dgcr8−/− ES cells, the authors identified the miR-290 cluster as being capable of rescuing the proliferation defect in these cells.82 Experimental approaches of direct miRNA–mRNA target identification include co-immunoprecipitation of the RISC with target mRNAs.94 This procedure was recently enhanced by applying the cross-linked immunoprecipitation (CLIP) approach for identifying RISC binding sites more precisely.95,96 By expanding the analyses, miRNA–target interactions can be elaborated into regulatory networks that can be combined with other types of regulatory interactions. Specifically, miRNAs can be represented as nodes with outgoing (mostly) negative links to their target mRNAs, although there are exceptions.97 MicroRNA expression can be used to explain discrepancies between mRNA expression and protein levels, and because miRNAs are regulated by the transcriptional machinery, the input links to miRNA nodes originate from the transcriptional regulatory machinery which is discussed next.
GENE REGULATION BY TRANSCRIPTION FACTORS AND CO-REGULATOR COMPLEXES
Experiments
Transcription, the initial step in gene expression, is regulated by the transcriptional machinery involving transcription factors and co-regulatory complexes. Transcription factors bind to cis-regulatory elements in proximity to gene coding sequences. Uncovering the dynamics and complexity of transcriptional regulation through transcriptional-factor binding to DNA remains an enormous challenge. Large-scale experimental methods can be applied to profile the global interactions of transcription factors with DNA. Such methods include chromatin immunoprecipitation (ChIP) combined with DNA microarrays, ChIP-chip,98 or deep sequencing, ChIP-seq,99 or ChIP-PET. 100 A recently developed alternative method, called DamID,101 is based on the expression of a fusion protein consisting of the protein of interest and DNA adenine methyltransferase (Dam). Methylation of adenines by Dam near the protein–DNA interacting sites marks the sites of interest. DamID is applied to eukaryotic cells where adenine methylation does not happen endogenously. DamID relies on selective PCR amplification via adapter oligonucleotides ligated to DNA fragments sequentially digested with DpnI (GAmeTC) and DpnII (GATC) followed bymicroarray hybridization. In addition, protein/DNA arrays such as those offered by Panomics can be used to measure the activity for canonical transcription factors.102 Protein–DNA interaction profiling using large-scale methods has been widely used by the stem cell research community when compared with other similar experimental systems and cell types.47,87,102,103,103–111
Databases
Facilitated by high-throughput protein/DNA interaction studies as well as results from low-throughput studies such as gel-shift assays, tools and databases are being developed for organizing transcription factor/DNA interaction information. Leading transcription-factor-binding-site-resources include JASPAR,112 TRANSFAC,113 and TRED.114 These databases contain collections of transcription factor binding profiles together with information on conserved regulatory elements stored in binding site matrices. Other, organism-specific databases are YEASTRACT115 for Saccharomyces cerevisiae, RegulonDB116 for Escherichia coli, and DBTBS117 for Bacillus subtilis. Recently, MacArthur et al.118 collected results from various ChIP experiments performed using ES cells and created a consolidated ChIP profiling dataset for over 20 transcription factors and their putative targets. Such a dataset can be used as part of ongoing efforts of data consolidation for hypotheses generation and multi-layered regulatory network reconstruction.
Tools for transcription factor binding site prediction
One approach to summarize the results from protein/DNA interactions is to develop consensus binding site sequences for individual transcription factors.119,120 Leading databases that provide such consensus binding site sequences as matrices, or logo-motifs, for mammalian cells are JASPAR112 and TRANSFAC.113 Once such matrices have been developed, it is straightforward to use these to map potential transcription factor binding sites across entire genomes. Representative tools that use such data include: PASTAA,121 P-Match,122 and Pscan.123 The complete list ismuch longer. PASTAA implements the TRAP124 method utilizing TRANSFAC to predict the affinity of transcription factors to gene promoters. Because such computational predictions have been proven to not be highly reliable, combining predictions with additional experimental data, such as data from ChIP and mRNA expression experiments, significantly improves predictability performance.
Applications to study ES cells
As mentioned above, high-throughput ChIP experiments have been widely applied to studying transcription-factor-targets in ES cells.47,87,102,103,103–111 For example, Hu et al.47 performed a genome-wide siRNA screen combined with ChIP-chip to elucidate the regulatory networks controlled by the two transcription factors Cnot3 and Trim28, discovering a new role for these factors in ES cell pluripotency/self-renewal; Marson et al.125 used ChIP-seq technology to link important self-renewal transcription factors to promoters of genes and miRNAs. They discovered that miRNAs are highly regulated by self-renewal transcription factors, and are highly expressed in ES cells, while specific groups of miRNAs are only expressed in differentiated cells. Applying ChIP-PET to profile protein/DNA interactions for several transcription factors, Loh et al.104 used an optimized algorithm named NestedMICA126 to predict the motif binding sites for transcription factor pairs such as Oct4/Sox2. This analysis helped to further characterize how heterodimers regulate mouse ES cell self-renewal. Their findings were consistent with two other studies that reported common Sox2/Oct4 heterodimer binding sites upstream of many genes important for self-renewal or differentiation.105,127 In another study, Sharov et al.128 applied ChIP experiments with pull-downs of Oct4, Sox2, and Nanog combined with time-course microarray analyses. They used an inducible Oct4-depletion ES cell line called ZHBTc4 and a Sox2 line called 2TS22C. Combining ChIP experiments with mRNA expression can be used to assign signs (activation or inhibition) to links from transcription factors to the genes they regulate.
SIGNALING THROUGH PROTEIN PHOSPHORYLATION
Experiments
Protein phosphorylation is a critical post-translational modification used to transfer information from the extracellular environment by affecting protein activity. Phosphorylation is used to regulate various biological processes including pluripotency/self-renewal as well as differentiation of ES cells. Classical experiments to identify phosphorylated sites include radioactive labeling and affinity chromatography. These methods are labor intensive and can only be performed on a small scale. Recently, large-scale phosphorylation data (phospho-proteomics) was made possible with approaches such as tandem mass spectrometry (MS) and antibody phosphoarrays.129,130 For example, a popular strategy in recent years is using stable isotope labeling by amino acids in cell culture (SILAC)131,132 combined with liquid-chromatography and mass spectrometry (LC)-MS/MS. With SILAC, the whole proteome of a given cell is labeled with stable heavy and light (normal) isotope variants. In this approach, the relative levels of protein phosphorylation from different samples are measured simultaneously by the ratio of intensities of light/heavy amino acids labeled with the distinct isotopes. Although SILAC quantitation is done at the MS level, an alternative method called iTRAQ uses stable isotope labeling at the tag-level. This allows multiplexing of up to eight samples, and as such, quantitation is done at the MS/MS level.133,134 An alternative to MS-based proteomics is antibody arrays. For example, Kinexus is a specialized method for analyzing the phosphoproteome using antibody-based microarrays (Kinex™ antibody microarrays), kinase substrate, and inhibitor profiling. Kinex is an antibody-based method that relies on sodium dodecyl sulfate (SDS)-polyacrylamide mini-gel electrophoresis and multilane immunoblotters to permit the specific and quantitative simultaneous detection of protein kinases or other signal transduction proteins.135
Databases
Protein phosphorylation-centered databases mostly report phopho-sites identified using techniques such as SILAC, whereas some databases also list the kinases that are likely responsible for the phosphorylation. Hence, there are many orphan phospho-sites, i.e., the kinases that are responsible for the phosphorylation of most sites are not known. Phosphorylation-centered databases can be developed manually by retrieving identified phosphorylations from results reported in low-throughput studies, as well as by consolidating results from high-throughput assays. Such phosphorylation repositories include Swiss-Prot,136 phospho.ELM,137,138 HPRD,139 PhosphoPoint,140 and PhosphoSitePlus.141 A comprehensive list of phosphorylation-centered databases is provided in Table 2.
TABLE 2.
Phosphorylation Sites and Kinase–Substrate Tools and Databases
Kinase–substrate identification and prediction tools
For data generated from SILAC phospho-proteomics, there are computational tools developed for data processing and analysis. For example, ASCORE189 and Colander190 were developed to process the raw data for phospho-site identification. Databases that record phospho-sites and predictive tools that utilize such data as training sets to predict additional potential phospho-sites are emerging. For instance, NetPhos,152 DISPHOS,154 NetPhosYeast,155 and PHOSIDA142 are such databases. Furthermore, tools to predict the protein kinases that catalyze phosphorylation events on specific peptide motifs are also being developed. Predictors such as NetPhosK,167 PhoScan,172 and NetworKIN197 attempt to link phosphorylation sites to the kinases most likely to phosphorylate those sites. Some tools, such as kinase enrichment analysis (KEA),186 compute the likelihood of specific kinases to phosphorylate a set of proteins based on annotated kinase–substrate interactions. Most predictors are sequence-based and depend on an assortment of machine-learning algorithms. As such, these algorithms require training data from known examples. This category consists of NetPhosK,167 KinasePhos,171 Phosite,158 GPS,169 ScanSite,166 and PhoScan.172 Alternatively, some additional information apart from sequence has also been integrated to augment specificity of substrates, including disorder information implemented by DISPHOS,154 structure information implemented by NetPhos152 and Predikin,165 as well as contextual factors by NetworKIN.197 Different machine-learning algorithms are implemented for kinase–substrate prediction using different tools. For example, support vector machine (SVM) is implemented by PHOSIDA142 and PredPhospho,151 artificial neural networks (ANN) by NetPhos,152 and hidden Markov model (HMM) by KinasePhos.171 In addition, DISPHOS relies on a logistic regression-based linear predictor, whereas GPS169 uses a group-based scoring method. The same group that developed GPS also developed an additional strategy called PPSP170 which implements Bayesian Decisions. Integrating and comparing such complementary efforts is expected to improve predictive specificity and accuracy.
Application to study ES cells
Characterization of the phosphoproteome status of ES cells can provide understanding of the cellular signaling status at the pluripotency/self-renewal state, and how external stimuli drive ES cell signaling toward differentiation. Because phosphorylation is a key mechanism for the regulation of the transcriptional machinery, phosphoproteomic experiments are expected to bridge the gap between the transcriptome and proteome. Recently, Wang et al.198 studied the phosphorylation status of 42 receptor tyrosine kinases (RTKs) in human ES cells under conditional medium simultaneously by means of membrane arrays with a pan-anti-phosphotyrosine antibody. RTKs such as IGF1 and insulin receptors contribute to hESC pluripotency/self-renewal. Such approaches aid to define the contributions of ligands to maintain stem cell characteristics in heterogeneous culture conditions. More systematically, Brill et al.199 performed an MDLC-MS/MS-based phosphoproteomic study in human ES cells and their differentiated derivatives to identify differentially modified proteins potentially involved in self-renewal or differentiation. Similarly, using SILAC, Prokhorova et al.132 studied the phosphoproteome status of undifferentiated human ES cells and identified 527 unique phosphopeptides; while Van Hoof et al.200 unraveled the phosphoproteome status of hES cells during differentiation induced by BMP. Coupled with the phosphoproteomic results, they utilized the NetworKIN algorithm197 for predicting upstream kinases for these phosphorylated substrates and identified CDK1/2 as an overrepresented kinase during early differentiation of hES cells. In another study, Saxe et al.201 used prediction algorithms implemented by ScanSite to find potential phosphorylation sites on Oct4, a key component in regulating self-renewal. Subsequently, based on the computational predictions, they performed experiments to confirm that one of the predicted phosphorylation sites partially controls Oct4-mediated transcriptional activity.
PROTEIN–PROTEIN INTERACTIONS AND CELL SIGNALING PATHWAYS
Experiments
To improve our understanding of the cell signaling and transcriptional complexes that regulate ES cells, further characterization of protein–protein interactions in high-throughput is essential. Protein interactions and cell signaling pathways curated from the literature complement high-throughput techniques, such as yeast-two-hybrid (Y2H)202 and MS following co-immunoprecipitation. Besides binary interactions protein level changes are also important. The multiple reaction monitoring-mass spectrometry (MRM/MS) assay quantifies a specific tryptic peptide that is selected as a stoichiometric representative of the cleaved protein against an internal synthetic stable isotope-labeled peptide, giving rise to the absolute measure of protein concentration.203,204
Databases
Leading resources for collecting and merging protein–protein interactions identified using different experimental techniques include BioGIRD,205 HPRD,206 MINT,207 IntAct,208 and Reactome.209 These protein interaction databases are often organized into cell signaling pathways which also include small-messengers such as DAG, cyclic AMP, and calcium, as well as non-covalent interactions and post-translational modifications such as phosphorylation and dephosphorylation. Some protein–protein interaction databases also include computationally predicted interactions.210 For example, the STRING211 database contains interactions predicted from co-localization and phylogenetic profiles. Data exchange and compatibility is handled through standardization of protein IDs and exchange formats such as BioPAX.
Application to study ES cells
The most extensive study to characterize protein–protein interactions in mouse ES cells used affinity purification followed by MS proteomics to unravel the protein interactions associated with Nanog, a key regular transcription factor for ES cell pluripotency/self-renewal. 212 Up-to-date, other protein interaction (interactome) studies in ES cells have been small-scale and focused on individual complexes.
ENRICHMENT ANALYSIS
Genome-wide experimental approaches such as mRNA microarrays, proteomics, and ChIP-chip/PET/seq generate large datasets that can be summarized as lists of genes/proteins that have been identified or as displaying changes in expression or activity under different conditions. Such lists can be analyzed using enrichment analysis methods which report the overlap between the experimentally identified lists and previously annotated functionally labeled gene-sets. Systematic annotation of such gene-sets assists researchers in measuring the similarity among different experiments and in identifying the functional signatures or biological themes in newly generated datasets. The most common enrichment analysis applications are based on GO,213 which is a hierarchical tree-structured database of controlled vocabulary terms associated with genes for gene annotation. GO is a useful way of collecting and organizing biological knowledge that can be reused for data analysis. Besides GO annotation, genes/proteins have been grouped into gene-sets using several types of prior biological knowledge, for example: chromosomal location, expression regulation by upstream transcription factors, binding to specific metabolites, shared structural domains, involvement in canonical biological pathways, and association with specific diseases.
Tools
Enrichment analysis tools such as DAVID214 and GSEA215 have been developed to handle many types of prior biological knowledge gene-sets. Apart from these two leading enrichment analysis tools, other approaches exist. GeneTrail216 is based on categories such as TRANSFAC for gene regulation, Refseq, KEGG, and GO; GFINDer217 also covers OMIM, which is for association with disease. The FatiGO tool218 allows users to analyze two sets of genes by means of statistical tests based on various criteria, including functional criteria [GO, Biocarta (http://www.biocarta.com/genes/index.asp)], regulatory criteria (miRNA), and chromosomal location. A list containing additional similar enrichment analysis tools is provided in Table 3. Based on the different algorithms used, enrichment tools can be categorized into three classes219: singular enrichment analysis (SEA); gene-set enrichment analysis (GSEA); and modular enrichment analysis (MEA). Common statistical methods such as Fisher Exact, Chi-Squared, and Binomial Proportion tests are used when comparing sets without considering the ranks of the genes within the set. Most tests assume independence for the probability of genes to appear together in an input list, generally a naïve assumption because genes that belong to the same functional category tend to be co-expressed. Tools such as GSEA also consider the ranks of the genes in the input list. Statistical tests, such as Kolmogorov–Smirnov or Rank-Sum, are used to compute the enrichment for such input lists against sets of genes from different categories in an arguably more accurate manner.
TABLE 3.
Gene-Set Enrichment Tools
Application to study ES cells
Enrichment analysis has been widely applied to interpret high-throughput profiling results from ES cells. For example, using DAVID, Storm et al.237 identified downstream gene-sets of phosphoinositide 3-kinase (PI3K) that show over-representation of functional groups in transcriptional regulation and DNA binding, including Zfp36, Sox4, Dnmt3a/b. This implicates the PI3K signaling pathway as playing an important role in maintaining ES cell pluripotency. Another example is the over-representation of functional groups involved in biological processes such as cellular growth, proliferation, and embryonic development for targets of c-Myc and Stat3, two key transcriptional factors governing ES cell pluripotency.108
Taken together, functional enrichment analyses can generate hypotheses about potential links between groups of miRNAs and transcription factors with the genes they regulate, and identifying molecular complexes that function in a coordinated manner during differentiation. Enrichment analyses can also help in computing the significance of signals from high-throughput experiments. For example, if enrichment analysis shows that many phospho-sites for a specific kinase are increasingly phosphorylated during the course of a particular treatment, it is likely that this kinase is more active under the treated versus control conditions. This allows making reasonable assumptions that the phospho-sites that are marginally identified to be real can be upgraded to greater reliability if they are predicted to be a substrate of the up-regulated kinase. Such concepts can be applied to similar scenarios where high-throughput screens can be combined with prior knowledge.
NETWORK RECONSTRUCTION
Construction of networks from literature
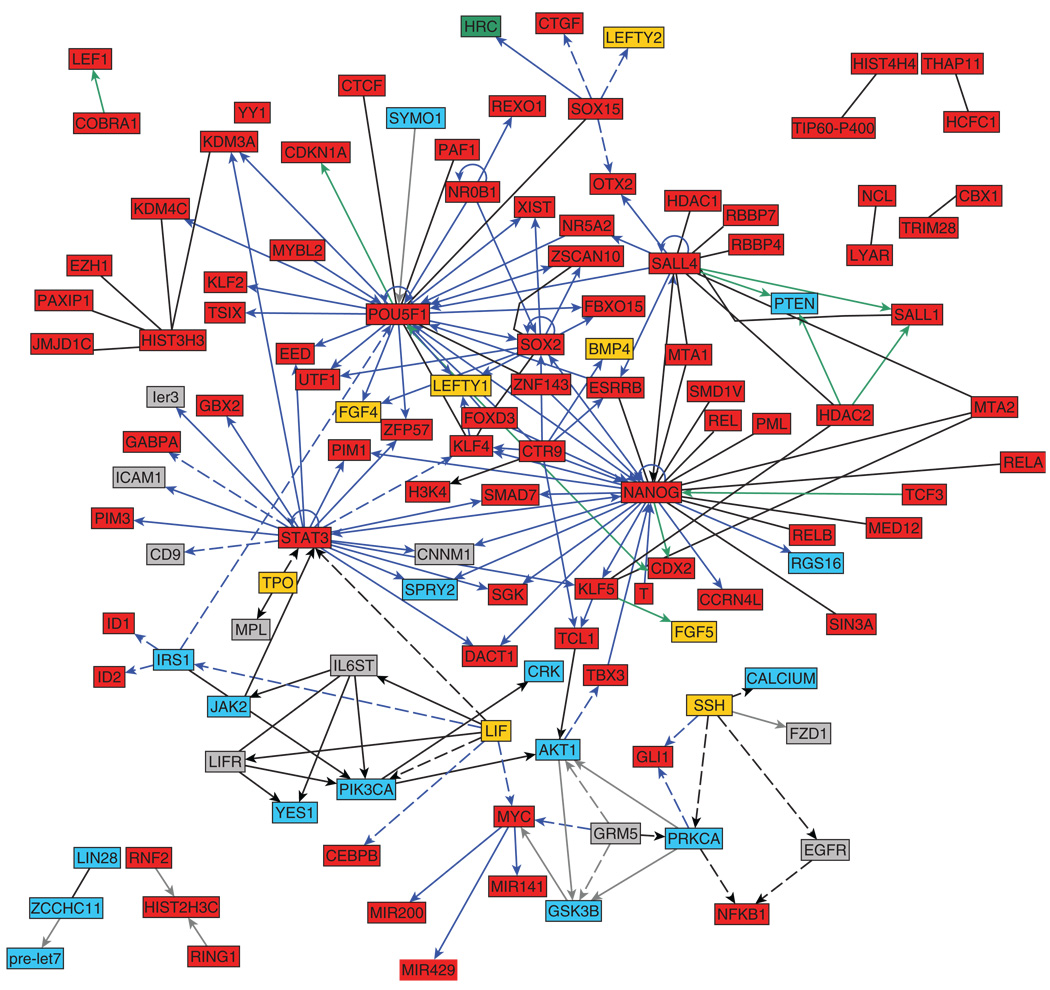
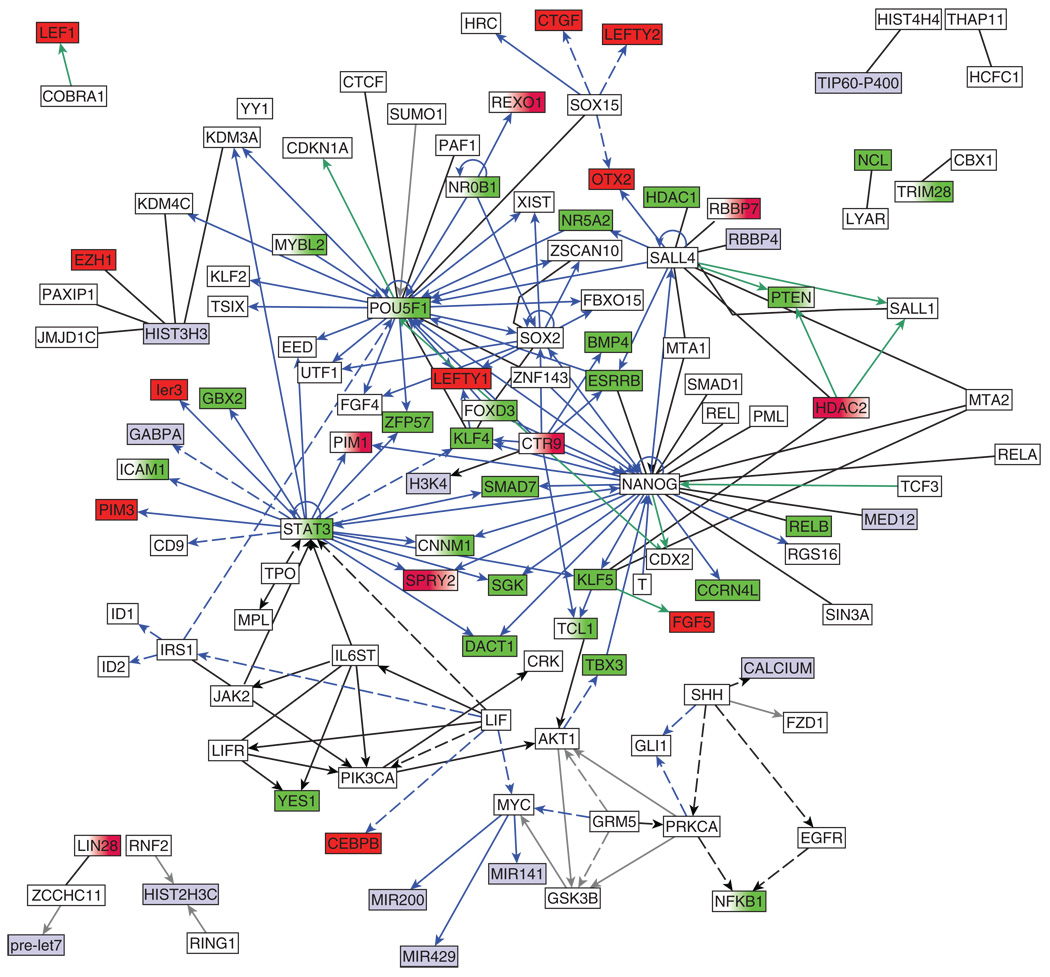
Representation of molecular intracellular biological systems as networks is useful for combining results from different studies and obtaining an overall bird’seye-view of the system.238–241 Network representation permits making predictions about undiscovered interactions and identifying functional modules. In principle, a molecular regulatory network consists of nodes and links, in which nodes represent molecular entities such as genes, gene products, miRNAs or metabolites, and links represent interactions/relationships that can be direct or indirect (influence or physical association), signed or unsigned (activation, inhibition, or neutral). One approach to reconstruct intracellular regulatory networks is to manually extract interactions from the literature. For example, Thiele et al.242 reconstructed a transcriptional machinery for E. coli by studying over 500 publications, whereas Ma’ayan et al.243 extracted over 1200 cell signaling interactions reported in mammalian neurons. For this review, we manually extracted binary interactions from 286 experimental studies specifically focused on ES cells. A regulatory pluripotency/self-renewal sub-network for mouse ES cells based on this manual extraction is presented in Figure 2, whereas the entire network is available online at http://amp.pharm.mssm.edu/ismid/literature. Note that the interactions on the website include binary interactions identified in mouse ES cells by Wang et al.,212 which were performed using high-throughput techniques. In contrast with previous studies, we combined direct and indirect interactions and included cell signaling interactions and gene regulatory interactions. Such accumulated-knowledge-based-networks provide a scaffold for further data integration and for the interrogation of data collected in new high-throughput and low-throughput studies. For example, changes in expression of mRNA levels detected in microarray experiments can be projected onto knowledge-based networks for inferring and expanding potential interactions among gene products at the mRNA and protein levels.244 Co-clustering245,246 can be applied to identify functional modules based on expression profiles combined with protein interaction networks. The known regulatory topology can also be used to begin to understand how information flows through the system over-time. Here, we projected a time-course mRNA expression profiling after Nanog knock-down in mouse ES cells247 with the literature-based network we developed for this review to illustrate this point (Figure 3).
FIGURE 2.
Initial literature-based regulatory network for mES cells self-renewal and pluripotency. The network contains 121 nodes representing genes or gene-encoding proteins with 187 edges involved in mouse ES cell self-renewal. Edge type: directed (positive/negative effects); undirected (neutral effect, i.e., protein–protein interactions). Edge color: dark blue (transcriptional positive); green (transcriptional negative); black (signaling positive); gray (signaling negative). Edge style: solid (direct); dashed (indirect). Node color: yellow (extracellular ligand); gray (membrane protein); cyan (cytosolic protein); red (nuclear proteins/genes); green (sarcoplasmic reticulum). A web-based system to navigate through components and their interactions is provided at http://amp.pharm.mssm.edu/iscmid/literature.
FIGURE 3.
Nodes from the literature-based network shown in Figure 2 are color coded based on changes in expression after Nanog down-regulation using shRNA in mES cells. Green represents down-regulation, red up-regulation. Data for gray nodes was not obtained.
Linking networks to disease
Having established networks from literature for specific biological processes, and linking such networks to genome-wide profiling studies, can lead to the identification of novel disease genes or to better understanding of the pathogenesis of previously less understood complex diseases. For example, a study that used mRNA expression microarrays at different developmental stages profiling a mouse model of prion disease, a disease caused by misfolding and aggregation of prion proteins, projected expression changes onto a network of protein interactions, similarly to what we did for Figure 3, to identify novel molecular origins of the disease.248 It is also possible to extract important disease-relevant information from networks just by examining the network topology, especially when the sign of the links (activation/inhibition) is known. A cross-disciplinary study by Abdi et al.249 showed how using circuit fault diagnosis commonly applied to study electrical circuits can identify vulnerable genes in literature-based mammalian cell signaling networks. Another example that falls within this category is a study by Mani et al.250 who developed an algorithm to characterize oncogenic mechanisms in B-cell lymphomas. They used a B-cell interaction map as a scaffold and then projected onto it a large set of microarray expression profiles. Another seminal study by Ideker and colleagues251 used a protein–protein interaction network combined with gene expression profiles to identifying markers of metastasis. Their method scored subnetworks based on mutual information with expression activity over tumor sample and class category. In summary, these approaches utilize prior knowledge network topology in conjunction with genome-wide expression profiling to unravel molecular mechanisms of disease.
Construction of networks from data
Reconstruction of regulatory networks can be completely data-driven. Approaches for gene regulatory network inference from microarray data can be categorized into three principle classes: (1) Linear models using differential equation to describe gene expression changes as a function of expression of other genes and external perturbations.252 (2) Information theory-based models, such as ARACNe253 or the CLR.254 With these methods, edges are weighted and filtered based on conditional mutual information. (3) Probabilistic-based graphical models such as Bayesian Networks255 and Module Networks.256 These methods treat expression of genes as random variables and embody the description of the joint probability distribution of these random variables. Commonly, research groups also attempt to integrate expression data with other data sources such as genome-scale ChIP results in order to augment the inference.257 Most network inference methods do not consider post-translational regulation which is controlling transcription factor activity. Post-translational modifications such as phosphorylation, acetylation, ubiquitination, or methylation may change the function and localization of transcription factors. With the limited experimental approaches for identifying PTMs, computational approaches for inferring this layer of regulation have been developed. For example, the MINDy algorithm258 infers candidate modulators of transcription factors using mutual information.
Application to study ES cells
Several studies used reverse engineering methods to characterize ES cell networks. For example, Chen and Zhong259 inferred a regulatory network in mouse ES cells from time-series microarray data by first finding delayed correlations and then inferring reaction rules that can recapitulate the dynamical changes observed in the data. For each of their identified transcription factor–target pair, ChIP-chip and RNAi data were used for model verification. Other probabilistic-based methods for network inference can be used.260–262 For example, Woolf et al.262 inferred a signaling network from a proteomics dataset. They reconstructed a self-renewal signaling network of mouse ES cells by applying a Bayesian-learning algorithm. With respect to defining a signed network, the links can be inferred from transcription factor–target gene interactions identified by ChIP experiments, whereas the signs of links can be drawn from transformation of raw expression data, or inversely related to knockout effects.118,261,263 For instance, Chavez et al.264 applied this methodology of integrating Oct4 ChIP-chip experiments and Oct4 RNAi in human ES cells. Similarly, Chen et al.105 constructed a regulatory network in ES cells inferred from integrated data of transcription factor binding sites and expression profiles in undifferentiated and differentiated ES cells. The resulting network shows high interconnectedness, which reflects the interwoven relationship between the core transcription factors responsible for maintaining the self-renewal state. Recently, Muller et al.265 combined microarray data, classification algorithms, and protein–protein interactions from MATISSE246 to construct a consensus stem cell network called PluriNet from many studies. Such network is undirected. Hence, more efforts are required for more complete reconstruction of ES cells regulatory networks expanding both depth and breadth around key transcription factors and cell signaling pathways responsible for stem cell development. Such networks will be then simulated using different dynamical modeling techniques. However, describing such computational methods is beyond the scope of this review.
MODELING THE DYNAMICS
Although we do not elaborate our discussion about dynamical computational modeling techniques, it is critical to emphasize that changes in cell fate occur over time where it will be necessary to explore regulatory network dynamics.266 In addition, a number of studies have documented the stochastic, intrinsically noisy nature of mRNA and protein expression in stem cells.267,268 Of course, stochastic components of stem cell regulation have been demonstrated more than 40 years ago in the hematopoietic system. In simpler biological systems such as the lytic versus lysogenic decision process of certain bacteriophages (lambda), stochasticity or noise have been shown to be vital for the biological decision process.269 In more complex systems, the problem comes down to the difficulty of discerning biologically important versus unimportant stochastic phenomena. A more general problem is that essentially most gene and protein expression studies provide data that are an average over a population of cells. Thus, intermediate levels of a given gene’s expression level can result from on or off expression in single cells, truly graded expression levels in single cells or a combination of both. Currently, robust technologies are emerging to quantitatively monitor gene expression in single cells, and the next several years will surely see additional insights into these issues. For example, single-cell behavioral complexity have been resolved in quantitative analyses of the Ras-MAPK and other biochemical signaling pathways. It is also noteworthy that during the development of PCR technologies, many of these types of ‘sensitivity’ issues have already been addressed; thus providing important paradigms. The bottom line is that all data collecting technologies have inherent limitations and these need to be considered when moving forward to the development of databases and computational analysis methods.
CONCLUSIONS
In this review, we summarized some of the systematic methods to profile intracellular regulatory molecular systems at different layers of regulation, focusing on ES cells in particular. We describe how utilization of high-throughput profiling approaches is paired with related databases and computational tools in a pipeline process that concludes in network reconstruction, data integration, and functional enrichment analyses (Figure 1). Clearly, the trend in the field is to combine several different types of high-throughput methods for interrogating cells at multiple layers of regulation. For example, in a recent study, Rong et al. measured mRNA expression, nuclear protein levels, and chromatin status markers as a time-series in mES cells after a defined perturbation: knock-down of the self-renewal essential transcription factor Nanog.247 Such approaches are gradually unraveling the molecular regulatory complexity of ES cells while requiring advanced computational analysis methods. Drilling down to further characterize the most interesting new components and interactions identified by high-throughput methods coupled with computational analyses should provide low-hanging-fruit for further functional experiments which are all expected to move the field rapidly forward.
REFERENCES
- 1.Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 2.Sharov AA, Piao Y, Matoba R, Dudekula DB, Qian Y, et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:e74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandenberger R, Wei H, Zhang S, Lei S, Murage J, et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat Biotechnol. 2004;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- 4.Anisimov SV, Tarasov KV, Tweedie D, Stern MD, Wobus AM, et al. SAGE identification of gene transcripts with profiles unique to pluripotent mouse R1 embryonic stem cells. Genomics. 2002;79:169–176. doi: 10.1006/geno.2002.6687. [DOI] [PubMed] [Google Scholar]
- 5.Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 6.Miura T, Luo Y, Khrebtukova I, Brandenberger R, Zhou D, et al. Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev. 2004;13:694–715. doi: 10.1089/scd.2004.13.694. [DOI] [PubMed] [Google Scholar]
- 7.Wei CL, Miura T, Robson P, Lim SK, Xu XQ, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 8.Hornshoj H, Bendixen E, Conley LN, Andersen PK, Hedegaard J, et al. Transcriptomic and proteomic profiling of two porcine tissues using high-throughput technologies. BMC Genomics. 2009;10:30. doi: 10.1186/1471-2164-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, et al. ArrayExpress–a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, et al. The mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res. 2007;35:D618–D623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda-Saavedra D, De S, Trotter MW, Teichmann SA, Gottgens B. BloodExpress: a database of gene expression in mouse haematopoiesis. Nucleic Acids Res. 2009;37 suppl 1:D873–D879. doi: 10.1093/nar/gkn854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter CJ, Palidwor GA, Sandie R, Krzyzanowski PM, Muro EM, et al. StemBase: a resource for the analysis of stem cell gene expression data. Methods Mol Biol. 2007;407:137–148. doi: 10.1007/978-1-59745-536-7_11. [DOI] [PubMed] [Google Scholar]
- 14.Hackney JA, Charbord P, Brunk BP, Stoeckert CJ, Lemischka IR, et al. A molecular profile of a hematopoietic stem cell niche. Proc Natl Acad Sci U S A. 2002;99:13061–13066. doi: 10.1073/pnas.192124499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toronen P, Kolehmainen M, Wong G, Castren E. Analysis of gene expression data using self-organizing maps. FEBS Lett. 1999;451:142–146. doi: 10.1016/s0014-5793(99)00524-4. [DOI] [PubMed] [Google Scholar]
- 18.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 19.Lepre J, Rice JJ, Tu Y, Stolovitzky G. Genes@Work: an efficient algorithm for pattern discovery and multivariate feature selection in gene expression data. Bioinformatics. 2004;20:1033–1044. doi: 10.1093/bioinformatics/bth035. [DOI] [PubMed] [Google Scholar]
- 20.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 21.Gardiner-Garden M, Littlejohn TG. A comparison of microarray databases. Brief Bioinform. 2001;2:143–158. doi: 10.1093/bib/2.2.143. [DOI] [PubMed] [Google Scholar]
- 22.Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, et al. EXPANDER–an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimers M, Carey VJ. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006;411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 25.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 26.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 27.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 28.Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westfall SD, Sachdev S, Das P, Hearne LB, Hannink M, et al. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. Stem Cells Dev. 2008;17:869–881. doi: 10.1089/scd.2007.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The International Stem Cell Initiative. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharya B, Cai J, Luo Y, Miura T, Mejido J, et al. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and differentiating embryoid bodies. BMC Dev Biol. 2005;5:22. doi: 10.1186/1471-213X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skottman H, Mikkola M, Lundin K, Olsson C, Stromberg AM, et al. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- 33.Mansergh FC, Daly CS, Hurley AL, Wride MA, Hunter SM, et al. Gene expression profiles during early differentiation of mouse embryonic stem cells. BMC Dev Biol. 2009;9:5. doi: 10.1186/1471-213X-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, et al. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- 35.Player A, Wang Y, Bhattacharya B, Rao M, Puri RK, et al. Comparisons between transcriptional regulation and RNA expression in human embryonic stem cell lines. Stem Cells Dev. 2006;15:315–323. doi: 10.1089/scd.2006.15.315. [DOI] [PubMed] [Google Scholar]
- 36.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 37.Sun YLH, Liu Y, Shin S, Mattson MP, Rao MS, et al. Cross-species transcriptional profiles establish a functional portrait of embryonic stem cells. Genomics. 2007;89:22–35. doi: 10.1016/j.ygeno.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharya B, Puri S, Puri RK. A review of gene expression profiling of human embryonic stem cell lines and their differentiated progeny. Curr Stem Cell Res Ther. 2009;4:98–106. doi: 10.2174/157488809788167409. [DOI] [PubMed] [Google Scholar]
- 39.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 40.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 41.Ren Y, Gong W, Xu Q, Zheng X, Lin D, et al. siRecords: an extensive database of mammalian siRNAs with efficacy ratings. Bioinformatics. 2006;22:1027–1028. doi: 10.1093/bioinformatics/btl026. [DOI] [PubMed] [Google Scholar]
- 42.Chalk AM, Warfinge RE, Georgii-Hemming P, Sonnhammer EL. siRNAdb: a database of siRNA sequences. Nucleic Acids Res. 2005;33:D131–D134. doi: 10.1093/nar/gki136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boese Q, Leake D, Reynolds A, Read S, Scaringe SA, et al. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 2005;392:73–96. doi: 10.1016/S0076-6879(04)92005-8. [DOI] [PubMed] [Google Scholar]
- 44.Chalk AM, Wahlestedt C, Sonnhammer EL. Improved and automated prediction of effective siRNA. Biochem Biophys Res Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 45.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Qian K, Tang ZP, Xing B, Liu N, et al. Bioinformatics and microarray analysis of microRNA expression profiles of murine embryonic stem cells, neural stem cells induced from ESCs and isolated from E8.5 mouse neural tube. Neurol Res. 2009 doi: 10.1179/174313209X455691. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 53.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 54.Maselli V, Di Bernardo D, Banfi S. CoGemiR: a comparative genomics microRNA database. BMC Genomics. 2008;9:457. doi: 10.1186/1471-2164-9-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pang KC, Stephen S, Engstrom PG, Tajul-Arifin K, Chen W, et al. RNAdb–a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 58.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 59.John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papadopoulos G, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou A. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 62.Yousef M, Jung S, Kossenkov AV, Showe LC, Showe MK. Naive Bayes for microRNA target predictions–machine learning for microRNA targets. Bioinformatics. 2007;23:2987–2992. doi: 10.1093/bioinformatics/btm484. [DOI] [PubMed] [Google Scholar]
- 63.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility inmicroRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 64.Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao F, Zuo Z, Cai G, Kang S, Gao X, et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW. GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res. 2009;37:W323–W328. doi: 10.1093/nar/gkp313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 72.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saetrom O, Snove O, Jr, Saetrom P. Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms. RNA. 2005;11:995–1003. doi: 10.1261/rna.7290705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, et al. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eran A, Kho A, Eisenberg I, Galdzicki M, Naxerova K, et al. Proceedings of ISCB2006. 2006 Poster L-38. [Google Scholar]
- 78.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nam S, Li M, Choi K, Balch C, Kim S, et al. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–W362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 85.Wu H, Xu J, Pang ZP, Ge W, Kim KJ, et al. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci U S A. 2007;104:13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cao H, Yang C-S, Rana TM. Evolutionary emergence of microRNAs in human embryonic stem cells. PLoS One. 2008;3:e2820. doi: 10.1371/journal.pone.0002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, Melen GJ, Sanchez L, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 91.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 92.Gu P, Reid JG, Gao X, Shaw CA, Creighton C, et al. Novel microRNA candidates and miRNA-mRNA pairs in embryonic stem (ES) cells. PLoS One. 2008;3:e2548. doi: 10.1371/journal.pone.0002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ciaudo C, Servant N, Cognat V, Sarazin A, Kieffer E, et al. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 2009;5:e1000620. doi: 10.1371/journal.pgen.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol. 2006;71:369–376. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 98.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 99.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 100.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 101.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 102.Bromberg KD, Ma’ayan A, Neves SR, Iyengar R. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 105.Chen X, Xu H, Yuan P, Fang F, Huss M, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 106.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 108.Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu X, Huang J, Chen T, Wang Y, Xin S, et al. Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res. 2008;18:1177–1189. doi: 10.1038/cr.2008.309. [DOI] [PubMed] [Google Scholar]
- 112.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao F, Xuan Z, Liu L, Zhang MQ. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:D103–D107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, et al. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishii T, Yoshida K, Terai G, Fujita Y, Nakai K. DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res. 2001;29:278–280. doi: 10.1093/nar/29.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Macarthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 120.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 121.Roider HG, Manke T, O’Keeffe S, Vingron M, Haas SA. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chekmenev DS, Haid C, Kel AE. P-Match: transcription factor binding site search by combining patterns and weight matrices. Nucleic Acids Res. 2005;33:W432–W437. doi: 10.1093/nar/gki441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zambelli F, Pesole G, Pavesi G. Pscan: finding overrepresented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Roider HG, Kanhere A, Manke T, Vingron M. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics. 2007;23:134–141. doi: 10.1093/bioinformatics/btl565. [DOI] [PubMed] [Google Scholar]
- 125.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Down TA, Hubbard TJ. NestedMICA: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res. 2005;33:1445–1453. doi: 10.1093/nar/gki282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pavesi G, Mauri G, Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics. 2001;17 suppl 1:S207–S214. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- 128.Sharov A, Masui S, Sharova L, Piao Y, Aiba K, et al. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hinsby AM, Olsen JV, Mann M. Tyrosine phospho-proteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J Biol Chem. 2004;279:46438–46447. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- 130.Gembitsky DS, Lawlor K, Jacovina A, Yaneva M, Tempst P. A prototype antibody microarray platform to monitor changes in protein tyrosine phosphorylation. Mol Cell Proteomics. 2004;3:1102–1118. doi: 10.1074/mcp.M400075-MCP200. [DOI] [PubMed] [Google Scholar]
- 131.Pimienta G, Chaerkady R, Pandey A. SILAC for global phosphoproteomic analysis. Methods Mol Biol. 2009;527:107–116. doi: 10.1007/978-1-60327-834-8_9. x. [DOI] [PubMed] [Google Scholar]
- 132.Prokhorova TA, Rigbolt KT, Johansen PT, Henningsen J, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol Cell Proteomics. 2009;8:959–970. doi: 10.1074/mcp.M800287-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y, Mulligan C, Denyer G, Delom F, Dagna-Bricarelli F, et al. Quantitative proteomics characterization of a mouse embryonic stem cell model of down syndrome. Mol Cell Proteomics. 2009;8:585–595. doi: 10.1074/mcp.M800256-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Williamson AJK, Smith DL, Blinco D, Unwin RD, Pearson S, et al. Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol Cell Proteomics. 2008;7:459–472. doi: 10.1074/mcp.M700370-MCP200. [DOI] [PubMed] [Google Scholar]
- 135.Pelech S, Sutter C, Zhang H. Kinetworks protein kinase multiblot analysis. Methods Mol Biol. 2003;218:99–111. doi: 10.1385/1-59259-356-9:99. [DOI] [PubMed] [Google Scholar]
- 136.Farriol-Mathis N, Garavelli JS, Boeckmann B, Duvaud S, Gasteiger E, et al. Annotation of posttranslational modifications in the Swiss-Prot knowledge base. Proteomics. 2004;4:1537–1550. doi: 10.1002/pmic.200300764. [DOI] [PubMed] [Google Scholar]
- 137.Diella F, Cameron S, Gemund C, Linding R, Via A, et al. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites– update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Keshava Prasad TS, Goel R, Kandasamy K, Keerthi-kumar S, Kumar S, et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang CY, Chang CH, Yu YL, Lin TC, Lee SA, et al. PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics. 2008;24:i14–i20. doi: 10.1093/bioinformatics/btn297. [DOI] [PubMed] [Google Scholar]
- 141.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 142.Gnad F, Ren S, Cox J, Olsen JV, Macek B, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, et al. PhosphoPep–a database of protein phosphorylation sites in model organisms. Nat Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, et al. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008;36:D1015–D1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gao J, Agrawal GK, Thelen JJ, Xu D. P3DB: a plant protein phosphorylation database. Nucleic Acids Res. 2009;37:D960–D962. doi: 10.1093/nar/gkn733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee TY, Huang HD, Hung JH, Huang HY, Yang YS, et al. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li H, Xing X, Ding G, Li Q, Wang C, et al. SysPTM - a systematic resource for proteomic research of post-translational modifications. Mol Cell Proteomics. 2009;8:1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, et al. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res. 2007;35:D229–D231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gong W, Zhou D, Ren Y, Wang Y, Zuo Z, et al. PepCyber:P~PEP: a database of human protein–protein interactions mediated by phosphoprotein-binding domains. Nucleic Acids Res. 2008;36:D679–D683. doi: 10.1093/nar/gkm854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ryu GM, Song P, Kim KW, Oh KS, Park KJ, et al. Genome-wide analysis to predict protein sequence variations that change phosphorylation sites or their corresponding kinases. Nucleic Acids Res. 2009;37:1297–1307. doi: 10.1093/nar/gkn1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 153.Mackey AJ, Haystead TA, Pearson WR. CRP: cleavage of radiolabeled phosphoproteins. Nucleic Acids Res. 2003;31:3859–3861. doi: 10.1093/nar/gkg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ingrell CR, Miller ML, Jensen ON, Blom N. Net-PhosYeast: prediction of protein phosphorylation sites in yeast. Bioinformatics. 2007;23:895–897. doi: 10.1093/bioinformatics/btm020. [DOI] [PubMed] [Google Scholar]
- 156.Miller ML, Soufi B, Jers C, Blom N, Macek B, et al. NetPhosBac - a predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics. 2009;9:116–125. doi: 10.1002/pmic.200800285. [DOI] [PubMed] [Google Scholar]
- 157.Tang YR, Chen YZ, Canchaya CA, Zhang Z. GAN-NPhos: a new phosphorylation site predictor based on a genetic algorithm integrated neural network. Protein Eng Des Sel. 2007;20:405–412. doi: 10.1093/protein/gzm035. [DOI] [PubMed] [Google Scholar]
- 158.Koenig M, Grabe N. Highly specific prediction of phosphorylation sites in proteins. Bioinformatics. 2004;20:3620–3627. doi: 10.1093/bioinformatics/bth455. [DOI] [PubMed] [Google Scholar]
- 159.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, et al. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, et al. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 2009;37:D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, et al. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 164.Brinkworth RI, Breinl RA, Kobe B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc Natl Acad Sci U S A. 2003;100:74–79. doi: 10.1073/pnas.0134224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Saunders NF, Brinkworth RI, Huber T, Kemp BE, Kobe B. Predikin and PredikinDB: a computational framework for the prediction of protein kinase peptide specificity and an associated database of phosphorylation sites. BMC Bioinformatics. 2008;9:245. doi: 10.1186/1471-2105-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 168.Kim JH, Lee J, Oh B, Kimm K, Koh I. Prediction of phosphorylation sites using SVMs. Bioinformatics. 2004;20:3179–3184. doi: 10.1093/bioinformatics/bth382. [DOI] [PubMed] [Google Scholar]
- 169.Xue Y, Ren J, Gao X, Jin C, Wen L, et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xue Y, Li A, Wang L, Feng H, Yao X. PPSP: prediction of PK-specific phosphorylation site with Bayesian decision theory. BMC Bioinformatics. 2006;7:163. doi: 10.1186/1471-2105-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, et al. KinasePhos 2.0: a web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35:W588–W594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li T, Li F, Zhang X. Prediction of kinase-specific phosphorylation sites with sequence features by a log-odds ratio approach. Proteins. 2008;70:404–414. doi: 10.1002/prot.21563. [DOI] [PubMed] [Google Scholar]
- 173.Neuberger G, Schneider G, Eisenhaber F. pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol Dir. 2007;2:1. doi: 10.1186/1745-6150-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dang TH, Van Leemput K, Verschoren A, Laukens K. Prediction of kinase-specific phosphorylation sites using conditional random fields. Bioinformatics. 2008;24:2857–2864. doi: 10.1093/bioinformatics/btn546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Plewczynski D, Tkacz A, Wyrwicz LS, Rychlewski L, Ginalski K. AutoMotif Server for prediction of phosphorylation sites in proteins using support vector machine: 2007 update. J Mol Model. 2008;14:69–76. doi: 10.1007/s00894-007-0250-3. [DOI] [PubMed] [Google Scholar]
- 176.Wan J, Kang S, Tang C, Yan J, Ren Y, et al. Meta-prediction of phosphorylation sites with weighted voting and restricted grid search parameter selection. Nucleic Acids Res. 2008;36:e22. doi: 10.1093/nar/gkm848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li L, Wu C, Huang H, Zhang K, Gan J, et al. Prediction of phosphotyrosine signaling networks using a scoring matrix-assisted ligand identification approach. Nucleic Acids Res. 2008;36:3263–3273. doi: 10.1093/nar/gkn161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Huang H, Li L, Wu C, Schibli D, Colwill K, et al. Defining the specificity space of the human SRC homology 2 domain. Mol Cell Proteomics. 2008;7:768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 179.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yoo PD, Ho YS, Zhou BB, Zomaya AY. SiteSeek: post-translational modification analysis using adaptive locality-effective kernel methods and new profiles. BMC Bioinformatics. 2008;9:272. doi: 10.1186/1471-2105-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 182.Schwartz D, Chou MF, Church GM. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol Cell Proteomics. 2009;8:365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ritz A, Shakhnarovich G, Salomon AR, Raphael BJ. Discovery of phosphorylation motif mixtures in phosphoproteomics data. Bioinformatics. 2009;25:14–21. doi: 10.1093/bioinformatics/btn569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang Y, Klemke RL. PhosphoBlast, a computational tool for comparing phosphoprotein signatures among large datasets. Mol Cell Proteomics. 2008;7:145–162. doi: 10.1074/mcp.M700207-MCP200. [DOI] [PubMed] [Google Scholar]
- 185.Yuan X, Hu ZZ, Wu HT, Torii M, Narayanaswamy M, et al. An online literature mining tool for protein phosphorylation. Bioinformatics. 2006;22:1668–1669. doi: 10.1093/bioinformatics/btl159. [DOI] [PubMed] [Google Scholar]
- 186.Lachmann A, Ma’ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ren J, Wen L, Gao X, Jin C, Xue Y, et al. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 188.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J Proteome Res. 2008;7:3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 190.Lu B, Ruse CI, Yates JR., 3rd Colander: a probability-based support vector machine algorithm for automatic screening for CID spectra of phosphopeptides prior to database search. J Proteome Res. 2008;7:3628–3634. doi: 10.1021/pr8001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lu B, Ruse C, Xu T, Park SK, Yates J., 3rd Automatic validation of phosphopeptide identifications from tandem mass spectra. Anal Chem. 2007;79:1301–1310. doi: 10.1021/ac061334v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jiang X, Han G, Feng S, Ye M, Yao X, et al. Automatic validation of phosphopeptide identifications by the MS2/MS3 target-decoy search strategy. J Proteome Res. 2008;7:1640–1649. doi: 10.1021/pr700675j. [DOI] [PubMed] [Google Scholar]
- 193.Payne SH, Yau M, Smolka MB, Tanner S, Zhou H, et al. Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J Proteome Res. 2008;7:3373–3381. doi: 10.1021/pr800129m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Du X, Yang F, Manes NP, Stenoien DL, Monroe ME, et al. 2nd Linear discriminant analysis-based estimation of the false discovery rate for phosphopeptide identifications. J Proteome Res. 2008;7:2195–2203. doi: 10.1021/pr070510t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kocher T, Savitski MM, Nielsen ML, Zubarev RA. PhosTShunter: a fast and reliable tool to detect phosphorylated peptides in liquid chromatography Fourier transform tandem mass spectrometry data sets. J Proteome Res. 2006;5:659–668. doi: 10.1021/pr0503836. [DOI] [PubMed] [Google Scholar]
- 196.Wan Y, Cripps D, Thomas S, Campbell P, Ambulos N, et al. PhosphoScan: a probability-based method for phosphorylation site prediction using MS2/MS3 pair information. J Proteome Res. 2008;7:2803–2811. doi: 10.1021/pr700773p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang L, Schulz TC, Sherrer ES, Dauphin DS, Shin S, et al. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, et al. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, et al. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 201.Saxe JP, Tomilin A, Scholer HR, Plath K, Huang J. Post-translational regulation of Oct4 transcriptional activity. PLoS One. 2009;4:e4467. doi: 10.1371/journal.pone.0004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Walhout AJ, Vidal M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 203.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 204.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phospho-proteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, et al. Human protein reference database–2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, et al. A bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 211.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang J, Rao S, Chu J, Shen X, Levasseur DN, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 213.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 215.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, et al. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Masseroli M, Galati O, Pinciroli F. GFINDer: genetic disease and phenotype location statistical analysis and mining of dynamically annotated gene lists. Nucleic Acids Res. 2005;33:W717–W723. doi: 10.1093/nar/gki454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 219.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 221.Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Martinez-Cruz LA, Rubio A, Martinez-Chantar ML, Labarga A, Barrio I, et al. GARBAN: genomic analysis and rapid biological annotation of cDNA microarray and proteomic data. Bioinformatics. 2003;19:2158–2160. doi: 10.1093/bioinformatics/btg291. [DOI] [PubMed] [Google Scholar]
- 224.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kokocinski F, Delhomme N, Wrobel G, Hummerich L, Toedt G, et al. FACT–a framework for the functional interpretation of high-throughput experiments. BMC Bioinformatics. 2005;6:161. doi: 10.1186/1471-2105-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wrobel G, Chalmel F, Primig M. goCluster integrates statistical analysis and functional interpretation of microarray expression data. Bioinformatics. 2005;21:3575–3577. doi: 10.1093/bioinformatics/bti574. [DOI] [PubMed] [Google Scholar]
- 228.Newman JC, Weiner AM. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Vencio RZ, Koide T, Gomes SL, Pereira CA. BayGO: Bayesian analysis of ontology term enrichment in microarray data. BMC Bioinformatics. 2006;7:86. doi: 10.1186/1471-2105-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Breitling R, Amtmann A, Herzyk P. Iterative Group Analysis (iGA): a simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics. 2004;5:34. doi: 10.1186/1471-2105-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Henegar C, Cancello R, Rome S, Vidal H, Clement K, et al. Clustering biological annotations and gene expression data to identify putatively co-regulated biological processes. J Bioinform Comput Biol. 2006;4:833–852. doi: 10.1142/s0219720006002181. [DOI] [PubMed] [Google Scholar]
- 233.Kim SB, Yang S, Kim SK, Kim SC, Woo HG, et al. GAzer: gene set analyzer. Bioinformatics. 2007;23:1697–1699. doi: 10.1093/bioinformatics/btm144. [DOI] [PubMed] [Google Scholar]
- 234.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Minguez P, et al. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Alibes A, Canada A, Diaz-Uriarte R. PaLS: filtering common literature, biological terms and pathway information. Nucleic Acids Res. 2008;36:W364–W367. doi: 10.1093/nar/gkn251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Prifti E, Zucker JD, Clement K, Henegar C. Fun-Net: an integrative tool for exploring transcriptional interactions. Bioinformatics. 2008;24:2636–2638. doi: 10.1093/bioinformatics/btn492. [DOI] [PubMed] [Google Scholar]
- 237.Storm MP, Kumpfmueller B, Thompson B, Kolde R, Vilo J, et al. Characterization of the phosphoinositide 3-kinase-dependent transcriptome in murine embryonic stem cells: identification of novel regulators of pluripotency. Stem Cells. 2009;27:764–775. doi: 10.1002/stem.3. [DOI] [PubMed] [Google Scholar]
- 238.Ma’ayan A, Blitzer RD, Iyengar R. Toward predictive models of mammalian cells. Annu Rev Biophys Biomol Struct. 2005;34:319–349. doi: 10.1146/annurev.biophys.34.040204.144415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ma’ayan A, Iyengar R. From components to regulatory motifs in signalling networks. Brief Funct Genomic Proteomic. 2006;5:57–61. doi: 10.1093/bfgp/ell004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ma’ayan A. Insights into the organization of biochemical regulatory networks using graph theory analyses. J Biol Chem. 2009;284:5451–5455. doi: 10.1074/jbc.R800056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ma’ayan A. Network integration and graph analysis in mammalian molecular systems biology. IET Syst Biol. 2008;2:206–221. doi: 10.1049/iet-syb:20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Thiele I, Jamshidi N, Fleming RM, Palsson BO. Genome-scale reconstruction of Escherichia coli’s transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput Biol. 2009;5:e1000312. doi: 10.1371/journal.pcbi.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ma’ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, et al. Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science. 2005;309:1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Berger S, Posner J, Ma’ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hanisch D, Zien A, Zimmer R, Lengauer T. Co-clustering of biological networks and gene expression data. Bioinformatics. 2002;18 suppl 1:S145–S154. doi: 10.1093/bioinformatics/18.suppl_1.s145. [DOI] [PubMed] [Google Scholar]
- 246.Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hwang D, Lee IY, Yoo H, Gehlenborg N, Cho J-H, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Abdi A, Tahoori MB, Emamian ES. Fault diagnosis engineering of digital circuits can identify vulnerable molecules in complex cellular pathways. Sci Signal. 2008;1:ra10–ra10. doi: 10.1126/scisignal.2000008. [DOI] [PubMed] [Google Scholar]
- 250.Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Chuang H-Y, Lee E, Liu Y-T, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:141. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Yeung MK, Tegnér J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc Natl Acad SciU S A. 2002;99:6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 254.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, et al. Large-scale mapping and validation of <named-content xmlns:xlink=“http://www.w3.org/1999/xlink” content-type=“genus-species” xlink:type=“simple”>Escherichia coli</named-content>transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 256.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 257.Liao JC, Boscolo R, Yang Y-L, Tran LM, Sabatti C, et al. Network component analysis: Reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, et al. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009;27:829–837. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chen CC, Zhong S. Inferring gene regulatory networks by thermodynamic modeling. BMC Genomics. 2008;9 suppl 2:S19. doi: 10.1186/1471-2164-9-S2-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Marcotte EM, Xenarios I, Eisenberg D. Mining literature for protein-protein interactions. Bioinformatics. 2001;17:359–363. doi: 10.1093/bioinformatics/17.4.359. [DOI] [PubMed] [Google Scholar]
- 261.Mason MJ, Fan G, Plath K, Zhou Q, Horvath S. Signed weighted gene co-expression network analysis of transcriptional regulation in murine embryonic stem cells. BMC Genomics. 2009;10:327. doi: 10.1186/1471-2164-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Woolf PJ, Prudhomme W, Daheron L, Daley GQ, Lauffenburger DA. Bayesian analysis of signaling networks governing embryonic stem cell fate decisions. Bioinformatics. 2005;21:741–753. doi: 10.1093/bioinformatics/bti056. [DOI] [PubMed] [Google Scholar]
- 263.Yeang CH, Ideker T, Jaakkola T. Physical network models. J Comput Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- 264.Chavez L, Bais AS, Vingron M, Lehrach H, Adjaye J, et al. In silico identification of a core regulatory network of OCT4 in human embryonic stem cells using an integrated approach. BMC Genomics. 2009;10:314. doi: 10.1186/1471-2164-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.MacArthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci U S A. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 269.Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 270.Lachmann A, Ma’ayan A. Lists2Networks: Integrated analysis of gene/protein lists. BMC Bioinformatics. 2010;11:87. doi: 10.1186/1471-2105-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]