Abstract
The heart forms as a linear heart tube that loops and septates to produce a mature four-chambered structure. The single vessel emerging from the embryonic heart, the truncus arteriosus, divides into the aorta and the pulmonary artery as part of this septation process, and a series of additional morphogenetic events result in the proper alignment and orientation of the cardiac outflow tract. Recent evidence indicates that this process involves the complex interactions of multiple cell types including primary and secondary heart fields, neural crest, pharyngeal mesenchyme, endoderm, and endothelium. Among the many signals that mediate tissue–tissue interactions during the formation of the outflow tract, we have focused on the role of the Notch signaling pathway. Here, we focus on recent advances in our understanding of Notch-mediated regulation of cardiac development with specific attention to the formation of the cardiac outflow tract.
Keywords: Congenital heart disease, Notch, Outflow tract, Neural crest, Second heart field
Introduction
Congenital heart disease is among the most common of all birth defects occurring as often as 1 in every 100 live births. About one-third of medically significant congenital cardiac defects involve the outflow tract of the heart, and these frequently require surgical repair. Both genetic and environmental influences affecting the occurrence of outflow tract defects have been implicated to cause disruption of the normal complex series of morphogenetic events that occur during embryonic cardiac development.
A significant advance in our understanding of how the cardiac outflow tract normally forms was provided by Margaret Kirby and colleagues in the 1980s when they demonstrated that neural crest cells contribute to the cardiac outflow tract and are required for septation of the truncus arteriosus [3, 19]. Ablation of pre-migratory neural crest results in a spectrum of cardiovascular disorders including persistent truncus arteriosus (PTA), double outlet right ventricle (DORV), and ventricular septal defect (VSD). Also included in this spectrum is tetralogy of fallot (TOF), a common congenital heart disorder that has four major components: valvular or subvalvular pulmonary stenosis, a VSD, a misaligned aorta, and right ventricular hypertrophy. In humans, these forms of congenital defects are seen both in isolation, and as part of more complex syndromes involving defects in other organs and tissues. Amongst the more common human syndromes involving outflow tract defects is DiGeorge syndrome, commonly associated with micro- or macro-deletions on chromosome 22q11 usually including deletion of TBX1, which has been strongly implicated in the etiology of this disorder [6]. Interestingly, TBX1 is not expressed in neural crest cells. Rather, it is expressed in second heart field derived myocardium, endoderm, and pharyngeal mesenchyme. In mouse models, loss of Tbx1 leads to secondary neural crest defect, underscoring the importance of tissue–tissue interactions during outflow tract formation [20]. Indeed, an ever-expanding number of mouse models of cardiac outflow tract defects have been described, and it is now clear that tissue-specific gene inactivation in a variety of tissues can result in quite similar forms of congenital heart disease. For example, while deletion of a bone morphogenetic protein receptor in the neural crest can cause outflow dysmorphology, an overlapping spectrum of defects can also be produced by deletion of the semaphorin receptor PlexinD1 in endothelial cells [17, 41]. As described in more detail below, manipulation of gene expression in the second heart field can also result in similar abnormalities. Hence, significant recent attention has focused on the interactions among these various cell types and the signaling pathways that mediate tissue–tissue communication.
Notch and Human Cardiovascular Disease
Among the many mechanisms by which cells and tissues communicate with one another, the Notch pathway has emerged as a potent mediator of cell fate determination and organogenesis. In humans, mutations in various components of the Notch signaling pathway have been associated with various cardiovascular disorders, including Alagille syndrome and cerebral autosomal dominant arteriopathy with subcortical infarct and leukoencephalopathy (CADASIL) syndrome [34, 38]. Alagille syndrome is a human disorder involving outflow tract cardiac defects. This syndrome is characterized by a spectrum of anomalies including congenital heart defects, such as peripheral pulmonary artery stenosis, aortic constriction, semilunar valve defects, and TOF, as well as impaired differentiation of intrahepatic bile ducts, skeletal defects, eye abnormalities, and kidney anomalies. Human mutations in Alagille syndrome have been identified in components of the Notch signaling pathway including NOTCH2 and JAGGED1, a ligand of the Notch receptor [38]. Recently, in a genome-wide survey of over 100 TOF patients and their unaffected parents, 11 de novo CNVs at ten unique loci were identified, including those at the NOTCH1 and JAGGED1 loci. This provides the first direct evidence that NOTCH1 mutations are associated with human TOF [10].
CADASIL syndrome is an autosomal dominant, vascular degenerative disease caused by mutations in the NOTCH3 receptor. It is characterized by non-atherosclerotic, amyloid-negative, angiopathy resulting in thickening of the small arteries of the brain, heart, and other visceral organs. Patients with CADASIL syndrome suffer from recurrent subcortical ischemic strokes, migraine headaches, and cognitive impairment. Other clinical features include early myocardial infarction, peripheral neuropathy as well as defects in renal function, eyesight, and hearing. Interestingly, the clinical course and prognosis of CADASIL, even within the same family, is quite variable. Studying the genetic modifiers of this disease is necessary to better understand vascular development and disease. Most mutations in the NOTCH3 gene, a gene with 33 exons, are in exons 3–6, 11, and 18–23 [34].
Notch mutations have also been associated with human aortic stenosis and bicuspid aortic valve [8]. The incidence of aortic stenosis increases with age in adults, and the incidence is also increased in the 2% of the population that have a bicuspid aortic valve [14]. NOTCH1 haploinsufficiency is associated with aortic valve disease including early calcification and bicuspid aortic valve disease, and heterozygous NOTCH1 mutations were associated with aortic valve calcification and aortic aneurysms [8]. However, two genome-wide linkage studies for valve calcification susceptibility loci suggest that NOTCH1 mutations do not account for all cases of calcific valve disease, and instead found several other loci that are associated with this phenotype [2, 23].
Notch Signaling
In humans and mice, there are 4 Notch receptors that are cell surface molecules capable of undergoing proteolytic cleavage upon ligand-mediated activation, which results in translocation of the intracellular domain to the nucleus and subsequent downstream gene activation. Ligands for Notch receptors include members of the Jagged and Delta families, which are membrane bound. Hence, Notch mediates communication between adjacent cells. Notch is a highly conserved pathway that plays fundamental roles in fate specification in organisms as diverse as flies and man. One important function of Notch signaling is to mediate lateral inhibition, in which one cell prevents neighboring cells from adopting similar fates by activating Notch. Notch is also important for lineage determination and boundary formation, and Notch plays fundamental roles in differentiation and multipotency in stem cells [12].
Upon activation, the intracellular domain of Notch (NICD) interacts with a number of nuclear proteins, including Mastermind-like protein (MAML) and recombination signal binding protein for immunoglobuluin J-kappa region (RBP-J), which are required to form an active transcription complex. Among the transcriptional targets of Notch are the Hrt and Hes family of genes and c-Myc [12, 39].
Downstream of Notch
Recent work using murine models has implicated Notch as a key mediator of signaling networks involved in outflow tract development. Deletion of the Notch ligand Jagged1, or inhibition of Notch signaling using a dominant negative MAML transgenic construct within cells of the second heart field, resulted in outflow tract abnormalities including PTA, DORV, and aortic arch artery patterning defects. Interestingly, inhibition of Notch signaling in cells of the second heart field affected morphogenesis of neighboring tissues, including faulty migration of cardiac neural crest cells and defective epithelial–mesenchymal transformation (EMT) within outflow tract endocardial cushions. Moreover, in these mutants we saw a down-regulation of Fgf8 and Bmp4 in second heart field derived myocardium. We were able to rescue defective EMT in an ex vivo assay by the addition of recombinant Fgf8, thereby implicating Notch as a critical mediator of Fgf8 signaling in the second heart field [13].
Fgf8, a member of the fibroblast growth factor (Fgf) family, is a soluble protein expressed in the cardiac crescent and in multiple tissues throughout development including pharyngeal endoderm, ectoderm, and second heart field [5, 18]. Fgf8 hypomorphs demonstrate defects in outflow tract formation and aortic arch patterning [1, 7, 25]. Loss of Fgf8 specifically in the second heart field results in similar abnormalities to those seen in global hypomorphs, suggesting a cell autonomous role for Fgf8 within this tissue [15, 28]. Inactivation of both Fgf receptor (Fgfr) 1 and 2, or loss of an adaptor protein FRS2α that is utilized by Fgf receptors, in second heart field precursors results in outflow tract abnormalities nearly identical to those seen in Fgf8 global hypomorphs, while unexpectedly inactivation of the receptors in neural crest does not result in a phenotype [29, 40]. These data suggest that Fgf8 signals in an autocrine loop within the second heart field, which may result in expression of another soluble factor that subsequently signals to the neural crest and/or endothelium to mediate outflow tract morphogenesis.
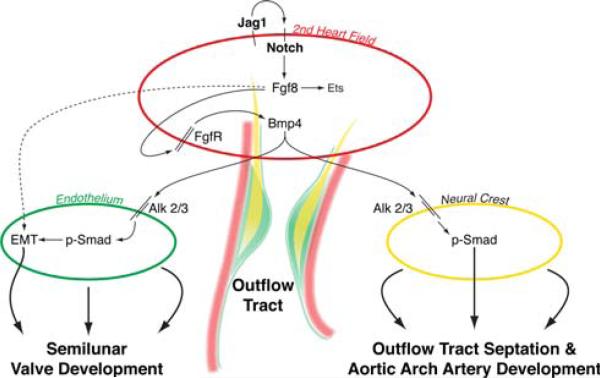
Bmp4 is one of the most strongly expressed members of the bone morphogenic protein family within the developing outflow tract, and recent studies suggest it may be responsible for transmitting the Fgf8 signal to the neural crest and endocardium [17]. Cardiac-specific deletion of Fgf8 results in a down-regulation of Bmp4 in the developing outflow tract, suggesting Bmp4 is downstream of Fgf8 [15, 22, 24, 29, 40]. Consistent with this observation, Bmp4 deletion in the second heart field results in PTA [24]. Deletion of the Bmp4 receptors Alk2 or Alk3 specifically in the neural crest results in outflow tract and aortic arch artery defects nearly identical to those described in the aforementioned Fgf8 mutants [17, 33]. The role of Bmp signaling to the neural crest is further supported by conditional deletion of Smad4, which functions downstream of activated Bmp receptors, resulting in PTA and hypocellular outflow tract cushions [16]. Evidence also exists to support a role for Bmp signaling to the outflow tract endocardium. Deletion of either Alk2 or Alk3 receptors in endocardium results in abnormal cellularization and defective EMT of the AV cushions [4, 9, 37]. We have developed a working model for the tissue-specific roles of various signaling pathways involved in outflow tract development (Fig. 1).
Fig. 1.
Tissue–tissue interactions during outflow tract development. The model depicts second heart field myocardium, endothelium undergoing EMT, and cardiac neural crest. Jagged1/Notch signaling in the second heart field is proposed to stimulate Fgf8, which functions within the second heart field to regulate downstream cascades including Bmp4, which in turn signals to endothelium and neural crest. Reprinted with permission from the Journal of Clinical Investigation [13]
The aforementioned studies do not elucidate whether Fgf8 is a direct target of Notch signaling, but recent studies raise the possibility that Hes1 may be an intermediary in a Notch-Fgf8 pathway. Rochais and colleagues discovered through enhancer trap analysis that Hes-1 is expressed in the pharyngeal mesoderm, including the region of the second heart field at 8.5 dpc. They went on to show that a significant percentage of Hes-1 mutant mice have outflow tract defects, including overriding aortas and ventricular septal defects. The authors note a shorter and straighter OFT in Hes1-/- embryos, as well as early reduction in proliferation of second heart field progenitors. Interestingly, this study also demonstrates a paucity of neural crest cells within the distal OFT, similar to what we describe in mutants with inactivated Notch signaling in the second heart field [31]. Tissue-specific deletion of Hes1 will further elucidate whether the effect on neural crest cells is indeed non-cell autonomous. Moreover, as Hes1 most often functions as a transcriptional repressor, if it is functioning as an intermediary in a Notch-Fgf8 pathway it may not be directly acting to up-regulate Fgf8 signaling, but rather may be repressing an unknown downstream repressor of Fgf8.
Notch and Valve Homeostasis
As mentioned earlier, Notch mutations have been implicated in human aortic stenosis. A mechanism for the premature calcification seen in NOTCH1 haploinsufficiency has been elucidated in a murine model. Notch is thought to repress an osteogenic program in murine aortic valves through repression of Bmp2 and Runx2, a transcriptional regulator of osteoblast cell fate [8, 26]. Nigam et al. found that knockdown of Bmp2 in sheep aortic valve interstitial cells blocked the calcification induced by Notch inhibition. Excitingly, periostin, which previously has been implicated in osteoblast differentiation and is expressed by valve precursors, has been implicated in regulating expression of the Notch ligand delta-like 1 homolog (Dlk1). In periostin null mice, Dlk1 is over-expressed in the embryonic OFT, and this over-expression negatively regulates Notch1 signaling leading to an induction of the osteoblast cell fate through Runx2 expression [36]. Therefore, periostin signaling through Notch represses a default osteogenic program in the OFT cushion mesenchyme and promotes development of the fibrogenic lineage.
In addition to a role in preventing valvular calcification, Notch is involved in normal valvular morphogenesis. Global Notch1 and RBP-J mutants have abnormal valvular morphogenesis as reflected by hypocellular endocardial cushions, defective EMT, and down-regulation of expression of Snail and Slug, two important mediators of EMT [27, 35]. Jagged1 ligand stimulation of endothelial cells is sufficient to induce EMT, whereas expression analysis suggests that Notch1 and Delta-like 4 may be active in the endothelium. Further analysis using conditional alleles will be instructive to determine the cellular requirements for Notch1 and RBP-J signaling in valve formation [27, 35].
Notch and Wnt
Crosstalk between Notch and Wnt signaling has been demonstrated in multiple contexts, and the two pathways are so often interconnected that the term “Wntch” has been proposed to describe the combined signaling module [11]. Nevertheless, the interactions between Notch and Wnt during cardiovascular morphogenesis are not well understood. Recent publications give tantalizing clues to the undoubtedly complex interplay of these pathways in the cardiovascular system. One such recent publication described the balance of Notch and Wnt signaling during angiogenesis. In endothelial stalk cells, Delta-like 4 induces expression of Notch-regulated ankyrin repeat (Nrarp) which serves to further limit Notch signaling and promote Wnt signaling in these cells. Ultimately, the balance between Notch and Wnt signaling helps to determine whether to make or break new vessel connections, thereby regulating retinal vessel density [30].
Both Notch and Wnt signaling are important regulators of cardiac development and OFT development. Some evidence to suggest that interplay in fact exists comes from deletion of Lrp6, an essential coreceptor for canonical Wnt signaling. Homozygous deletion of Lrp6 results in outflow tract defects and ventricular septal defects, with a reduction of Fgf8 signaling and hypocellular outflow tract cushions. The neural crest is clearly affected as well, as evidenced by blunting of Pax3 expression and loss of Msx1 and Msx2 expression [32]. Though the tissue-specific role of Lrp6 remains to be demonstrated, this phenotype is strikingly similar to the aforementioned studies where Notch signaling in the second heart field was perturbed, and both pathways appear to converge on Fgf8. It will be interesting to assess for perturbation of Notch signaling in Lrp6 mutant mice.
Further evidence for interaction between Notch and Wnt in outflow tract formation comes from studies in which Notch1 was deleted in the second heart field using Islet1-Cre. The absence of Notch1 in this region results in an expansion of cardiac progenitor cells in the ventral pharynx, reminiscent of the phenotype seen in mice with constitutively active β-catenin [21]. However, there are some differences between the two phenotypes, including a hypoplastic right ventricle in Notch1 mutants versus right ventricular enlargement in the constitutively active β-catenin mice, which may reflect a role for Notch in migration of the cells into the primitive right ventricle. Kwon et al. [21] further tested the interplay of Notch/Wnt pathways by inactivating Notch1 using siRNA in ES cells, and confirmed a concomitant increase in active phosphorylated β-catenin. Therefore, they suggest that β-catenin signaling promotes expansion of cardiac progenitor cells, while Notch1 activity may negatively regulate this process through control of phosphorylated β-catenin. Thus, emerging work is beginning to dissect the interplay of Notch/Wnt in cardiac outflow tract formation and this will likely be an area of much future investigation.
In this review, we have attempted to highlight some of the key cell types and signaling pathways involved in tissue–tissue interactions during outflow tract development. These studies provide initial insights into how second heart field precursors are communicating to neural crest and endothelium to orchestrate outflow tract formation. Continued efforts will be required to understand the temporal and spatial components of tissue–tissue interactions that must take place for proper cardiogenesis.
Acknowledgments
We would like to thank the members of the Epstein laboratory for many helpful discussions. This work was supported by the American Heart Association Physician-Scientist/Post-Doctoral fellowship (AHA0825548D) to R.J., the University of Pennsylvania, Division of Cardiology T-32 and Benjamin & Mary Siddons Measey Foundation to S.R., and NIH P01 HL075215 and funds from the WW Smith Endowed Chair for Cardiovascular Research to J.A.E.
Footnotes
Stacey Rentschler and Rajan Jain contributed equally to this work.
References
- 1.Abu-Issa R, Smyth G, Smoak I, Yamamura K-i, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 2.Bella JN, Tang W, Kraja A, Rao DC, Hunt SC, Miller MB, Palmieri V, Roman MJ, Kitzman DW, Oberman A, Devereux RB, Arnett DK. Genome-wide linkage mapping for valve calcification susceptibility loci in hypertensive sibships: the Hypertension Genetic Epidemiology Network Study. Hypertension. 2007;49:453–460. doi: 10.1161/01.HYP.0000256957.10242.75. [DOI] [PubMed] [Google Scholar]
- 3.Bockman DE, Kirby ML. Dependence of thymus development on derivatives of the neural crest. Science. 1984;223:498–500. doi: 10.1126/science.6606851. [DOI] [PubMed] [Google Scholar]
- 4.Choi M, Stottmann RW, Yang YP, Meyers EN, Klingensmith J. The bone morphogenetic protein antagonist noggin regulates mammalian cardiac morphogenesis. Circ Res. 2007;100:220–228. doi: 10.1161/01.RES.0000257780.60484.6a. [DOI] [PubMed] [Google Scholar]
- 5.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 6.Epstein JA. Developing models of DiGeorge syndrome. Trends Genet. 2001;17:S13–S17. doi: 10.1016/s0168-9525(01)02450-7. [DOI] [PubMed] [Google Scholar]
- 7.Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 9.Gaussin V, Van de Putte T, Mishina Y, Hanks MC, Zwijsen A, Huylebroeck D, Behringer RR, Schneider MD. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proceedings of the National Academy of Sciences. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck Mde L, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayward P, Kalmar T, Arias AM. Wnt/Notch signalling and information processing during development. Development. 2008;135:411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 12.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 13.High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest. 2009;119:1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 15.Ilagan R, Abu-Issa R, Brown D, Yang Y-P, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 16.Jia Q, McDill BW, Li SZ, Deng C, Chang CP, Chen F. Smad signaling in the neural crest regulates cardiac outflow tract remodeling through cell autonomous and non-cell autonomous effects. Dev Biol. 2007;311:172–184. doi: 10.1016/j.ydbio.2007.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- 18.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 19.Kirby ML. Cardiac morphogenesis—recent research advances. Pediatr Res. 1987;21:219–224. doi: 10.1203/00006450-198703000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Kochilas L, Merscher-Gomez S, Lu MM, Potluri V, Liao J, Kucherlapati R, Morrow B, Epstein JA. The role of neural crest during cardiac development in a mouse model of DiGeorge syndrome. Dev Biol. 2002;251:157–166. doi: 10.1006/dbio.2002.0819. [DOI] [PubMed] [Google Scholar]
- 21.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc Natl Acad Sci USA. 2004;101:4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin LJ, Ramachandran V, Cripe LH, Hinton RB, Andelfinger G, Tabangin M, Shooner K, Keddache M, Benson DW. Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Hum Genet. 2007;121:275–284. doi: 10.1007/s00439-006-0316-9. [DOI] [PubMed] [Google Scholar]
- 24.McCulley DJ, Kang J-O, Martin JF, Black BL. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev Dyn. 2008;237:3200–3209. doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 26.Nigam V, Srivastava D. Notch1 represses osteogenic pathways in aortic valve cells. J Mol Cell Cardiol. 2009;47:828–834. doi: 10.1016/j.yjmcc.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noseda M, McLean G, Niessen K, Chang L, Pollet I, Montpetit R, Shahidi R, Dorovini-Zis K, Li L, Beckstead B, Durand RE, Hoodless PA, Karsan A. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ Res. 2004;94:910–917. doi: 10.1161/01.RES.0000124300.76171.C9. [DOI] [PubMed] [Google Scholar]
- 28.Park EJ, Ogden LA, Talbot A, Evans S, Cai C-L, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phng LK, Potente M, Leslie JD, Babbage J, Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G, Gerhardt H. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev Cell. 2009;16:70–82. doi: 10.1016/j.devcel.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rochais F, Dandonneau M, Mesbah K, Jarry T, Mattei MG, Kelly RG. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS One. 2009;4:e6267. doi: 10.1371/journal.pone.0006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song L, Li Y, Wang K, Zhou CJ. Cardiac neural crest and outflow tract defects in Lrp6 mutant mice. Dev Dyn. 2009;239:200–210. doi: 10.1002/dvdy.22079. [DOI] [PubMed] [Google Scholar]
- 33.Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang SC, Jeng JS, Lee MJ, Yip PK. Notch signaling and CADASIL. Acta Neurol Taiwan. 2009;18:81–90. [PubMed] [Google Scholar]
- 35.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tkatchenko TV, Moreno-Rodriguez RA, Conway SJ, Molkentin JD, Markwald RR, Tkatchenko AV. Lack of periostin leads to suppression of Notch1 signaling and calcific aortic valve disease. Physiol Genomics. 2009;39:160–168. doi: 10.1152/physiolgenomics.00078.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Sridurongrit S, Dudas M, Thomas P, Nagy A, Schneider MD, Epstein JA, Kaartinen V. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev Biol. 2005;286:299–310. doi: 10.1016/j.ydbio.2005.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warthen DM, Moore EC, Kamath BM, Morrissette JJ, Sanchez P, Piccoli DA, Krantz ID, Spinner NB. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27:436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- 39.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Lin Y, Zhang Y, Lan Y, Lin C, Moon AM, Schwartz RJ, Martin JF, Wang F. Frs2{alpha}-deficiency in cardiac progenitors disrupts a subset of FGF signals required for outflow tract morphogenesis. Development. 2008;135:3611–3622. doi: 10.1242/dev.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Singh MK, Degenhardt KR, Lu MM, Bennett J, Yoshida Y, Epstein JA. Tie2Cre-mediated inactivation of plexinD1 results in congenital heart, vascular and skeletal defects. Dev Biol. 2009;325:82–93. doi: 10.1016/j.ydbio.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]