SUMMARY
More than 700 bacterial species have been detected in human oral cavity. They form highly organized microbial communities and are responsible for many oral infectious diseases, such as dental caries and periodontal disease. The prevention and treatment of these diseases require a comprehensive knowledge of oral microbial communities, which largely relies on culture-dependent methods to have detailed phenotypic and physiological analysis of these communities. However, most of the currently available lab media can only selectively support the growth of a limited number of bacterial species within these communities, and fail to sustain the original oral microbial diversity. In this study, using denaturing gradient gel electrophoresis (DGGE) as an index to systematically survey and analyze the selectivity of commonly used lab media, we developed a new medium (SHI medium) by combining the ingredients of several selected media which can support different sub-populations within the original oral microbial community derived from pooled saliva. DGGE and 454 pyrosequencing analysis showed that SHI medium was capable of supporting a more diversified community with a microbial profile closest to that of the original oral microbiota. Furthermore, 454 pyrosequencing revealed that SHI medium supported the growth of many oral species that have not been cultured so far. Crystal violet assay and the CLSM (confocal laser scanning microscope) analysis indicated that, compared with other media, SHI medium is able to support more complex saliva-derived biofilm with higher biomass yield and more diversified species. This DGGE-guided method could also be used to develop novel media for other complex microbial communities.
Keywords: oral microbial community, growth medium, 454 pyrosequencing
INTRODUCTION
Oral microbiota is one of the most complex bacterial communities associated with human body. So far, more than 700 different bacterial species have been identified from human oral cavity (Aas et al., 2005; Paster et al., 2001; Paster et al., 2006) and the majority of them are associated with dental plaque, forming highly organized microbial communities (Kuramitsu et al., 2007; Marsh, 2005). These diverse microbial residents within the community display extensive interactions while forming organized biofilm structures, carrying out sophisticated physiological functions, and inducing microbial pathogenesis under certain conditions (Kolenbrander et al., 2002; Kolenbrander et al., 2010; Kuramitsu et al., 2007).
Oral microflora has been implicated in many oral infectious diseases, including dental caries and periodontitis (Dahlen, 1993; Marsh, 1994; Sigmund, 1979; Tatsuji & Takeyoshi, 2004). Current concept proposed that in the case of complex microbiota, it is not merely the presence of a single organism in a complex community which determines the properties of a microflora, but it is the interactions between the resident bacteria which is crucial (Marsh, 1994; Marsh, 2005). Such a microbial community-based pathogenic theory serves as a new concept for understanding the oral microbial community and the host in health and disease, as well as suggesting new strategies for disease treatment and prevention. With this new concept, it is crucial to study oral microbial flora as a whole, analyzing its microbial composition as well as its physiological and biological properties.
The application of culture-independent, PCR-based high-throughput methods, such as DGGE (Fujimoto et al., 2003; Li et al., 2005), terminal restriction fragment length polymorphism (T-RFLP) (Liu et al., 1997), denaturing high-performance liquid chromatography (DHPLC) (Barlaan et al., 2005) and metagenomic approach (Lazarevic et al., 2009) have greatly facilitated community analysis by revealing its genetic diversity. However, conventional culture-dependent, in vitro methods are still indispensable in providing valuable information, including both phenotypic and genetic characterization of individual bacterium, as well as new physiological functions resulted from interactions among different microbial inhabitants within the community.
Many in vitro systems have been developed to study oral microbial community, such as constant depth film fermentor (CDFF) (Kinniment et al., 1996), saliva-conditioned flow cell (Cook et al., 1998; Foster & Kolenbrander, 2004) and artificial mouths (Sissons et al., 1991). Modified basal medium mucin (BMM), a peptone yeast extract-based medium supplemented with mucin (Sissons et al., 1991; Sissons et al., 1995) and defined medium mucin (DMM) (Wong & Sissions, 2001), a chemically defined saliva analogue with mucin are commonly used in these systems to support the growth of oral microbial community. Although the rates of growth in wet weight and the pattern in biofilm growth are similar to natural dental plaque (Sissons et al., 1995), no detailed analysis has been done to determine how the media may affect microbial community compositions due to their selectivity, and the bias that might have been introduced because of the ill represented microbial composition within cultivated microflora. Currently, even with the advanced anaerobic cultivation technique and available complex media for bacterial cultivation, less than 50 percentage of oral bacterial species can be successfully cultivated (Kolenbrander, 2000; Wade, 2002), which has hampered our detailed understanding of the oral microbial world. A better medium with low selectivity would greatly facilitate our in vitro study of oral microbial community.
Traditionally, the development of new medium has been largely relied on a trial-and-error methodology, which can be very time-consuming and costly. In this study, we used DGGE as an index to help us with the medium development. DGGE is a PCR-based approach for microbial community analysis. It provides “fingerprints” for each microbial flora and has been widely used to analyze environmental and human-associated microbial communities (Nakatsu, 2007; Zijnge et al., 2003). Using DGGE technique, we first analyzed different profiles of oral microbial community cultivated by commonly-used lab media. By comparing the profiles with that of the original oral sample, we selected a few media that were able to support the growth of certain salivary microbial subpopulation, and their combined profiles can best represent the profiles of original salivary microflora. The main active components from those selected media were then chosen as the ingredients for developing the new medium for cultivating saliva-derived oral microbial flora.
This report describes the development of a new medium (SHI medium) for cultivating saliva-derived oral microbial flora via the systematic applications of PCR-DGGE as an index. We demonstrated that, compared with other commonly used media, the newly developed SHI medium can sustain an in vitro microbial community with high diversity and similar microbial profiles to original saliva-derived oral microflora.
METHODS
Saliva collection
Saliva samples were collected from 6 healthy subjects, age 25~35. None was being treated for any systemic disease or taking any prescription or non-prescription medications. Subjects were asked to refrain from any food or drink 2 hours prior to donating saliva and spit directly into the saliva collection tube, 5 ml of saliva was collected from each person. Saliva samples were pooled together and centrifuged at 2,600g for 10 mins to spin down large debris and eukaryotic cells. The supernatant was referred to as pooled saliva and used throughout this study. 5 ml of pooled saliva was centrifuged again at 14,000 g for 5 mins and the pellet was collected for DNA extraction and PCR-DGGE analysis to obtain the bacterial profile of original saliva. The rest of the pooled saliva would be used for coating wells, seeding planktonic culture and starting biofilm.
Cultivating saliva-derived microbial flora using commonly used lab media
0.5 ml of above pooled saliva was inoculated into 5 ml of each of the following medium: CYE (casitone-yeast extract) broth (Difco), LB (Luria-Bertani) broth(Difco), TH (Todd-Hewitt) broth (Difco), BHI (brain heart infusion) broth (Difco), ASS (artificial saliva solution) defined medium (He et al., 2008), BMM (basal medium mucin), TYGVS (tryptone-yeast extract gelatin- volatile fatty acids-serum) (Ohta et al., 1986), sheep blood supplemented NAM (N-acetyl muramic Acid) broth, Columbia(Difco) broth (Teles et al., 2008), cooked meat medium (Difco), PYG (peptone-yeast extract-glucose medium), and chopped meat medium (Difco). The cultures were incubated under anaerobic condition (nitrogen 85%, carbon dioxide 5%, and hydrogen 10%) at 37°C for 24 hours. Bacteria were collected by centrifugation at the speed of 14,000 g for 3 mins and total genomic DNA was extracted using the MasterPure™ DNA purification kit (EPICENTRE, Madison, WI, USA) for PCR-DGGE analysis.
SHI medium
The SHI medium has the following composition: proteose peptone (Difco) 10 g/L; trypticase peptone (Difco) 5.0 g/L; yeast extract (Difco) 5.0 g/L; KCl 2.5 g/L; sucrose 5 g/L; haemin 5 mg/L; VitK 1 mg/L; urea 0.06 g /L, arginine 0.174 g/L; mucin (type III, porcine, gastric, Sigma Chemical Co., St Louis, Mo) 2.5 g/L; sheep blood (Colorado serum company) 5% and N -acetylmuramic acid (NAM) 10 mg/L.
Growth of saliva-derived biofilms
1) For crystal violet assay
2 ml pooled saliva was mixed with equal volume of PBS and was centrifuged at 14,000 g for 3 minutes. 200 μl of supernatant was added to each well of the 24-well plate to pre-coat the wells and plates were incubated at 37 ° C with lid open for 1 hour to dry the saliva coating. Plates were then sterilized under UV light for 1 hour before 150 μl of pooled saliva was inoculated into pre-coated well containing 850 μl of BHI, TH broth or SHI medium. Plates were incubated at 37°C under anaerobic condition to allow biofilm formation. After overnight growth, biofilms were evaluated by crystal violet assay. Three replicates were performed.
2) For CLSM microscopy
2 ml pooled saliva was mixed with equal volume of PBS and was centrifuged at 14,000 g for 3 minutes. 100 μl of supernatant was added to each well of the 8-well chamber of the Lab-Tek ® II Chamber Slide™ System (Nalge Nunc International; Naperville, IL, USA). The objective slide was replaced by a thin cover-slide for proper CLSM microscopy. Chambers were incubated at 37 ° C with lid open for 1 hour to dry the saliva coating, and sterilized under UV light for 1 hour. 100 μl of pooled saliva was seeded into each pre-coated 8-well chamber. Each chamber contained 300 μl of TH or SHI medium. The chamber was incubated at 37°C under anaerobic condition overnight before biofilm was analyzed by confocal laser scanning microscopy (CLSM). Three replicates were performed.
Crystal violet assay
Quantification of biofilms was achieved by staining with crystal violet. Overnight biofilms were rinsed 3 times with PBS to remove the unattached bacteria. 500 μl 0.5% crystal violet was added into each well and incubated at room temperature for 20 mins before solution was poured off and wells were washed two times with PBS. 300 μl of 95% ethanol was added into each well and plates were incubated at room temperature with gentle shaking until no crystal violet was released from biofilm. Ethanol solution was then transferred to a new 24-well plate and biofilm mass was evaluated at an optical density of 595 nm using a micro-plate reader. Three replicates were performed.
The confocal laser scanning microscopy (CLSM) and image analysis
Overnight biofilms were rinsed 3 times with PBS to remove the unattached bacteria, biofilms were labeled using the LIVE/DEAD BacLight™ Bacterial Viability staining kit (Invitrogen) according to the manufacturer's instructions. Briefly, biofilms were labeled with 1.67 μM SYTO9 (a green fluorescent dye that can cross intact membranes) and 10 μM propidium iodide (a red fluorescent dye that can only penetrate into cells that have lost membrane integrity). The biofilms were monitored through a 40x oil-immersion lens with a PASCAL LSM5 confocal laser scanning microscope (Zeiss, Germany). Green and red fluorescence was imaged sequentially to avoid cross-contamination of fluorescent signals. Image stacks of five randomly chosen spots were collected for each experimental sample, representative images were shown in the result section.
CLSM image were analyzed by the computer program COMSTAT (Heydorn et al., 2000). Image stacks were converted to individual grayscale Tiff images for each slice. Gray scale images were converted into black and white, and compared with the original image to determine the threshold for the images, and the best value is chosen to give the most accurate conversion of the gray-scale to the black/white picture. The threshold value is fixed and then used for all image stacks. The image stacks of biofilm grown in TH and SHI medium were averaged and compared.
PCR-DGGE analysis
Total genomic DNA of bacterial samples were isolated using the MasterPure™ DNA purification kit (EPICENTRE, Madison, WI, USA). DNA quality and quantity were measured by a UV spectrophotometer at 260 nm and 280 nm. (Spectronic Genesys™, Spectronic Instrument, Inc. Rochester, New York, USA)
Amplification of bacterial 16S rRNA genes by PCR was carried out as described previously by Li et al (Li et al., 2005). Briefly, the universal primer set, Bac1 (5’ – CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GAC TAC GTG CCA GCA GCC – 3’) (Sheffield et al., 1989) and Bac2 (5' - GGA CTA CCA GGG TAT CTA ATC C - 3') was used to amply an approximately 300-bp internal fragment of the 16s rRNA gene. Each 50-μl PCR reaction contains 100ng of purified genomic DNA, 40 pmol of each primer, 200 μM of each dNTP, 4.0 mM MgCl2; 5 μl of 10X PCR buffer; and 2.5 U of Taq DNA polymerase (Invitrogen). Cycling conditions were 94°C for 3 mins, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 2 mins, with a final extension period of 5 mins at 72°C. The resulting PCR products were evaluated by electrophoresis in 1.0 % agarose gels.
Polyacrylamide gels at an 8% concentration were prepared with a denaturing urea/formamide gradient between 40% (containing 2.8 mol/L urea and 16 % (v/v) formamide) and 70% (containing 4.9 mol/L urea and 28 % (v/v) formamide). Approximately 300 ng of the PCR product were applied per well. The gels were submerged in 1 X TAE (Tris-Acetate-EDTA) buffer (40 mmol/L Tris base, 40 mmol/L glacial acid acetic, 1 mmol/L EDTA) and the PCR products were separated by electrophoresis for 17 hours at 58°C using a fixed voltage of 60 V in the Bio-Rad DCode System (Bio-Rad laboratories, Inc. Hercules, CA, USA). After electrophoresis, the gels were rinsed and stained for 15 mins in 1 X TAE buffer containing 0.5 μg/ml ethidium bromide, followed by 10 mins of de-staining in 1 X TAE buffer. DGGE profile images were digitally recorded using the Molecular Imager Gel Documentation system (Bio-Rad Laboratories, Hercules, CA, USA).
454 Pyrosequencing
Barcoded (approximately 10-12 bp long) 16S primers 27F [AGAGTTTGATYMTGGCTCAG] (Edwards et al., 1989) and 534R [ATTACCGCGGCTGCTGG] (Muyzer et al., 1993) were used to PCR amplify the 16S rRNA V3 region from the extracted DNA. The degenerate bases Y (nucleotide C or T) and M (nucleotide A or C) were incorporated into the primers. PCR products were cleaned up using Qiaquick column purification kit (Qiagen Inc, USA). The amplicons were normalized, pooled, and library construction was completed using Titanium chemistry (Roche Diagnostics, Inc. Basel, Switzerland). The average length of sequencing products were about 253 bp, SPRI clean up was performed using Ampure XP beads (Beckmann Coulter, Inc, USA) to ensure that the target amplicons with size of 507 bp were used for library preparation. The adaptor primer A (5’ CCATCTCATCCCTGCGTCTCTCCGAC 3’) and B (5’ CCTATCCCCTGTGTGCCTTGGCAGTC 3’) were ligated to both the 5’ and 3’ ends of the amplicons, before they were attached onto an emulsion PCR (emPCR) bead for amplification prior to 454 pyrosequencing. Using each sample's individual barcodes, the 454 sequence data were deconvolved into the respective samples. After trimming the barcodes and removing low quality sequences, taxonomic assignments of the sequences were made using the Ribosomal Database Project classifier (Wang et al., 2007; Cole et al., 2009) which classifies sequences to the genus level. After filtering, a total of 11,692 reads for the original saliva and 9,971 from the in-vitro grown sample were used in the analyses. Those 454 sequences classified as belonging to the Streptococcus genus were analyzed further to identify their nearest neighbors in the RDP database. This was done by searching these sequences against all Streptococcus sequences in the RDP database using BLAST.
Statistical Analysis
Significance of differences between average values was analyzed by t tests using MS Excel.
RESULTS
Survey of the commonly used lab media for their ability in cultivating oral microbiota
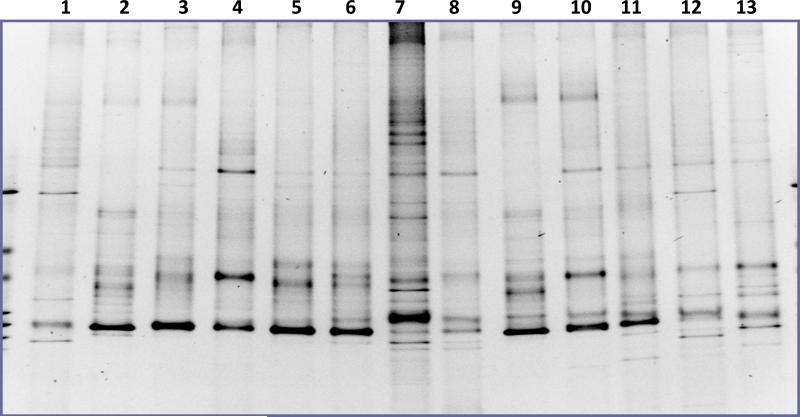
One of the limiting factors in in vitro study of oral microbial community was the selectivity of culture media, which often resulted in the ill-represented microbial diversity of oral community. In an effect to systematically survey the ability of different lab culture media in supporting the growth of oral microbiota, 12 current commonly-used lab media were chosen to cultivate human pooled salivary samples, including BMM and NAM, two of the frequently used media in sustaining oral microbial community. The microbial diversity within each medium was analyzed using PCR-DGGE technique as a profiling tool. Result showed that, compared with original saliva, bacterial samples from all the tested media display various levels of reduction in the number of bands, suggesting a decreased microbial diversity when using these media to cultivate salivary microbes. At the same time, there was striking difference in the banding pattern between samples from different medium, indicating each medium had selectivity for certain bacterial sub-population within the community (Fig.1).
Figure 1.
PCR-DGGE analysis showing the bacterial profiles of saliva-derived overnight planktonic culture in different media. 1, PYG; 2, ASS; 3.columbia; 4, TYGVS; 5, TH; 6, BHI; 7, Original saliva; 8, LB; 9, CYE; 10, Chopped meat medium; 11, Cooked meat medium; 12, BMM; 13, Sheep blood supplemented NAM broth. Three replicates were performed and a representative gel image is shown.
SHI medium displayed the least selectivity in cultivating oral microbiota from pooled human saliva
PCR-DGGE analysis indicated that all the tested media showed various level of selectivity in supporting the growth of saliva-derived oral microbiota. Figure 1 also revealed that although the individual banding pattern from PYG, BMM and NAM is different, if combined, the cumulated banding pattern was very close to that of the original saliva. We reasoned that combining the main nutritional ingredients from those 3 media could result in a new culture medium that might be able to support more diversified bacterial growth. Using this as a guideline, we developed a new medium—named SHI medium, for cultivating oral microbiota.
Using proteose peptone, trypticase peptone and yeast extract as the basic components, we developed SHI medium by supplementing the basic components with mucin, haemin, VitK, urea, and arginine which are the active ingredients of BMM medium. Two main supplements of the NAM broth, sheep blood and N -acetylmuramic acid were also included in the new medium.
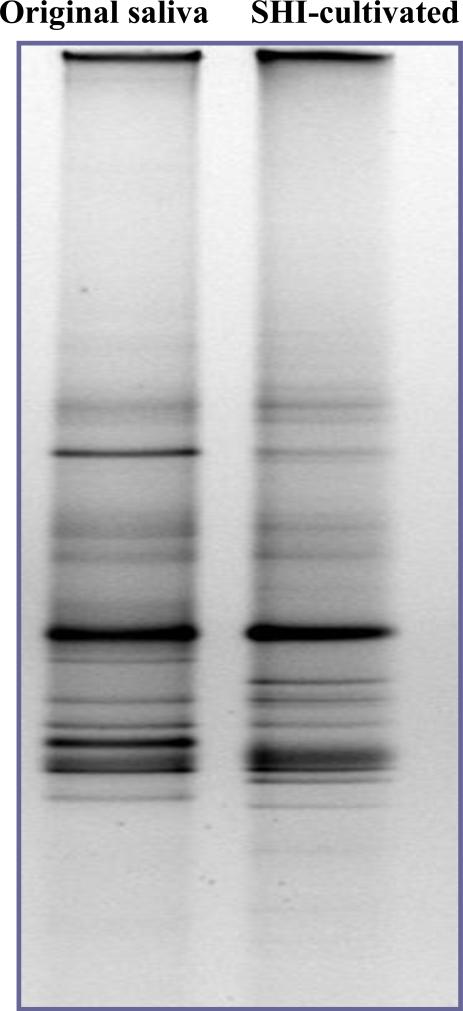
PCR-DGGE analysis revealed that SHI medium was able to sustain a microbial community with a profile very similar to that of the original saliva, suggesting that compared with other commonly used lab media, SHI medium had the least selectivity in cultivating saliva-derived oral microbiota (Fig.2).
Figure 2.
The bacterial profile of original pooled saliva and the microbial profile of saliva-derived overnight planktonic culture in newly developed SHI medium. Three replicates were performed and a representative gel image is shown.
454 pyrosequencing analysis
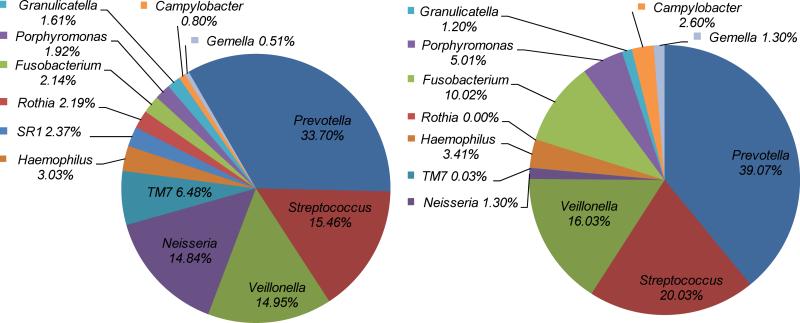
In order to further confirm that the newly developed SHI medium was indeed able to support the growth of diversified bacterial species within salivary samples, we performed 454 pyrosequencing analysis on SHI medium-cultivated and original salivary samples. Result showed that, bacterial species of at least 40 different genera, which fall into 8 bacterial divisions, were detected from the original saliva samples, with Firmicutes, Proteobacteria and Bacteroidetes being the most dominant phyla. Most excitingly, the microbial profile of SHI medium cultivated salivary flora displayed a striking similarity to that of the original sample at the genus as well as at the species level, including microbes within the phylum TM7 (Fig.3 and 4).
Figure 3.
Comparison of original salivary and SHI medium cultivated salivary bacterial species distribution using 454 pyrosequencing. Taxonomic assignments of the sequences were made using the Ribosomal Database Project which classifies sequences to the genus level. The community profile of the dominant members from A) the original pooled saliva and B) the in vitro grown community in SHI medium.
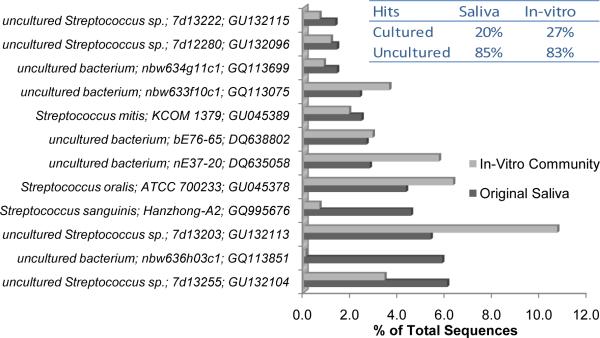
Figure 4.
Comparison of dominant sequences falling within the Streptococcus genus for the original saliva and the in vitro SHI medium cultivated community. For this analysis, all Streptococcus 16S rRNA gene sequences of cultured and uncultured strains were downloaded from RDP. Each 454 sequence that was classified as belonging to the Streptococcus genus, was assigned to the best matching Streptococcus RDP sequence. A total of 295 and 296 unique sequences for Streptococcus were observed for the original saliva and in vitro grown sample respectively. Total Streptococcus sequences matching the cultured and uncultured strains for each sample are shown in the inset.
Comparison of dominant sequences falling within the Streptococcus genus for the original saliva and the SHI medium cultivable saliva flora showed that all the streptococcus spp. within saliva sample, both cultured and previously listed as uncultured streptococcus spp. could be recovered by cultivating with newly developed SHI medium (Fig. 4).
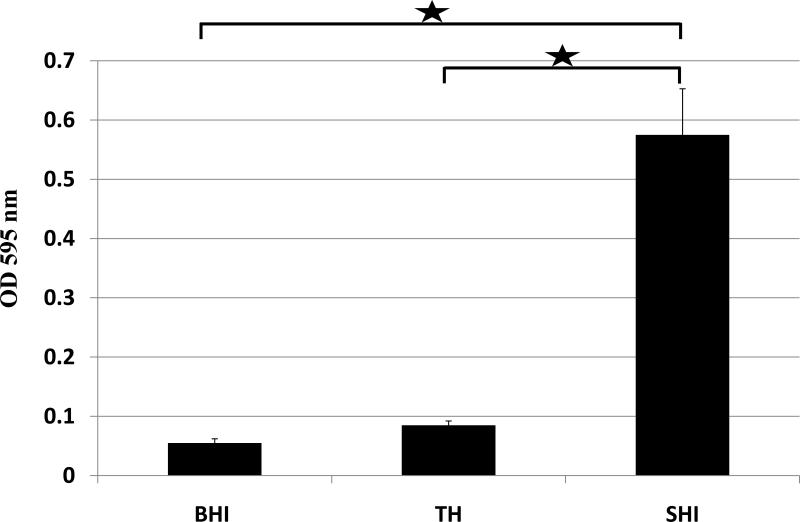
Shi medium allowed better biofilm fromation of saliva-derived microbes than TH and BHI medium
Above studies showed that SHI medium was able to support more diversified microbial growth than other commonly used lab media. We also wanted to assess the biofilm formation of salivary microbes when cultivated in different media. Crystal violet staining showed that biofilm in SHI medium gave the highest OD595 reading, which was significantly higher than those grown in BHI or TH media (p<0.05); while there was no difference between biofilm samples in BHI and TH (p>0.1) (Fig. 5). The result indicated that compared with BHI and TH media, SHI medium was able to allow better biofilm formation of saliva-derived microbes.
Figure 5.
The crystal violet assay to assess overnight biofilm formation of saliva-derived microbes cultivated in BHI, TH or SHI medium. Three replicates were performed for each assay. Average values ± SD are shown.  indicates significant difference (P< 0.05) between the two values.
indicates significant difference (P< 0.05) between the two values.
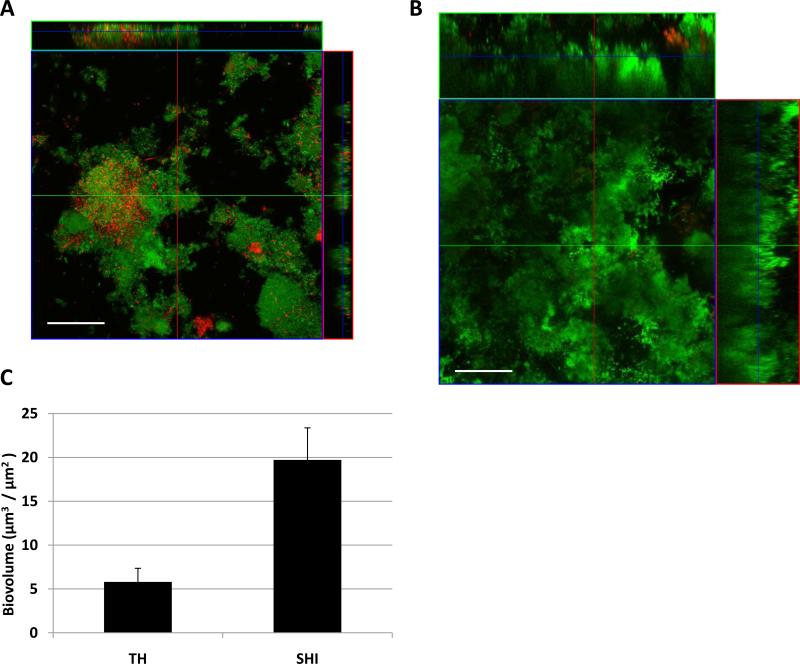
CLSM (confocal laser scanning microscopy) imaging and COMSTAT analysis of saliva-derived biofilms cultivated in different media
Results presented above implied that, compared with other commonly used lab media, SHI medium was superior in cultivating saliva-derived oral microbiota in terms of microbial diversity and biofilm it was able to support. To confirm this observation, we performed the confocal laser scanning microscopy imaging on the saliva-derived biofilm cultivated in different medium. CLSM imaging revealed that, saliva-derived biofilm grown in TH media was sparse with less surface coverage and much thinner compared with biofilm grown in SHI medium (Fig. 6A and 6B). COMSTAT analysis showed that, the biofilm grown in TH medium had an average biovolume of 5.8 ± 1.67 μm3 / μm2; while the average biovolume of SHI medium cultivated biofilm was 19.71 ± 3.42 μm3 / μm2 (Fig. 6C). Statistical analysis indicated that the difference in biovolume between the two media was significant (P < 0.001). Furthermore, after overnight growth in TH medium, more than 10% of the bacterial cells within biofilm were dead (red-stained), while the dead cells made up less than 5% of the total bacterial population within SHI medium cultivated biofilm. The result is consistent with crystal violet staining data, suggesting SHI medium was able to sustain the growth of saliva-derived biofilm with higher biomass.
Figure 6.
Comparison of saliva-derived biofilms grown in TH or SHI medium visualized by CLSM. Z-stack image of saliva-derived biofilm (live/dead stained) grown in (A) TH medium, and (B) SHI medium. Biovolume of biofilms formed using different media was calculated (C) using the software COMSTAT. Image of five randomly chosen spots were collected for each experimental sample, representative images are shown. Error bar in the calculated biovolume was standard deviation. Scale bar = 50 μm.
DISCUSSION
Developing medium for cultivating oral microbial flora has always been a challenge. This is largely due to the diversified bacterial species with different nutritional requirements residing within the oral cavity (Kolenbrander, 2000).
Saliva has been used in many in vitro systems as a nutrient source for sustaining the oral microbial flora (De Jong & Van Der Hoeven, 1987; Foster & Kolenbrander, 2004; Palmer et al., 2001). However, the use of natural saliva has many disadvantages: it is difficult to collect and sterilize in large quantities, and is variable in composition and properties. Furthermore, due to its limited contents of carbohydrates, saliva is not efficient in supporting the growth of complex oral microbiota. Several media have been developed to try to enhance the growth of complex oral microbial communities. In one effort to increase the growth yield of in vitro cultivation of dental plaque-derived oral microbial community, mucin, the principle glycoprotein in saliva, was added to the BM broth, a medium that had originally been developed for cultivating Bacteroides spp. isolated from human oral cavity (Shah et al., 1976). The mucin-containing new medium (BMM) greatly increased the growth yield, largely due to the increased number of anaerobes (Glenister et al., 1988). However, the recovery of certain oral species, such as veillonella decreased. Later on, Wong et al developed a chemically defined analogue of saliva (defined medium mucin, DMM) in an effort to substitute for BMM complex medium (Wong & Sissions, 2001). They demonstrated that DMM-grown dental plaque can achieve realistic growth rates and patterns similar to that obtained by BMM. Another frequently used medium in cultivating oral microbes is NAM (N-acetyl muramic acid) broth. It contains NAM, a monosaccharide derivative of N-acetylglucosamine, which has been shown to facilitate the growth of certain subgingival anaerobic bacteria, such as Tannerella forsythia and Porphyromonas gingivalis (Teles et al., 2008).
However, no detailed analysis has been done to compare the microbial profiles between medium-cultivated and original dental plaque or saliva samples, thus it is not clear how selective these media are in supporting oral community and whether the cultivated microbial flora can best represent the genetic diversity within plaque or saliva samples. In an effort to survey the ability of different media in sustaining the growth of oral microbes, we chose commonly used lab media, including BMM and NAM, to cultivate salivary microbial flora. PCR-DGGE analysis revealed that none of the tested media is able to support a community which maintains high diversity and similar community profile to the original microbial flora within saliva samples (Fig.1).
The traditional trial-and-error method in developing new medium also made the process very time-consuming and less efficient. In this study, we used DGGE as an index to help us developing new medium for best sustaining the genetic diversity of the oral community. By comparing the DGGE profiles of microbial flora cultivated by tested media with that of the original samples, we found that although the individual cultivated microbial pattern of PYG, BMM and sheep blood supplemented NAM medium is different, if combined, the cumulative pattern is very close to that of the original saliva sample. Based on this observation, we proposed that by adding the critical ingredients of the three media, we might be able to develop a new medium that better meet the nutritional requirement of more diversified salivary microbes, thus yield a cultivable flora which can better represent the microbial diversity within the salivary sample.
Using this as a guideline, we developed SHI medium by adding critical supplements from these broths to the base ingredients, which include peptone and yeast extract, the main components of the chosen media. Mucin and haemin from BMM were included in the new medium. Mucin is the principle glycoprotein of saliva and has been shown to be an important growth limiting substrate for the complex oral microflora (Glenister et al., 1988); while haemin has been demonstrated to be able to stimulate the growth of a variety of oral species, including cocci, rods and filaments bacteria (Gilmour & Poole, 1970). As a nutrient rich supplement, sheep blood was also added to the new medium to facilitate the growth of fastidious and slow growing, obligate anaerobic bacteria within the oral flora. Another main supplement is N-acetyl muramic acid from NAM broth, which has been shown to enhance the growth of certain gram-negative, anaerobic subgingival bacterial such as Tannerella forsythia (Wyss, 1989). Lastly, sucrose was used as a substitute for glucose, an ingredient from PYG, and it was shown to achieve a better recovery of the streptococci from the salivary samples than glucose (data not shown).
DGGE analysis showed that the new SHI medium sustained the growth of a microbial community with the closest profile to the original samples (Fig.2). 454 pyrosequencing results strongly support that SHI medium is able to maintain an in vitro salivary microbial community with high diversity and similar microbial profiles at the genus as well as at the species level (Fig.3). Importantly, the in vitro community, like the saliva sample, has a high percentage of uncultured representatives (85% and 83% respectively) (Fig. 4). Cristal violet staining and CLSM analysis of the biofilm showed that compared with commonly used lab media, such as TH broth, SHI medium is superior in yielding more biomass when used to cultivate saliva-derived biofilm (Fig.5 and Fig.6).
The in vitro community also contains several of the candidate phylum TM7 (0.06% of total) which have been referred to as biology's “dark matter” problem (Marcy et al., 2007). This phylum is a major focus of study because although they have been identified (via clone sequences) in a wide variety of habitats, researchers have yet to obtain a stable culture of any isolate. A number of novel approaches have been used to investigate TM7 organisms, including the use of microfluidic devices (Marcy et al., 2007) and cell separation by fluorescent in situ hybridization and flow cytometry (Podar et al., 2007) to obtain single cells for whole genome sequencing. The data presented here show that representatives of TM7 are actively growing in vitro environment and may provide for the first time the ability to study their biological function in a stable mixed community.
As with all other media, the newly developed SHI media also display certain level of selectivity. For example, based on 454 pyrosequencing analysis, sequences with hits in SR1 division, which accounted for 2.3% of the total sequence hits in original saliva samples were lost when cultivated in SHI medium. At the same time, some minor species within the original saliva samples, such as Treponema, Caenibacterium, Oribacterium, which accounted for less than 0.2% of the total sequence hits, also couldn’t be recovered from in vitro cultivation (data not shown). Nevertheless, the high degree of overlap between the two profiles at genus as well as species level indicated that SHI medium could be useful in establishing a representative model oral community, and facilitating the study of previously un-culturable oral bacterial species, such as TM7. Finally it is reasonable to suggest that this DGGE-guided method could be used to develop novel media not only for oral microbial communities but also for other complex microbial communities.
ACKNOWLEDGEMENTS
The authors acknowledge support from NIH (GM54666 to WS).
REFERENCES
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlaan EA, Sugimori M, Furukawa S, Takeuchi K. Profiling and monitoring of microbial populations by denaturing high-performance liquid chromatography. J Microbiol Meth. 2005;61:399–412. doi: 10.1016/j.mimet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acid Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GS, Costerton JW, Lamont RJ. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J Periodontal Res. 1998;33:323–327. doi: 10.1111/j.1600-0765.1998.tb02206.x. [DOI] [PubMed] [Google Scholar]
- Dahlen G. Role of suspected periodontopathogens in microbiological monitoring of periodontitis. Adv Dent Res. 1993;7:163–174. doi: 10.1177/08959374930070020701. [DOI] [PubMed] [Google Scholar]
- De Jong MH, Van Der Hoeven JS. The growth of oral bacteria on saliva. J Dent Res. 1987;66:498–505. doi: 10.1177/00220345870660021901. [DOI] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JS, Kolenbrander PE. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl Environ Microbiol. 2004;70:4340–4348. doi: 10.1128/AEM.70.7.4340-4348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto C, Maeda H, Kokeguchi S, Takashiba S, Nishimura F, Arai H, Fukui K, Murayama Y. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J Periodontal Res. 2003;38:440–445. doi: 10.1034/j.1600-0765.2003.02607.x. [DOI] [PubMed] [Google Scholar]
- Gilmour MN, Poole AE. Growth stimulation of the mixed microbial flora of human dental plaques by haemin. Arch oral biol. 1970;15:1343–1353. doi: 10.1016/0003-9969(70)90022-1. [DOI] [PubMed] [Google Scholar]
- Glenister DA, Salamon KE, Smith K, Beighton D, Keevil CW. Enhanced growth of complex communities of dental plaque bacteria in mucin-limited continuous culture. Microb Ecol Health Dis. 1988;1:31–38. [Google Scholar]
- He X, Wu C, Yarbrough D, Sim L, Niu G, Merritt J, Shi W, Qi F. The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol Microbiol. 2008;70:112–126. doi: 10.1111/j.1365-2958.2008.06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Neilsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiol. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Kinniment SL, Wimpenny JWT, Adams D, Marsh PD. Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiol. 1996;142:631–638. doi: 10.1099/13500872-142-3-631. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems1. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr. Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010 doi: 10.1038/nrmicro2381. Advance online publication. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, Schrenzel J, Francois P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Meth. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ku CYS, Xu J, Saxena D, Caufield PW. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res. 2005;84:559–564. doi: 10.1177/154405910508400614. [DOI] [PubMed] [Google Scholar]
- Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Nati Acad Sci. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- Marsh PD. Dental plaque: biological significance of a biofilm and community life-style. J Clin Periodontol. 2005;32:7–15. doi: 10.1111/j.1600-051X.2005.00790.x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu CH. Soil microbial community analysis using denaturing gradient gel electrophoresis. Soil Sci Soc Am J. 2007;71:562–571. [Google Scholar]
- Ohta K, Makinen KK, Loesche WJ. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun. 1986;53:213–220. doi: 10.1128/iai.53.1.213-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RJ, Jr., Kazmerzak K, Hansen MC, Kolenbrander PE. Mutualism versus independence: strategies of mixed-species oral biofilms In vitro using saliva as the sole nutrient source. Infect Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA, Holland T, Cotton D, Hauser L, Keller M. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah HN, Bowden GH, Hardie JM, Williams RAD. Comparison of the biochemical properties of Bacteroides melaninogenicus from human dental plaque and other sites. J Appl Microbiol. 1976;41:473–492. doi: 10.1111/j.1365-2672.1976.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Cox DR, Lerman LS, Myers RM. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Nati Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund SS. Criteria for the infectious agents in dental caries and periodontal disease. J Clin Periodontol. 1979;6:16–21. doi: 10.1111/j.1600-051x.1979.tb02114.x. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Cutress TW, Hoffman MP, Wakefield JSJ. A multi-station dental plaque microcosm (Artificial Mouth) for the study of plaque growth, metabolism, pH, and mineralization. J Dent Res. 1991;70:1409–1416. doi: 10.1177/00220345910700110301. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Wong L, Cutress TW. Patterns and rates of growth of microcosm dental plaque biofilms. Oral Microbiol Immunol. 1995;10:160–167. doi: 10.1111/j.1399-302x.1995.tb00137.x. [DOI] [PubMed] [Google Scholar]
- Tatsuji N, Takeyoshi K. Microbial etiology of periodontitis. Periodontology 2000. 2004;36:14–26. doi: 10.1111/j.1600-0757.2004.03671.x. [DOI] [PubMed] [Google Scholar]
- Teles FR, Haffajee AD, Socransky SS. The reproducibility of curet sampling of subgingival biofilms. J Periodontol. 2008;79:705–713. doi: 10.1902/jop.2008.070424. [DOI] [PubMed] [Google Scholar]
- Wade W. Unculturable bacteria--the uncharacterized organisms that cause oral infections. J R Soc Med. 2002;95:81–83. doi: 10.1258/jrsm.95.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, Sissions CH. A comparison of human dental plaque microcosm biofilms grown in an undefined medium and a chemically defined artificial saliva. Arch oral biol. 2001;46:477–486. doi: 10.1016/s0003-9969(01)00016-4. [DOI] [PubMed] [Google Scholar]
- Wyss C. Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect Immun. 1989;57:1757–1759. doi: 10.1128/iai.57.6.1757-1759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, Harmsen HJM, Kleinfelder JW, Rest ME, Degener JE, Welling GW. Denaturing gradient gel electrophoresis analysis to study bacterial community structure in pockets of periodontitis patients. Oral Microbiol Immunol. 2003;18:59–65. doi: 10.1034/j.1399-302x.2003.180110.x. [DOI] [PubMed] [Google Scholar]