Abstract
Direct inhibition of transcription factor complexes remains a central challenge in the discipline of ligand discovery. In general, these proteins lack surface involutions suitable for high-affinity binding by small molecules. Here we report the design of synthetic, cell-permeable, stabilized α-helical peptides that target a critical protein–protein interface in the NOTCH transactivation complex. We demonstrate that direct, high-affinity binding of the hydrocarbon-stapled peptide SAHM1 prevents assembly of the active transcriptional complex. Inappropriate NOTCH activation is directly implicated in the pathogenesis of several disease states, including T-cell acute lymphoblastic leukaemia (T-ALL). The treatment of leukaemic cells with SAHM1 results in genome-wide suppression of NOTCH-activated genes. Direct antagonism of the NOTCH transcriptional program causes potent, NOTCH-specific anti-proliferative effects in cultured cells and in a mouse model of NOTCH1-driven T-ALL.
Transcription factors are master regulators of cell state. Commonly activated by genetic events or upstream signalling pathways, they mediate the neoplastic phenotype and maintain tissue specification in cancer. As such, they are highly desirable targets for ligand discovery1. Owing to expansive protein–protein interfaces and a general absence of hydrophobic pockets, transcription factors have proven among the most chemically intractable of all therapeutic targets. With the exception of nuclear hormone receptors that have evolved the ability to bind natural small-molecule ligands, potent and specific inhibitors of human transcription factors have not been realized. Here we report the successful development of a direct-acting antagonist of an oncogenic transcription factor, NOTCH1.
NOTCH proteins participate in conserved pathways that regulate cellular differentiation, proliferation and death2,3. Mammalian NOTCH receptors (NOTCH1–4 in humans) are single-pass transmembrane proteins that transmit juxtacrine signals initiated by ligands of the Delta, Serrate or Lag-2 family. Ligand binding to the extracellular domain of NOTCH1 initiates sequential proteolytic processing events catalysed respectively by an ADAM family metalloprotease and a γ-secretase complex, resulting in cytoplasmic release of the intracellular domain of NOTCH1 (ICN1)4–6. ICN1 then translocates to the nucleus and loads onto the DNA-bound transcription factor CSL7. The engagement of ICN1 with CSL creates a long, shallow groove along the interface of the two proteins that serves as a binding surface for co-activator proteins of the mastermind-like (MAML) family8,9. The resulting ICN–CSL–MAML ternary complex then recruits the core transcription machinery, effecting activation of NOTCH-dependent target genes.
The duration and strength of NOTCH signalling is normally tightly controlled. Whereas loss-of-function mutations have been observed in a variety of diseases10–12, gain-of-function mutations in the NOTCH pathway are causally linked with cancer. Indeed, human NOTCH1 was first discovered owing to its involvement in a t(7;9) chromosomal translocation observed in patients with T-ALL13. Subsequently, various activating mutations in NOTCH1 have been discovered in >50% of patients with T-ALL14. Recently, further aberrations that potentiate NOTCH signalling have been identified, including loss-of-function of the NOTCH1 E3 ubiquitin ligase FBXW7 and the intracellular NOTCH inhibitor NUMB, in T-ALL and breast adenocarcinoma, respectively15,16. Activated NOTCH signalling has also been observed in cancers of the lungs, ovaries, pancreas and gastrointestinal tract as well as in melanoma, multiple myeloma and medulloblastoma17–23.
Efforts to antagonize the NOTCH pathway have relied on blocking the generation of ICN using small-molecule inhibitors of the γ-secretase complex (GSIs)24,25. These molecules are not strictly NOTCH-specific, as they indiscriminately block the many signalling pathways downstream of γ-secretase26. T-ALL patients treated with GSIs suffer dose-limiting gastrointestinal toxicity, the origin of which is uncertain but it may result from chronic blockade of NOTCH1 and NOTCH2 proccessing27. Some cell lines containing activating NOTCH1 mutations are resistant to GSIs, and those that do respond commonly undergo growth arrest rather than apoptosis. These observations underscore the mechanistic utility and potential therapeutic value of NOTCH antagonists that act by directly targeting the NOTCH transactivation complex.
Stapled α-helical peptides targeting the NOTCH complex
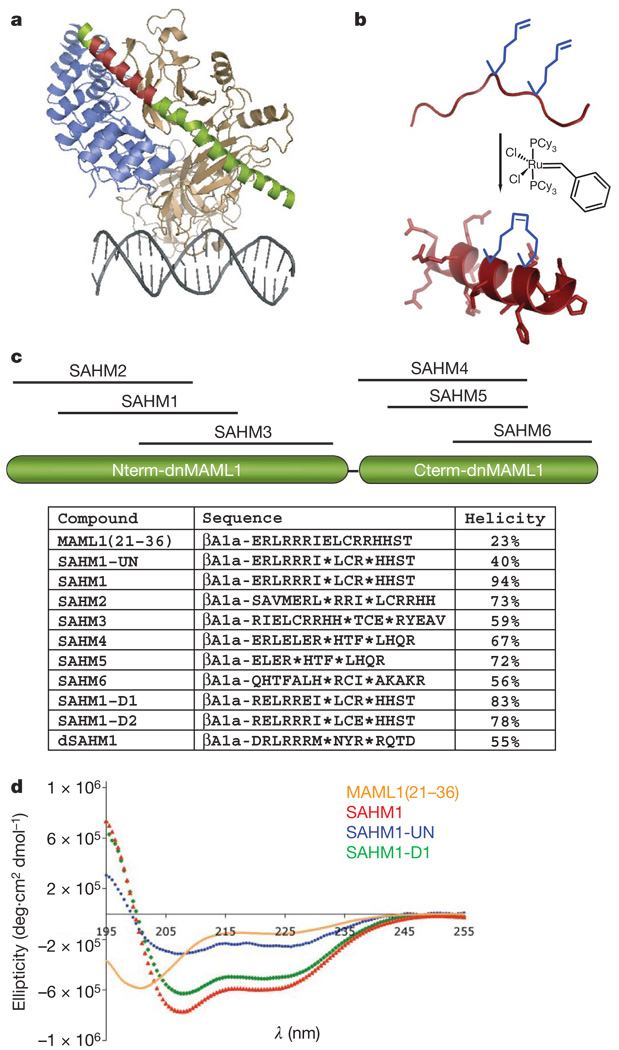
A dominant-negative fragment of MAML1 (residues 13–74; termed dnMAML1) has been shown to antagonize NOTCH signalling and cell proliferation when expressed in T-ALL cell lines28,29. In X-ray structures of the Homo sapient30 and Caenorhabditis elegans31 core ICN–CSL–MAML complexes, this dnMAML1 polypeptide forms a nearly continuous α-helix that engages an elongated groove formed by the assembly of ICN1 and CSL (Fig. 1a). This α-helical interaction motif suggested that the NOTCH transactivation complex might be suitable for targeting by helix-mimetics such as hydrocarbon-stapled α-helical peptides, which in other systems have proven capable of targeting intracellular protein–protein interactions32,33. Specifically, we reasoned that a stapled fragment of dnMAMLl might prevent binding of full-length MAML1 to the ICN1–CSL complex, thereby depriving the complex of its transcriptional activation function despite the presence of an upstream NOTCH signal. Relative to unmodified peptides, hydrocarbon-stapled peptides have shown improved binding affinity, metabolic stability and serum half-life. Operationally, peptide stapling is accomplished by co-synthetic incorporation of a non-natural amino acid, S5, at neighbouring positions along one face of the α-helix (i and i + 4 positions), followed by ring-closing olefin metathesis (Fig. 1b).
Figure 1. Design of MAML1-derived stapled peptides targeting NOTCH1-CSL.
a, Structure of the NOTCH1 ternary complex (Protein Data Bank (PDB) accession 2F8X): CSL (tan), DNA (grey), dnMAML1 (green) and ICN1 (blue). The 16-amino-acid stretch of MAML1 targeting ICN1 and CSL is shown in red, and was used to design the stapled peptide SAHM1. b, Schematic of peptide stapling. A non-natural alkenyl amino acid (S5) is incorporated at two positions in the peptide chain and then cross-linked by ring-closing olefin metathesis. c, Schematic, sequences and helical character of MAML1-derived SAHM peptides. βAla denotes a β-alanine spacer. Asterisks denote the location of S5 residues, which are cross-linked in all SAHM peptides except SAHM1-UN. d, Circular dichroism spectroscopy of four MAML1 derived peptides illustrating the incremental effects of synthetic modification.
The structure of the human NOTCH1 ternary complex was used as the basis for the design of a series of stapled α-helical peptides derived from MAML1 (SAHMs). Six candidate peptides were designed that together scan the entire contact surface of dnMAML1 with ICN1–CSL (Fig. 1c). Functional evaluation of this panel led to the selection of a stapled peptide designated SAHM1, which spans residues Glu 21 to Thr 36. As shown in Fig. 1d and Supplementary Fig. 1, stapling conferred upon SAHM1 a marked increase in helical character, as compared with its unmodified and modified but unstapled counterparts (MAML1(21–36) and SAHM1-UN, respectively). As specificity controls for functional studies34, we generated mutant peptides SAHM1-D1 and SAHM1-D2, as well as dSAHM1, the corresponding peptide from the Drosophila Mastermind (MAM) protein.
Analysis of cell penetration using fluorescein isothiocyanate (FITC)-labelled peptides and quantitative epifluorescence microscopy showed robust and roughly equivalent cellular uptake by FITC–SAHM1, FITC–SAHM2, FITC–SAHM3, FITC–SAHM6 and the negative control peptides FITC–SAHM-D1, FITC–SAHM-D2 and FITC–dSAHM1. Cell penetration was less efficient for FITC–SAHM4 and negligible for FITC–-SAHM5 (Supplementary Fig. 2a, b). The uptake of FITC–SAHM1 was diminished at 4 °C relative to 37 °C, and was unaffected by rhodamine conjugation, consistent with an active, endocytic peptide import mechanism as observed previously with stapled peptides (Supplementary Fig. 2c, d). The molecules in this study are considerably smaller than the ~25-kilodalton (kDa) exclusion size of the nuclear pore complex, and thus as expected, FITC–SAHM1 was distributed throughout the cytoplasm and nucleus (Supplementary Fig. 2e).
SAHMs bind the ICN1–CSL complex competitively with MAML1
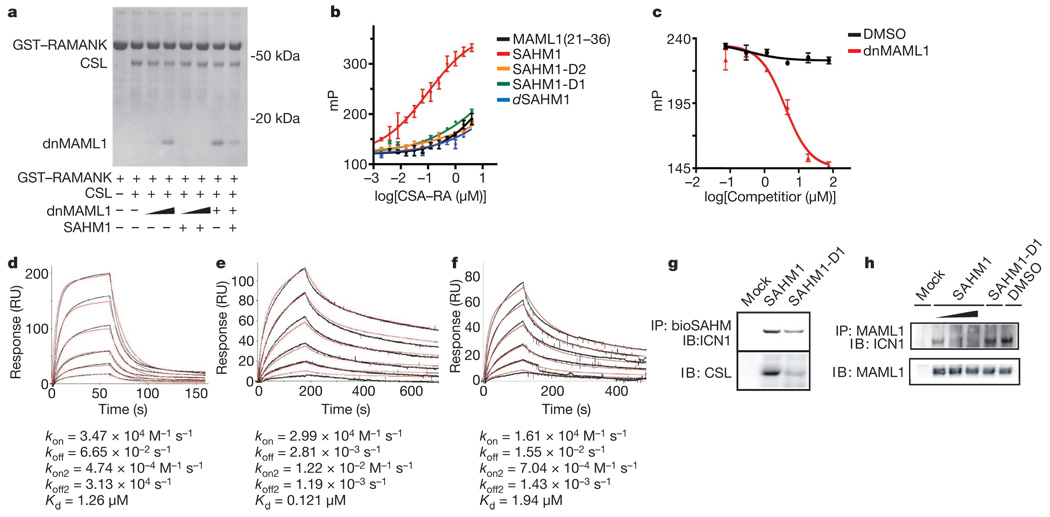
SAHM binding to the NOTCH complex was first investigated using an in vitro pull-down assay. In brief, a glutathione-S-transferase (GST)-tagged fragment of ICN1 bearing only the CSL-binding domain (GST–RAMANK) was immobilized on glutathione-labelled agarose beads and used to precipitate CSL and dnMAML1 in the presence and absence of SAHM1 (Fig. 2a). SAHM1 inhibited dnMAML1 binding competitively, but did not perturb binding of CSL to RAMANK.
Figure 2. SAHM1 specifically engages the NOTCH1 transactivation complex.
a, In vitro assembly of the NOTCH1 complex. Bead-immobilized RAMANK protein was incubated as indicated with CSL (0.5 µM), dnMAML1 (0.5 µM for lanes 3 and 5 (from left); 2.5 µM for lanes 4 and 6; 5 µM for lanes 7 and 8), and SAHM1 (10 µM). Bound proteins were washed, eluted and resolved by gel electrophoresis (Coomassie). b, Fluorescence polarization of FITC–SAHM peptides binding to RAMANK–CSL. c, Direct competition between unlabelled dnMAML1 and FITC–SAHM1. Concentrations of FITC–SAHM1 (15 nM) and RAMANK–CSL (0.6 µM) were held constant. dnMAML1 IC50 = 3.9 ± 0.9 µM. d, CSL binding to immobilized RAMANK by SPR. Black curves represent sensogram data and the red curve denotes fit to a two-step kinetic model. Binding constants are shown. kon, association rate; koff, dissociation rate; RU, response units. e, f, Binding of RAMANK–CSL complexes to immobilized bioSAHM1 (e) and bioSAHM1-D1 (f). g, bioSAHM1 and bioSAHM1-D1 pull-down assays in KOPT-K1 lysates. Bound protein fractions were probed with antibodies specific for ICN1 (top) and CSL (bottom). h, Competitive co-immunoprecipitation of endogenous ICN1 by MAML1 in the presence of vehicle, SAHM1 (0.5, 1 and 10 µM from left to right) or SAHM1-D1 (10 µM). Unless noted otherwise data represent the mean ± s.d. (n = 3).
SAHM binding was next measured quantitatively using fluorescence polarization spectroscopy. Soluble FITC–SAHM1 was found to bind the pre-formed RAMANK–CSL complex with a dissociation constant (Kd) = 0.12 ± 0.02 µM, whereas the unmodified FITC–MAML1(21–36) peptide bound the complex with markedly diminished affinity (Fig. 2b). Thus, helix stabilization promotes target binding. The mutant peptides FITC–SAHM1-D1, FITC–SAHM1-D2 and FITC–dSAHM1 bound RAMANK–CSL considerably less avidly than FITC–SAHM1, prompting the use of these peptides as negative controls in subsequent functional studies. Notably, fluorescence polarization of FITC–SAHM1 was decreased by unlabelled competitor dnMAML1, confirming the overlap between the SAHM1 and the dnMAML binding sites on RAMANK–CSL (Fig. 2c).
Kinetic insights into complex assembly and SAHM binding were gained using surface plasmon resonance (SPR). A constant amount of GST–RAMANK was immobilized on an immunoglobulin (anti-GST) biosensor surface and exposed to increasing concentrations of CSL. The resulting dose-dependent response data were best fit to a two-step association with rapid on- and off-rates and overall micromolar affinity (Fig. 2d), and are consistent with a published model supporting a two-step association34. Next, SPR was used to analyse binding of SAHMs to the pre-assembled RAMANK–CSL complex. A SAHM1 derivative containing an amino-terminal biotin moiety (bioSAHM1) was immobilized on a streptavidin sensor surface. Pre-incubated, equimolar RAMANK and CSL were introduced in increasing concentrations. Curve-fitting of these kinetic data indicated that bioSAHM1 bound RAMANK–CSL with an apparent Kd closely matching the value observed by fluorescence polarization (Fig. 2e). Again, SAHM1-D1 showed substantially decreased affinity (Fig. 2f).
To determine whether SAHMs bind endogenous NOTCH proteins present in human T-ALL cells, we performed pull-down assays in KOPT-K1 cellular lysates. BioSAHM1 beads were found to pull down full-length ICN1 and CSL, whereas bioSAHM1-D1 was markedly less avid (Fig. 2g). Next we examined the ability of SAHMs to compete with endogenous MAML1, again in KOPT-K1 cellular lysates. After immunoprecipitation of MAML1, the presence of ICN1 was detected by immunoblot. This association was diminished in a dose-dependent manner by SAHM1 but not SAHM-1D (Fig. 2h). Together, these experiments demonstrate conclusively that SAHM1 binds ICN1 and CSL and thereby directly antagonizes recruitment of the MAML1 co-activator to the ICN1–CSL complex.
SAHM1 specifically represses NOTCH1 target gene expression
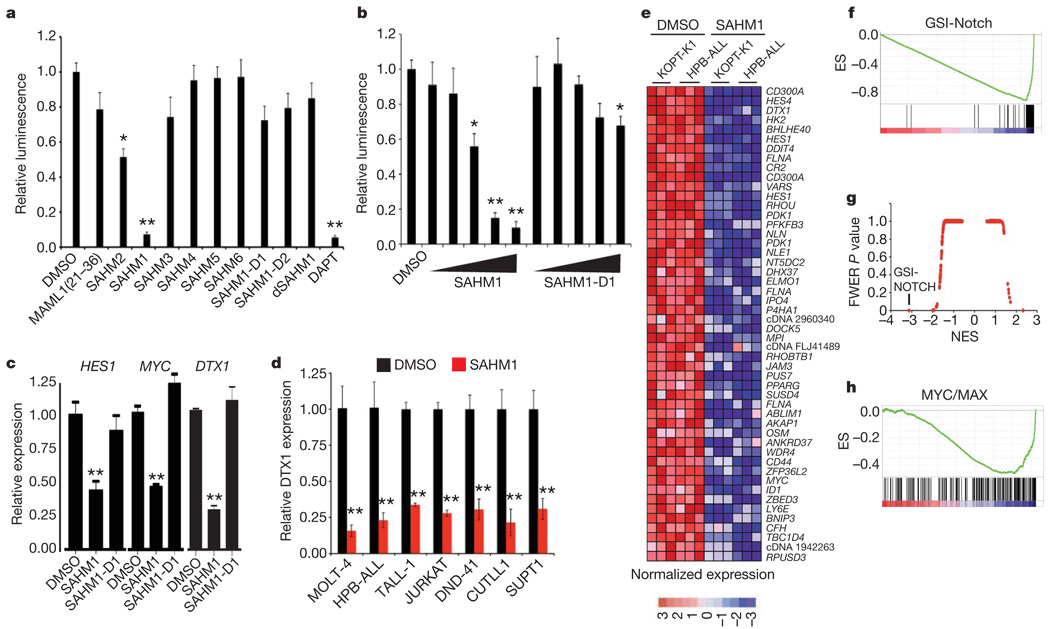
To explore the effects of SAHMs on transcriptional activity, we first used an established reporter-gene assay in which firefly luciferase is transcriptionally regulated by constitutively activated NOTCH1 (ref. 35). SAHM1 treatment resulted in near complete repression of luciferase activity, comparable to a known GSI (DAPT)25. SAHM2 conferred a twofold signal reduction. SAHM4, SAHM5, SAHM6, SAHM1-D1, SAHM1-D2 and dSAHM1 were inactive (Fig. 3a). Reporter gene repression by SAHM1 was observed to be dose-dependent, with a half-maximum inhibitory concentration (IC50) of 6.5 ± 1.6 µM (Fig. 3b). In a second reporter-gene assay monitoring β-lactamase transcriptionally regulated by ICN1 (ref. 36), SAHM1 C dose-dependent repression compared to SAHM1-D1 or vehicle alone (Supplementary Fig. 3a).
Figure 3. SAHM1 represses NOTCH1 target gene expression.
a, Inhibition of a NOTCH1-dependent luciferase reporter by SAHM peptides. Signal was normalized to Renilla luciferase control. b, Dose-dependent effects of SAHM1 and SAHM1-D1 in the dual-luciferase assay, using threefold dilutions (0.55–45 µM) of ligand compared to vehicle alone. c, qRT–PCR analysis of the HES1, MYC and DTX1 mRNA levels in KOPT-K1 cells treated for 24 h with SAHM1 or SAHM1-D1 (20 µM) relative to dimethylsulphoxide (DMSO) control. d, qRT–PCR analysis of DTX1 mRNA levels in a panel of human T-ALL cell lines. e, Heat map representation of the top 50 downregulated genes (P < 0.001), induced by SAHM1 in KOPT-K1 and HPB-ALL cells. f, Quantitative comparison of genes downregulated by GSI (GSI-NOTCH gene set) with the SAHM1 gene expression profile in KOPT-K1 and HPB-ALL cells by GSEA. g, Comparison of all transcription factor target gene sets in the Molecular Signatures Database to the GSI-NOTCH gene set for enrichment in the SAHM1 expression profile by GSEA. Data are plotted as the family-wise error rate (FWER) P value versus the NES. GSI-NOTCH is marked as the most enriched gene set. h, GSEA of the second most enriched gene set (MYC/MAX), applied to the SAHM1 expression profile. Unless noted otherwise data represent the mean ± s.d. (n = 3). *P < 0.05, **P < 0.01.
We next studied the effect of SAHMs on the expression of NOTCH target genes in NOTCH1-dependent T-ALL, quantified by PCR with reverse transcription (RT–PCR). A human T-ALL cell line (KOPT-K1) containing activating mutations in the heterodimerization (HD) and degradation domains (PEST) of NOTCH1 was treated with SAHM1, SAHM1-D1 and vehicle. Decreased expression of the NOTCH1 target genes HES1, MYC and DTX1 was uniquely observed after treatment with SAHM1 (Fig. 3c). A consistent repressive effect of SAHM1 on NOTCH1-dependent gene expression was observed across a panel of human T-ALL cell lines (Fig. 3d), containing diverse mutant NOTCH1 alleles (Supplementary Table 1). SAHM1 had no effect on ICN1 protein stability compared to DAPT (Supplementary Fig. 3b), supporting transcriptional inhibition downstream of ICN1 production.
SAHM1 triggers global suppression of NOTCH1 signalling
Transcriptional profiling has recently been demonstrated to establish mechanistic connectivity between dissimilar compounds acting on common targets or target pathways37. Gene set enrichment analysis (GSEA) has also emerged as a robust method for comparing expression profiles corresponding to defined cellular states38. Integrating these two methodologies, we performed gene expression profiling as multidimensional phenotyping to first measure the global transcriptional effects of SAHM1 and then compare these effects with those produced by GSI treatment. First, triplicate data sets were generated from SAHM1 and vehicle-treated KOPT-K1 and HPB-ALL cells and analysed on Affymetrix oligonucleotide microarrays. Supervised hierarchical clustering and rank-ordering identified numerous canonical NOTCH1 target genes including HES4, DTX1, HES1 and MYC among the top downregulated genes by SAHM1 (Fig. 3e). Next, a set of transcripts downregulated by GSI in T-ALL cell lines was curated from a published expression profile (referred to here as the GSI-NOTCH gene set)39. Enrichment for this gene set within the SAHM1 expression profile was studied by GSEA, revealing a strong, statistically significant correlation (enrichment score (ES) = −0.89, normalized ES (NES) = −3.66, P < 0.0001, Fig. 3f). Leading-edge analysis identified further enrichment for transcripts annotated as NOTCH1 targets in the scientific literature (Supplementary Fig. 4a). As a measure of biological specificity, enrichment analysis was conducted on all transcription factor target gene sets available in the Molecular Signatures Database (MSigDB). Taken together, the GSI-NOTCH gene set emerged as a statistical outlier as the most enriched in the SAHM1 profile (Fig. 3g and Supplementary Table 2). Interestingly, analysis of SAHM1-repressed genes also identified significant enrichment for gene sets regulated by transcriptional activators such as MYC/MAX and E2F, which have been previously identified as downstream targets of NOTCH activation (Fig. 3h and Supplementary Table 2)40,41. Taken together, these data establish that SAHM1 exerts a specific antagonistic effect on gene expression driven by NOTCH.
NOTCH inhibition halts the proliferation of T-ALL cells
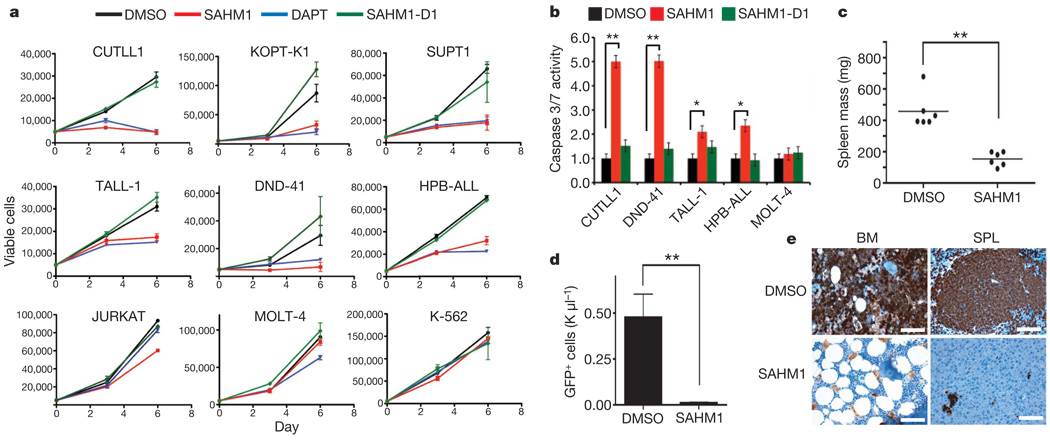
Previous work has shown that chemical (GSIs) or genetic (dnMAML1 expression) inhibition of NOTCH induces cell-cycle arrest, apoptosis and decreased proliferative capacity in a subset of T-ALL cell lines29,42. We assembled a panel of genetically annotated T-ALL cell lines, which have previously demonstrated sensitivity to GSI (CUTLL1, SUPT1, HPB-ALL, TALL-1, DND-41 and KOPT-K1; Supplementary Table 1). SAHM1 treatment in these cells produced a marked reduction in cell proliferation whereas SAHM1-D1 was inactive (Fig. 4a). SAHM1 treatment did not affect the proliferation of K562 cells, an erythroleukaemia cell line without dependency on NOTCH1 for growth, or JURKAT and MOLT-4 cells, both of which are T-cell lines bearing activating NOTCH1 mutations but are insensitive to NOTCH inhibition owing to the loss of PTEN15,41,43. In sensitive T-ALL cell lines, SAHM1 exposure prompted activation of caspase 3 and 7, consistent with the induction of apoptosis (Fig. 4b).
Figure 4. SAHM1 reduces T-ALL proliferation and leukaemic initiation potential.
a, Growth effects of SAHMs on a panel of human T-ALL cell lines of known mutational status. Cells were incubated with 15 µM DAPT (blue triangles), SAHM1 (red squares), SAHM1-D1 (green diamonds) or DMSO (black circles) and monitored for proliferation after 3 and 6 days of culture. Data points are mean ± s.d. (n = 3). b, Effects of SAHMs on apoptosis of T-ALL cells monitored using Capase-glo 3/7 (Promega) in cultures carried out as in a. Error bars, s.d. c, d, Ex-vivo treatment of L1601PΔP cells with SAHM1 (5 µM, 12 h) limits leukaemia initiation in secondary murine recipients. Reduction of spleen weight in the SAHM1 cohort (n = 6) compared to vehicle (n = 6) at the first sign of disease toxicity (23 days) (P = 0.001) (c). Circulating GFP-positive cell count in the blood is reduced by ~100-fold in the SAHM1 cohort (n = 6) relative to vehicle (n = 6) (P = 0.0026) (d). Error bars, s.d. Statistical analyses performed with a two-tailed t-test. *P < 0.01, **P < 0.005. e, An immunohistochemical stain for GFP that imparts a brown colour shows extensive leukaemic infiltration of bone marrow (BM) and spleen (SPL) in representative mice receiving vehicle-treated transplants relative to SAHM1. SPL, spleen. Scale bars,
These findings suggested that even brief exposure of T-ALL cells to SAHM1 might be sufficient to prevent the establishment of leukaemia in vivo. To recapitulate the human disease genotype and phenotype, we devised a murine model of T-ALL induced by the NOTCH1 allele L1601PΔP, which bears dual NOTCH1 mutations originally identified in human T-ALL cells (Supplementary Fig. 5). Retroviral infection of the L1601PΔP allele into whole bone marrow followed by transplantation into lethally irradiated syngeneic recipient mice resulted in development of a T-ALL phenotype; primary cells demonstrated sensitivity to SAHM1 treatment in vitro (Supplementary Fig. 6a–c). To determine whether SAHM1 treatment could curb leukaemic engraftment, we pre-treated primary L1601PΔP cells with SAHM1 or vehicle before inoculation into secondary recipient mice (Supplementary Fig. 6d). At study end point, the SAHM1-treated cohort showed a statistically significant reduction in spleen weight (Fig. 4c) and the absolute number of donor-derived circulating lymphoblasts (Fig. 4d). Histopathological examination of bone marrow and spleen demonstrated a marked reduction in disease burden and leukaemic infiltration among animals administered SAHM1-treated cells (Fig. 4e).
SAHM1 inhibits leukaemic progression and NOTCH1 signalling
To evaluate whether SAHM1 treatment could attenuate the growth of established T-ALL in vivo, we developed a bioluminescent murine model of T-ALL. Either MIG-L1601PΔP or the control MIG retrovirus was used to infect bone marrow isolated from C57BL/6 mice in which firefly luciferase was constitutively expressed from the ubiquitin C promoter44. Infected cells were then transplanted into isogenic C57BL/6-TyrC/C albino recipients to facilitate non-invasive bioluminescence imaging45. Animals transplanted with Luc-L1601PΔP cells developed progressive T-ALL (Fig. 5a) that was quantified by serial imaging (Fig. 5b), whereas no measurable disease developed in mice receiving control-transduced cells (Fig. 5a, b).
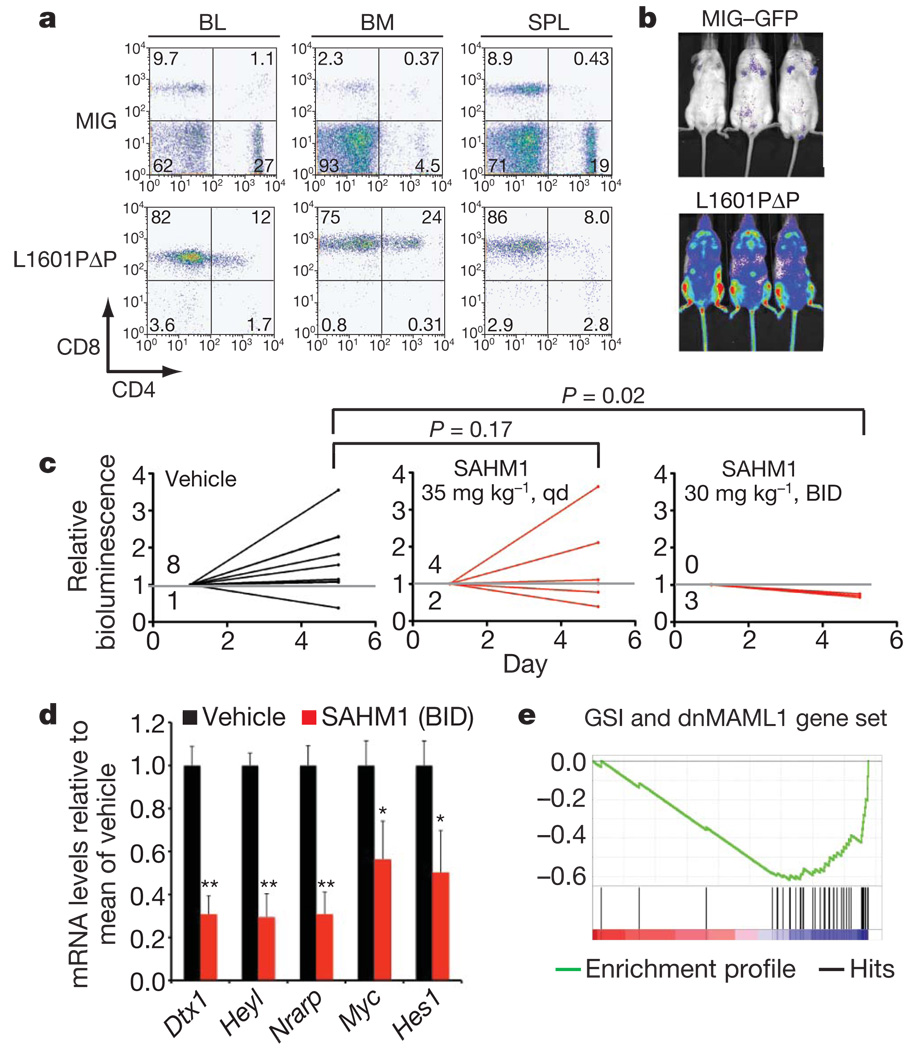
Figure 5. SAHM1 treatment inhibits NOTCH signalling and leukaemic progression in vivo.
a, Flow cytometric analysis of cells isolated from C57BL/6-TyrC/C mice reconstituted with luciferase-expressing haematopoietic stem cells transduced with an L1601PΔP NOTCH1 allele or empty vector (MIG). BL, blood. b, Bioluminescence imaging of primary recipients of Luc-L1601PΔP cells or control cells 2 months postreconstitution. c, Bioluminescence quantification of tumour burden in mice with established disease treated with vehicle, 35 mg kg−1 SAHM1 per day (QD; P = 0.17), or 30 mg kg−1 SAHM1 twice daily (BID; P = 0.02). A Fisher’s Exact test was used to compare disease progression between the cohorts. d, qRT–PCR analysis reveals repression of Hesl (P = 0.0187), Myc (P = 0.023), Nrarp (P = 0.001), Heyl (P = 0.0006) and Dtxl (P = 0.0006) mRNA levels in blood collected at day five from vehicle (n = 3) and SAHM1 (30 mgkg−1 BID, n = 3)-treated mice. e, Enrichment of GSI and dnMAML1 downregulated transcripts in the gene expression profile of leukocytes isolated from SAHM1-treated mice. Data in d represent the mean ± s.d. of triplicate measurements. Unless otherwise noted statistical analyses were performed with a two-tailed t-test. *P < 0.05, **P < 0.005.
Balanced cohorts of secondary recipient mice with established leukaemia were treated with vehicle, daily SAHM1 (35 mg kg−1), or twice-daily SAHM1 (30 mg kg−1) by intraperitoneal injection. Vehicle-treated mice showed progressive disease, with eight out of nine mice showing increased bioluminescence over 5 days of treatment (Fig. 5c). Mice receiving daily SAHM1 treatments showed a slightly lower mean change in bioluminescence and included fewer animals with progressive disease (four out of six). All mice treated with twice-daily SAHM1, however, showed a significant dose-dependent regression of tumour as evidenced by a decrease in bioluminescence.
To confirm that the anti-leukaemic effect observed with SAHM1 treatment was associated with attenuated NOTCH1 signalling, mononuclear cells from vehicle- and SAHM1-treated (twice-daily) animals were collected for NOTCH1 target gene transcriptional analysis. A significant decrease in messenger RNA levels was evident for Heyl, Hes1, Myc, Dtx1 and Nrarp in mice treated with SAHM1 compared to vehicle (Fig. 5d). Gene expression profiling also revealed significant pharmacodynamic repression of the NOTCH transcriptional program in vivo. A gene set of murine NOTCH1 targets was curated from a published report in which NOTCH1-dependent murine T6E cells were profiled after treatment with GSI or ectopic expression of dnMAML1 (ref. 40). Significant enrichment for the murine NOTCH1 gene set was observed in isolated cells from SAHM1-treated animals (Fig. 5e and Supplementary Fig. 7). Together, these data provide a direct link between inhibition of the NOTCH pathway and the anti-leukaemic activity of SAHM1 in vivo.
Discussion
Chemical intractability has limited the discovery of synthetic entities targeting transcription factors. Here we set out to assess whether a new type of targeting molecule, a stapled peptide derived from MAML1, could target a seemingly intractable transcription factor, human NOTCH1. We demonstrated direct binding to the pre-assembled form of the NOTCH1–CSL complex and competitive inhibition of MAML1 co-activator binding. Analysis of direct NOTCH1 target gene levels and the global expression profile induced by SAHM1 confirmed specific repression of the NOTCH signalling program in human and murine T-ALL cells. Given the complexities of transcriptional responses and the state of their characterization, it is impossible to rule out conclusively any off-target activity by SAHM1. In the context of human and murine T-ALL however, GSEA provides a striking correlation between the expression effects of SAHM1 and a GSI. Thus, it is clear that the NOTCH pathway is the major target of SAHM1. Direct transcriptional repression was subsequently found to enact NOTCH-specific anti-proliferative effects in annotated T-ALL cell lines. Finally, SAHM1 treatment curbed leukaemic progression and inhibited NOTCH1 signalling in a relevant murine model of T-ALL. Previous studies in mice and preliminary data in humans have shown that treatment with GSI alone is toxic owing to on-target effects on intestinal crypts, suggesting that different targeting strategies or drug combinations are needed; indeed recent reports are showing promise46. Our studies so far have not observed gastrointestinal toxicity at necropsy in treated animals (Supplementary Fig. 8a, b). Although expanded evaluation of SAHM1 as a possible therapeutic agent is needed, these early results indicate a potential therapeutic window.
As a direct transcriptional antagonist, SAHM1 should prove broadly useful in further determining the role of NOTCH in both normal tissues and disease processes, and presents a starting-point for the development of a targeted therapeutic agent to treat NOTCH-driven cancers and non-malignant conditions. Furthermore, we expect the approach described here will prove broadly applicable to several other transcription factor complexes previously considered beyond the reach of ligand discovery.
METHODS SUMMARY
SAHM peptides were synthesized manually using standard 9-fluorenylmethoxy-carbonyl (Fmoc)-peptide chemistry on MBHA rink amide resin, and cross-linked using Grubbs-I catalyst (benzylidene-bis(tricyclohexylphosphine)dichlororuthenium). Recombinant human CSL (amino acids 9–435), GST–RAMANK and RAMANK (amino acids 1761–2127 of NOTCH1) and dnMAML1 (amino acids 13–74) were expressed and purified as previously reported47. GST pull-down and fluorescence polarization experiments were performed in 20 mM Tris, pH 8.4, containing 150 mM NaCl, 1 mM EDTA and 1 mM dithiothreitol (DTT). SPR experiments were performed in the identical buffer with 187 mM NaCl and 0.01% P-20. Fluorescence polarization was measured at λex = 485 nm and λem = 525 nm on a Perkin Elmer Spectramax-M5 multi-label plate reader. SPR experiments were performed on a Biacore S51 SPR instrument using anti-GST and streptavidin functionalized CM5 chips. Luciferase and β-lactamase reporter gene assays were performed as described35,36. For quantitative RT–PCR (qRT–PCR), RNA was extracted from T-ALL cells, reverse transcribed to complementary DNA, and amplified using standard primers and probes specific for human and murine transcripts. Expression values were calculated by the 2−ΔΔCt method relative to the β-actin gene. RNA from triplicate SAHM- and vehicle-treated KOPT-K1 and HPB-ALL samples was used for analysis on Affymetrix U133 Plus 2.0 expression arrays, processed using GenePattern software. Gene set enrichment analysis was performed using GSEA software (http://www.broad.mit.edu/GSEA). Cell viability assays were performed in white, 96-well plates (Corning) in RPMI media containing 10% FCS. Fresh media and compounds were added every 3 days as needed. The L1601PΔP NOTCH1-dependent murine T-ALL cells were generated by reconstitution of lethally irradiated syngeneic mice with bone marrow cells transduced with the L1601PΔP NOTCH1 allele as described48. Luc-L1601PΔP animals were regularly monitored for bioluminescence, and euthanized at 2–3 months after transplant to obtain splenic T-ALL cells for secondary transplants and in vivo studies. Secondary recipients were monitored for leukaemia by bioluminescence and subsequently separated into treatment cohorts with matched disease burden.
Supplementary Material
Acknowledgements
We thank S. Schreiber and the Broad Institute Chemical Biology Program for discussions and access to instrumentation; J. Rocnik for assistance in the establishment of the T-ALL murine model; A. Ferrando for providing GSI microarray data; M. Hancock and Invitrogen for providing β-lactamase HeLa reporter clones; K. Ross and A. Subramanian for conversations and guidance about GSEA; and S. Gupta and the Broad Institute Microarray Core. This work was supported by a Specialized Center of Research grant from the Leukaemia & Lymphoma Society (to J.E.B., J.C.A., S.C.B.), an AACR Centennial Pre-doctoral Research Fellowship in Cancer Research (to R.E.M.), the American Society of Hematology (to J.E.B.), MCCB-NIH Training Grant No. 5T32GM007598 (to R.E.M.), the Human Frontier Science Program (to C.D.), and the Harvard & Dana Farber Program in Cancer Chemical Biology (to J.E.B., R.E.M., G.L.V.). The project has been funded in part with Federal funds from the National Cancer Institute’s Initiative for Chemical Genetics, National Institutes of Health, under Contract No. N01-CO-12400. Histology and immunohistochemical staining was performed in the Dana Farber/Harvard Cancer Center Specialized Histopathology Services Core Laboratory.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions R.E.M., G.L.V. and J.E.B. conceptualized the study, designed the experiments, interpreted data, and wrote the manuscript. Design, synthesis and biological characterization of SAHM peptides was performed by R.E.M. C.D.B., J.C.A. and S.C.B. contributed key reagents and analysed data. R.E.M., M.C., T.N.D., J.C.A., A.L.K., D.G.G. and J.E.B. established the bioluminescent T-ALL model, designed and performed in vivo experiments and analysed data.
Author Information All microarray data has been deposited to the Gene Expression Omnibus at the National Center for Biotechnology Information under accession numbers GSE18198 and GSE18351. Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nature Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Bray SJ. Notch signalling: a simple pathway becomes complex. Nature Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 4.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 5.Ye Y, Lukinova N, Fortini ME. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B, et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 7.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 8.Petcherski AG, Kimble J. Mastermind is a putative activator for Notch. Curr. Biol. 2000;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- 9.Wu L, et al. MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for NOTCH receptors. Nature Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 10.Li L, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nature Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 11.Joutel A, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 12.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 13.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 14.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 15.O’Neil J, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fre S, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 18.Uyttendaele H, et al. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nature Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 20.Konishi J, et al. γ-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 21.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 22.Park JT, et al. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto Y, et al. Notch mediates TGFα-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 24.Seiffert D, et al. Presenilin-1 and -2 are molecular targets for γ-secretase inhibitors. J. Biol. Chem. 2000;275:34086–34091. doi: 10.1074/jbc.M005430200. [DOI] [PubMed] [Google Scholar]
- 25.Dovey HF, et al. Functional γ-secretase inhibitors reduce β-amyloid peptide levels in brain. J. Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 26.Lleó A. Activity of γ-secretase on substrates other than APP. Curr. Top. Med. Chem. 2008;8:9–16. doi: 10.2174/156802608783334060. [DOI] [PubMed] [Google Scholar]
- 27.Riccio O, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maillard I, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 29.Weng AP, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 31.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 32.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J. Am. Chem. Soc. 2000;122:5891–5892. [Google Scholar]
- 33.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J. Mol. Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aster JC, et al. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol. Cell. Biol. 2000;20:7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallis TM, et al. An improved β-lactamase reporter assay: multiplexing with a cytotoxicity readout for enhanced accuracy of hit identification. J. Biomol. Screen. 2007;12:635–644. doi: 10.1177/1087057107301499. [DOI] [PubMed] [Google Scholar]
- 37.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feedforward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao SS, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69:3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 42.Lewis HD, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem. Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker CM, et al. A novel noninvasive model of endometriosis for monitoring the efficacy of antiangiogenic therapy. Am. J. Pathol. 2006;168:2074–2084. doi: 10.2353/ajpath.2006.051133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubbs MC, et al. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22:66–77. doi: 10.1038/sj.leu.2404951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Real PJ, et al. γ-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nature Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL·intracellular Notch1·Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 48.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J. Clin. Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.