Abstract
Relative quantification by normalization against a stably expressed reference gene is a widely used data analysis method in microarray and quantitative real-time polymerase chain reaction (qRT-PCR) platforms; however, recent evidence suggests that many commonly utilized reference genes are unstable in certain experimental systems and situations. The primary aim of this study, therefore, was to screen and identify stably expressed reference genes in a well-established rat model of vocal fold mucosal injury. We selected and evaluated the expression stability of nine candidate reference genes. Ablim1, Sptbn1 and Wrnip1 were identified as stably expressed in a model-specific microarray dataset and were further validated as suitable reference genes in an independent qRT-PCR experiment using 2-ΔCT and pairwise comparison-based (geNorm) analyses. Parallel analysis of six commonly used reference genes identified Sdha as the only stably expressed candidate in this group. Sdha, Sptbn1 and the geometric mean of Sdha and Sptbn1 each provided accurate normalization of target gene Tgfb1; Gapdh, the least stable candidate gene in our dataset, provided inaccurate normalization and an invalid experimental result. The stable reference genes identified here are suitable for accurate normalization of target gene expression in vocal fold mucosal injury experiments.
Keywords: gene expression, geNorm, housekeeping gene, inflammation, larynx, tissue repair, transcription, wound healing
Introduction
Quantification of gene expression has become increasingly important in biological and medical research and is now considered an essential tool for investigating the molecular mechanisms of disease under various environmental, biological and therapeutic conditions. Microarray offers a high throughput platform for identifying differentially expressed genes across the entire transcriptome, whereas quantitative real-time polymerase chain reaction (qRT-PCR) offers higher assay sensitivity and precision in studying specific transcripts of interest. There are primarily two methods used for analyzing qRT-PCR data: Absolute and relative quantification. Absolute quantification provides an exact transcript copy number or concentration following data transformation via a standard curve [1]. Although many normalization strategies can be employed for relative quantification [2], the comparative cycle threshold (CT) method, also known as the 2-ΔΔCT method [3-7], is the most widely used. In this approach, data obtained by qRT-PCR are normalized against an internal reference gene (often referred to as a housekeeping gene) to control technical and biological variation due to differences in source RNA integrity, cDNA input concentration, enzymatic efficiency, and overall transcription levels in cells or tissues of interest [8]. An ideal reference gene should exhibit constantly stable expression under various experimental conditions (such as different stages of injury or disease progression) as well as comparable transcript abundance to that of the target gene [9].
A number of genes, such as Gapdh, B2m and Actb, are commonly utilized as reference genes for qRT-PCR data normalization across a wide variety of experimental systems; however, many have been demonstrated to be unstable under certain conditions [10-15]. Moreover, several studies have shown that a reference gene that is stable in one experimental system might be unstable in another [11, 16, 17]. Therefore, selection and systematic validation of an experimental model-specific stable reference gene is a key requirement for reliable qRT-PCR normalization and accurate results. Despite this fact, careful validation is largely overlooked and countless studies continue to use arbitrarily selected reference genes, leading to inappropriate data normalization and potentially erroneous results. In response to this problem, several statistical approaches have been developed to identify stable reference genes (or a basket of reference genes) from a panel of candidate genes [4, 18-22] and have been applied across a number of experimental systems (for recent examples see [10, 23-25]). Additionally, evaluation of variance within microarray datasets has been utilized in an attempt to identify new candidate reference genes from the entire genome [25-29].
Vocal fold mucosal injury culminating in scar formation is a common cause of recalcitrant dysphonia and voice handicap [30, 31]. This challenging clinical disorder has spurred a body of work focused on improving understanding and management of the local inflammatory response, alteration in extracellular matrix production and degradation, and the effect of various therapeutic interventions on injury outcomes; much of the literature in this area has utilized qRT-PCR [32-40]. To date, the majority of these studies (and many others in mucosal biology) have been performed using traditional reference genes for qRT-PCR normalization, under the assumption that the expression of these genes is stable across experimental conditions. It is unclear, however, whether these putative reference genes are truly stably expressed, given that the system undergoes such dramatic physiological and pathological changes post-injury [41, 42].
Given these concerns, the identification and validation of stably expressed reference genes is critical to ensuring minimal analytical noise and valid data interpretation in widely-used models of mucosal injury, such as the vocal fold; however, there are currently no empirical reports of reference gene validation in any mucosal injury model, either in vivo or in vitro. The primary aim of this study, therefore, was to screen and identify stably expressed reference genes in a well-established rat model of vocal fold mucosal injury [41, 43-45]. First, we generated and screened a microarray dataset for candidate genes demonstrating minimal expression variance across a range of injury and non-injury conditions. We then implemented a previously reported pairwise comparison-based approach (geNorm [19]) to rank gene stability and selected three novel candidates representing different functional categories and transcript abundance levels. Next, these three candidates were combined with six commonly used reference genes identified in the primary biomedical research literature; the expression stability of all nine candidate genes was evaluated in an independent qRT-PCR experiment and data were analyzed using both 2-ΔCT and geNorm algorithms. Finally, in a third independent experiment, we used the least and most stable candidate genes identified in our primary analyses to normalize expression of a target gene (Tgfb1) at several post-injury time points, demonstrating that reference gene selection can have a significant influence on data interpretation in this model.
Materials and Methods
Animals and mucosal injury procedures
All animal experiments were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the Animal Welfare Act (7 U.S.C. et seq.); the animal use protocol was approved by the Institutional Animal Care and Use Committee of the University of Wisconsin-Madison.
Four-month-old Fischer 344 inbred male rats (Charles River, Wilmington, MA) were used for all experiments. Bilateral vocal fold mucosal stripping injuries were created under endoscopic guidance as previously reported [41, 43]; experimentally naïve rats were used as non-injury controls. Rats were euthanized via CO2 asphyxiation at each post-injury time point and vocal fold mucosae were harvested using microdissection in an RNAse-free environment. Bilateral mucosae from each animal were pooled and immersed in 10 μL RNAlater (Qiagen, Valencia, CA). Samples were stored at 4° C overnight and then at -80° C until RNA isolation.
Animals intended for the microarray experiment were euthanized at three time points to capture global expression profiles characteristic of the early inflammatory phase (3 days post-injury), active proliferation phase (14 days post-injury), and maturation/remodeling phase (60 days post-injury) of wound healing. Twenty post-injury rats (5 arrays, 4 pooled animals per array) and 12 age-matched naïve control rats (3 arrays, 4 pooled animals per array) were allocated to each time point.
Animals intended for the qRT-PCR experiment were euthanized at four time points during the inflammatory wound healing phase (1, 3, 5 and 7 days post-injury). Five post-injury rats were allocated to each time point. Five additional naïve control rats were age-matched to the 7 day post-injury time point.
RNA isolation
Total RNA was isolated using the RNeasy Micro (microarray experiment) and RNeasy Micro Plus (qRT-PCR experiment) kits (Qiagen) according to the manufacturer's instructions. RNA yield and integrity were initially evaluated using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). Samples with a concentration above 40 ng/mL, OD260:280 of 1.8-2.0, and OD260:230 above 1.8 were retained. Samples intended for microarray were further evaluated using the Agilent 2100 Bioanalyzer and RNA 6000 Pico kit (Agilent, Santa Clara, CA) according to the manufacturer's instructions. Samples with electropherograms exhibiting sharp 18S and 28S rRNA peaks and no evidence of degradation were retained.
Microarrays
Total RNA yield in the 60 day post-injury group was sufficient to run just four of the five arrays intended for this condition. All other arrays and conditions were run as planned. Biotinylated antisense cRNA was prepared by single round in vitro amplification of 1.2 μg input RNA using the MessageAmp II-Biotin Enhanced aRNA kit (Ambion, Austin, TX) according to the manufacturer's instructions (the in vitro transcription reaction was performed at 37° C for 14 h). Poly-A RNA controls (Affymetrix, Santa Clara, CA) were spiked into each reaction. Fragmented cRNA sample quality was confirmed using 2% agarose gel electrophoresis, Agilent 2100 Bioanalyzer analysis (Pico kit), and hybridization to Affymetrix GeneChip Test3 arrays. Samples were hybridized to Affymetrix GeneChip Rat Genome 230 2.0 arrays at 45° C for 16 h. Post-processing was performing using the GeneChip Fluidics Station 450, arrays were scanned using the GC3000 G7 scanner, and fluorescent intensity data were background-corrected and extracted using Expression Console software (Affymetrix). All hybridization, post-processing and scanning procedures were performed according to Affymetrix protocols; all control parameters for Test3 and Rat Genome arrays were within manufacturer guidelines.
qRT-PCR
Reverse transcription prior to qRT-PCR was performed using the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA) with 20 ng input total RNA per 20 μL reaction according to the manufacturer's instructions. Negative control samples were performed in parallel by omitting RNA template or reverse transcriptase.
Commercial rat specific primers used for PCR amplification were purchased from Qiagen with guaranteed 100% amplification efficiency. The following QuantiTect Primer Assays were used for each primer set: QT01801905 (Ablim1), QT00193473 (Actb), QT00176295 (B2m), QT00199633 (Gapdh), QT00365722 (Hprt), QT00195958 (Sdha), QT01578017 (Sptbn1), QT00183344 (Wrnip1), QT01798412 (Ywhaz), QT00187796 (Tgfb1). qRT-PCR was performed on a 7500 Fast Real-Time PCR system (Applied Biosystems, Foster, CA) using the QuantiTect SYBR Green PCR kit (Qiagen). Each 25 μL total volume reaction contained 12.5 μL 2× QuantiTect Master Mix, 2.5 μL 10× QuantiTect Primer Assay and 10 μL cDNA template diluted with nuclease free H2O. Amplifications were performed in MicroAmp Fast Optical 96-well reaction plates with optical adhesive film covers (Applied Biosystems) according to cycling conditions suggested for the Applied Biosystems 7500 instrument in the QuantiTect SYBR Green handbook (initial activation at 95° C for 15 min; 40 cycles of 94° C for 15 s, 55° C for 30 s, 72° C for 30 s).
All PCR reactions were performed in triplicate using cDNA synthesized from the same batch and starting amount of total RNA. Negative controls containing no cDNA template were included for each gene within each PCR run. To avoid variation in amplification conditions across runs, reactions for all experimental conditions (i.e., all non-injury controls and post-injury time points) for each gene of interest were performed in the same 96-well plate. Amplification specificity for each gene was confirmed by a single distinct melting curve. PCR products were separated using 1.5% agarose gel electrophoresis to confirm the presence of a single band at the expected amplicon size.
Data analysis
Microarray fluorescent intensity data were log2 transformed and sorted by coefficient of variation (CV) for each gene across experimental conditions. qRT-PCR data representing candidate reference gene expression stability over time were analyzed using the 2-ΔCT method; data representing normalization of target gene Tgfb1 were analyzed using the 2-ΔΔCT method (efficiency was maintained at 2 based on the 100% guaranteed efficiency of the primer sets used in this study) [4]. Mean CT values from triplicate runs were used as input data (CT value range for all triplicates was below 0.5).
Statistical comparisons of fold change in relative gene expression level were performed using one-way analysis of variance (ANOVA) with post-hoc pairwise comparisons (Fisher's least significant difference) between experimental groups of interest. Pairwise comparisons of candidate reference gene stability and calculation of geometric mean-based normalization factors were performed using the geNorm algorithm as previously described [19]. Briefly, the average expression stability measure (M-value) represents the average pairwise variation of one candidate reference gene compared to all other genes in the test panel. Genes with the lowest M-values are considered the most stably expressed. The stability ranking of each candidate gene is determined by stepwise exclusion of the gene with highest M-value, followed by recalculation of average expression stability for the remaining genes. The developers of the geNorm algorithm argued that target gene normalization is significantly improved by calculating a normalization factor based on the geometric mean of relative expression levels for multiple reference genes. To determine the optimal number of reference genes for accurate normalization, the pairwise variation (Vi/Vi + 1) between sequential normalization factors (NFi and NFi + 1) is calculated. Once Vi/Vi + 1 drops below 0.15 for a given basket of reference genes, inclusion of the (i + 1)th gene provides no gain in normalization accuracy.
The geNorm algorithm was implemented using the SLqPCR package for R (http://www.bioconductor.org/packages/2.2/bioc/html/SLqPCR.html) developed by Matthias Kohl (SIRS-Lab, Jena, Germany). ANOVA were implemented using SAS 9.1.3 for Windows (SAS Institute, Cary, NC). An alpha-level of .01 was used for all statistical comparisons; all p-values were two-sided. Data were graphed showing mean ± standard error. Heat maps were generated using Heat Map Builder 1.0 [46].
Results
Microarray-based selection of candidate reference genes
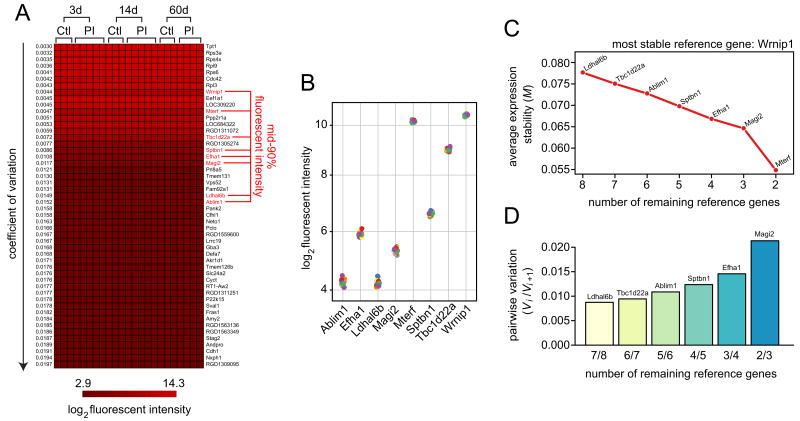
We generated and screened a vocal fold mucosal injury expression microarray dataset to identify stably expressed genes across a number of post-injury conditions. A total of 23 arrays containing 31,042 probe sets (∼28,000 genes) were screened, representing the early inflammatory phase (3 days post-injury; 5 arrays), active proliferative phase (14 days post-injury; 5 arrays), and maturation/remodeling phase (60 days post-injury; 4 arrays) of wound healing; nine age-matched non-injury control arrays were included in the dataset (three per post-injury time point). Background-corrected fluorescent intensity values were converted to the log2 scale and genes were sorted by CV across all arrays: The 50 genes exhibiting the lowest CVs in the dataset (0.0030-0.0197) were partitioned for further analysis (Figure 1A). These 50 stably expressed genes represented a wide range of transcript abundance levels and functional categories; notably, genes with low CVs tended to be high abundance, and vice versa. Six of the eight most stable genes transcribe for ribosomal proteins important to protein synthesis (Rps3a, Rps4×, Rpl9, Rps6, Rpl3).
Figure 1. Microarray-based selection of candidate reference genes.
An expression microarray dataset was generated representing rat vocal fold mucosa 3, 14 and 60 days post-injury. Age-matched non-injury controls were included at each time point. A total of 23 arrays (14 post-injury, 9 non-injury controls) were included in the dataset. (A) Heat map showing log2 fluorescent intensity for the 50 genes with the lowest coefficient of variation across all arrays representing both injury and non-injury experimental conditions. Genes are ranked by ascending coefficient of variation. Eight genes exhibited mean fluorescent intensity within the mid-90% of all genes on the arrays; these genes were subjected to further analysis. (B) Distribution of log2 fluorescent intensity for the eight genes of interest identified in panel A. Twenty-three data points are shown for each gene, inclusive of injury and non-injury experimental conditions. (C) Average expression stability of a basket containing the eight genes of interest during stepwise exclusion of the least stable gene (geNorm algorithm). The abscissa reflects the number of genes under consideration throughout the stepwise procedure; the ordinate reflects the average expression stability (M-value) of the remaining genes after exclusion of the least stable gene (indicated by name on the plot). (D) Pairwise variation (Vi/Vi+1) between two sequential normalization factors (NFi and NFi+1) calculated to determine the optimal number of reference genes for accurate normalization (geNorm algorithm). Ctl, non-injury control; PI, post-injury; d, day.
Next, we tightened our selection criteria to eliminate genes with mean fluorescent intensities in the upper or lower vigintiles across all arrays, in order to avoid potential qRT-PCR normalization issues in subsequent experiments due to extremely high or low abundance candidate reference genes. This filter eliminated 42 of the 50 most stable genes in the dataset; eight genes with mid-90% fluorescent intensities remained (Ablim1, Efha1, Ldhal6b, Magi2, Mterf, Sptbn1, Tbc1d22a, Wrnip1; Figure 1A). These eight genes exhibited tightly clustered abundance levels across arrays (Figure 1B). Analysis using the pairwise comparison-based geNorm algorithm [19] revealed Wrnip1 as the most stably expressed gene within the group, followed by Mterf and Magi2 (lowest M-values in the stepwise procedure presented in Figure 1C). Analysis of pairwise variation (Vi/Vi+1) between sequential geometric mean-based normalization factors (NFi and NFi+1) suggested that two reference genes (Wrnip1 and Mterf2) are sufficient to accurately normalize expression of a target gene in this dataset and experimental model (Figure 1D).
Given the relatively high stability of all genes identified in our array screen, we selected three candidate genes for further validation in qRT-PCR experiments based on relative abundance and functional categorization. Wrnip1 [47] was selected as the most stable gene exhibiting relatively high abundance (Figure 1B) within the mid-90% of our dataset. Ablim1 [48] and Sptbn1 [49] were selected as additional stably expressed candidate reference genes with relatively low and midrange abundance respectively (Figure 1B). The full name, accession number and functional category for each candidate gene are provided in Table 1.
Table1. Candidate reference genes evaluated in this study.
| Gene symbol | Gene product name | Accession number | Function |
|---|---|---|---|
| Ablim1† | Actin-binding LIM protein 1 | NM_001044394 | Mediates interaction between actin filaments and cytoplasmic targets |
| Actb§ | Actin, beta | NM_031144 | Cytoskeletal structural protein |
| B2m§ | Beta-2 microglobulin | NM_012512 | Beta-chain of MHC class I molecules |
| Efha1† | EF-hand domain family, member A1 | NM_134396 | Stimulates tubulin polymerization; calcium ion binding |
| Gapdh§ | Glyceraldehyde-3-phosphate dehydrogenase | NM_017008 | Oxidoreductase in glycolysis and gluconeogenesis |
| Hprt1§ | Hypoxanthine phosphoribosyltransferase 1 | NM_012583 | Catalytic enzyme in purine salvage pathway |
| Ldhal6b† | Lactate dehydrogenase A-like 6B | NM_183334 | Catalyzes lactate-pyruvate interconversion; involved in glycolysis |
| Magi2† | Membrane associated guanylate kinase, WW and PDZ domain containing 2 | NM_053621 | Synaptic scaffolding molecule |
| Mterf† | Mitochondrial transcription termination factor | NM_053499 | Regulates mtDNA transcription |
| Sdha§ | Succinate dehydrogenase complex, subunit A | NM_130428 | Catalyzes mitochondrial succinate oxidation |
| Sptbn1† | Spectrin, beta, non-erythrocytic 1 | NM_001013130 | Actin cross-linking and molecular scaffold protein |
| Tbc1d22a† | TBC1 domain family, member 22A | XM_001078492 | Regulates Rab GTPase activity |
| Wrnip1† | Werner helicase interacting protein 1 | NM_172332 | Interacts with Werner syndrome ATP-dependent helicase; potential role in replication and other nuclear transactions |
| Ywhaz§ | Tyrosine 3-monooxygenase | NM_013011 | Component of the mitochondrial import stimulation factor; regulates signal transduction by binding to phosphorylated serine residues on a variety of signaling molecules |
Candidate identified as mid-90% fluorescent intensity in microarray screen;
Candidate identified in literature review.
Literature-based selection of additional candidate reference genes
The three novel candidate reference genes identified in our array dataset were evaluated alongside six traditional reference genes identified in a review of the primary biomedical research literature in the areas of wound healing and tissue repair. Hprt1, Sdha and Ywhaz were selected based on a recent validation study reporting stable expression in an in vivo ischemic brain injury model in rat [23]. Actb, B2m and Gapdh were selected as they are commonly utilized reference genes in vocal fold mucosal injury studies [32-39] but have never undergone formal validation. The full name, accession number and functional category for each candidate gene are provided in Table 1.
Effect of vocal fold mucosal injury on the expression of candidate reference genes during the inflammatory wound healing phase
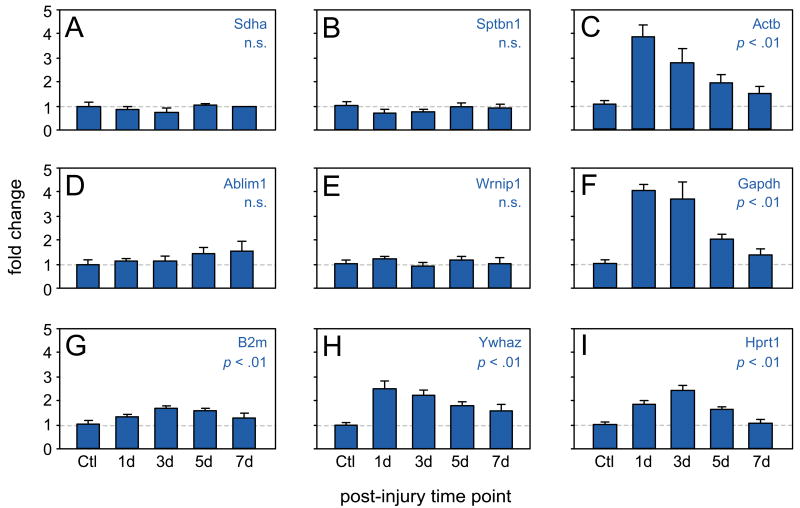
We used qRT-PCR to measure the mRNA expression of our nine candidate reference genes in rat vocal fold mucosa 1-7 days post-injury, and in non-injury controls. Relative expression levels were calculated using the 2-ΔCT method. Gapdh and Actb exhibited an immediate injury response, characterized by a 4-fold increase in expression at 1 day post-injury followed by gradual tapering of expression between 3 and 7 days post-injury (p < .01, Figure 2C and F). Ywhaz followed a similar time course to Gapdh and Actb with less dramatic upregulation (2.5-fold) at 1 day post-injury (p < .01; Figure 2H). Hprt and B2m also demonstrated significant perturbation following injury, with peak expression at 3 days (p < .01, Figure 2G and I). All five of these candidate reference genes trended toward baseline expression levels at the 7 day post-injury time point. Pairwise comparisons between the control and 7-day post-injury conditions revealed no significant differences for any candidate gene.
Figure 2. Effect of vocal fold mucosal injury on the expression of candidate reference genes.
RNA was extracted from rat vocal fold mucosa 1-7 days post-injury and non-injury controls and qRT-PCR was performed for nine candidate reference genes: (A) Sdha, (B) Sptbn1, (C) Actb, (D) Ablim1, (E) Wrnip1, (F) Gapdh, (G) B2m, (H) Hprt and (I) Ywhaz. Gene expression levels were calculated using the 2-ΔCT method and are presented as mean fold change ± standard error. n = 5 animals per time point. p-values reflect one-way ANOVA across all time points. Ctl, non-injury control; d, day post-injury; n.s., non-significant difference.
Four candidate reference genes, including the three novel candidates identified using our array dataset, exhibited no significant difference in expression across post-injury time points compared to control: Sdha (Figure 2A), Sptbn1 (Figure 2B), Ablim1 (Figure 2D), and Wrnip1 (Figure 2E). These four genes were therefore considered the most stably expressed over time, according to 2-ΔCT analysis.
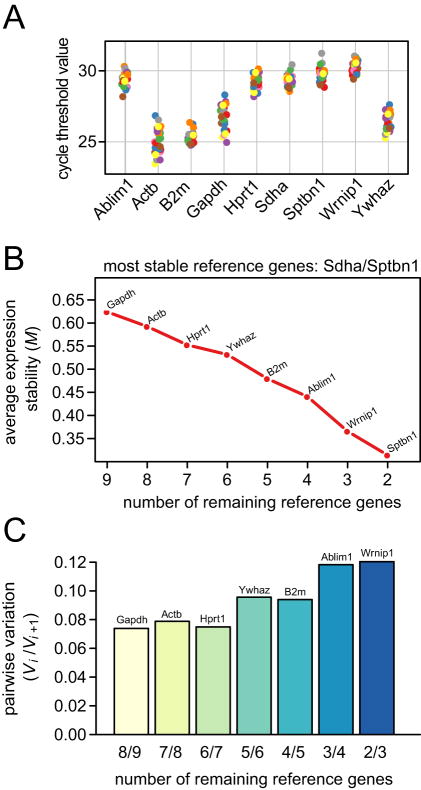
Pairwise comparison-based (geNorm) analysis and stability ranking of candidate reference genes
Evaluation of the distribution of CT values obtained from the amplification curves of all samples analyzed by qRT-PCR revealed a range of ∼24-31 across all candidate reference genes (Figure 3A). CT values were most tightly clustered for Wrnip1 and Sdha (CVs = 0.0159 and 0.0175 respectively) and least tightly clustered for Gapdh and Actb (CVs = 0.0344 and 0.0339 respectively). Gapdh and Actb were also identified as the least stable genes using the geNorm algorithm [19], whereas Sdha and Sptbn1 were tied as the two most stably expressed genes with identical M-values of 0.31 (Figure 3B). The overall stability ranking of the nine candidate reference genes was as follows (from most to least stable): Sdha/Sptbn1 (tied), Wrnip1, Ablim1, B2m, Ywhaz, Hprt, Actb and Gapdh. Analysis of pairwise variation (Vi/Vi+1) between sequential geometric mean-based normalization factors (NFi and NFi+1) suggested that the two most stably expressed genes (Sdha and Sptbn1) are sufficient to accurately normalize expression of a target gene in this dataset and experimental model (Figure 3C). The addition of Wrnip1 as a third reference gene yielded a pairwise variation value of 0.12, which is below the recommended cut-off value of 0.15, indicating that the addition of this gene to a geometric averaging basket provides no gain in normalization accuracy [19].
Figure 3. Pairwise comparison-based (geNorm) analysis of candidate reference genes.
(A) Distribution of expression levels (cycle threshold values) for all samples and all candidate reference genes. Twenty-five data points are shown for each gene, inclusive of injury and non-injury experimental conditions. (B) Average expression stability of a basket containing all candidate reference genes during stepwise exclusion of the least stable gene. The abscissa reflects the number of genes under consideration throughout the stepwise procedure; the ordinate reflects the average expression stability (M-value) of the remaining genes after exclusion of the least stable gene (indicated by name on the plot). (C) Pairwise variation (Vi/Vi+1) between two sequential normalization factors (NFi and NFi+1) calculated to determine the optimal number of reference genes for accurate normalization.
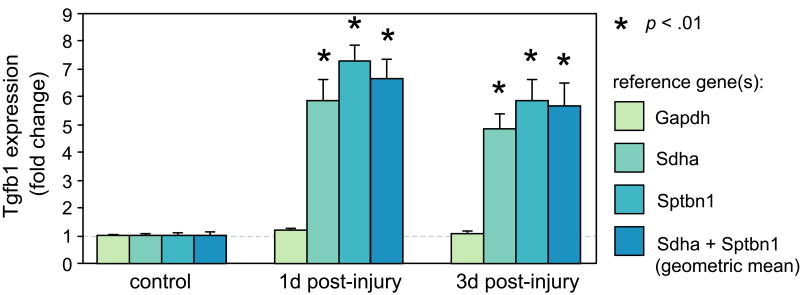
Relative expression of a target gene is significantly influenced by reference gene stability following vocal fold mucosal injury
Having validated and ranked the stability of our pool of candidate reference genes, we next investigated the consequence of using a stable or unstable reference gene for normalization of a target gene in our vocal fold mucosal injury model. We selected Tgfb1 as a target gene as it is a well characterized fibrotic cytokine with clearly documented upregulation in response to injury [50]. We used the 2-ΔΔCT method to examine the relative expression of Tgfb1 at 1 and 3 days post-injury, normalized to Gapdh, Sdha, Sptbn1, and the geometric mean of Sdha and Sptbn1 (the geometric mean normalization factor was derived using geNorm). Normalization against Gapdh (the least stable candidate reference gene) revealed no significant difference in Tgfb1 expression versus non-injury control at 1 or 3 days post-injury, whereas normalization against Sdha or Sptbn1 (tied as the most stable candidate reference genes) or their geometric mean revealed significant upregulation of Tgfb1 versus non-injury control at both post-injury time points (p < .01; Figure 4). Pairwise comparisons within each experimental time point revealed no significant difference between Tgfb1 expression normalized against Sdha, Sptbn1 or the geometric mean of Sdha and Sptbn1; however, each of these reference genes (and the geometric mean) yielded significantly different Tgfb1 expression values compared to the Gapdh normalized condition at the same time point (p < .01; Figure 4).
Figure 4. Relative expression of the target gene Tgfb1 is significantly influenced by reference gene stability following vocal fold mucosal injury.
Tgfb1 expression was normalized against reference genes Gapdh, Sdha, Sptbn1 and the geometric mean of Sdha and Sptbn1. Gene expression levels were calculated using the 2-ΔΔCT method and are presented as mean fold change ± standard error. n = 5 animals per time point. *, p < .01 versus both the non-injury control condition and Gapdh normalized condition at the same time point; d, day post-injury.
Discussion
qRT-PCR is an indispensable technique in biological and medical research, however the appropriate implementation of this technique is highly dependent upon normalization with a stably expressed reference gene (or basket of reference genes). Unfortunately, the importance of appropriate reference gene selection is often ignored in gene expression studies; therefore, systematic and experimental condition-specific validation of carefully selected candidate reference genes is necessary, especially in model systems such as vocal fold mucosal injury, where global gene expression may be dramatically altered in the context of rapid physiological and pathological changes [41, 42]. Here we report a microarray-driven systematic study of candidate reference gene stability in a rat vocal fold model of mucosal injury. To our knowledge, this is the first report of reference gene validation in any mucosal injury experimental system.
To select candidate reference genes for validation, we first conducted a genome-wide screen to identify stably expressed genes in a vocal fold mucosal injury microarray dataset generated in our laboratory. Stably expressed genes exhibiting mid-90% mean fluorescent intensity compared to the entire genome were evaluated using a pairwise comparison-based approach (geNorm). Three candidate genes (Ablim1, Sptbn1 and Wrnip1) representing distinct functional categories and low, midrange and high transcript abundance were selected for further validation. An independent qRT-PCR experiment using 2-ΔCT analysis confirmed that all three genes exhibited stable mRNA expression throughout the acute inflammatory wound healing period (1-7 days post-injury), with no significant differences compared to non-injury controls. Parallel analysis of a sub-panel of traditional reference genes identified just one stably expressed gene (Sdha) among six candidates. Hprt1 and Ywhaz, two candidates previously validated alongside Sdha as stably expressed in an ischemic brain injury model [23], were significantly upregulated following vocal fold mucosal injury. Actb, B2m and Gapdh, three candidates commonly employed (but previously unvalidated) as reference genes in vocal fold and other mucosal injury models [32-39, 51, 52], were also unstable post-injury. These findings support the predictive value of microarray screening for identifying suitable candidate genes for further validation, demonstrate that a reference gene that is stable in one in vivo tissue injury system may not necessarily be stable in another, and highlight the experimental noise that may accompany the adoption of a putative reference gene without careful validation.
Stepwise ranking of expression stability using the geNorm algorithm provided additional validation of our 2-ΔCT analysis: Sdha, Sptbn1, Wrnip1 and Ablim1 were ranked as the four most stable genes; Gapdh and Actb were ranked as the two least stable genes. Normalization of target gene Tgfb1 using stable (Sdha and Sptbn1) and unstable (Gapdh) reference genes led to significantly different experimental results. The erroneous interpretation of no change in Tgfb1 expression 1 and 3 days post-injury with Gapdh normalization (under-representing actual Tgfb1 upregulation by 5- to 7-fold) is a striking example of the potential impact of inappropriate reference gene selection in this model system.
The geNorm approach to reference gene selection is driven by the argument that geometric averaging using a basket of stably expressed genes results in superior normalization compared to use of a single gene [19]. Pairwise variation evaluation of our panel of candidate genes demonstrated that the geometric mean of Sdha and Sptbn1 was sufficient to provide accurate normalization of a target gene, without the addition of Wrnip1. Given the low cell density [41] and therefore low amount of recoverable RNA in the native rat vocal fold mucosa (∼150-300 ng per animal in this study), and our finding that Sdha and Sptbn1 were tied as the two most stably expressed genes in our dataset, we also evaluated the individual normalization performance of these two reference genes compared to their geometric mean. We found no significant difference in the expression of target gene Tgfb1 whether normalized against Sdha, Sptbn1 or the geometric mean of Sdha and Sptbn1, suggesting that in experiments with especially limited RNA (e.g., those involving a large number of target genes), accurate normalization and quantification of target gene expression can be practically obtained using a single (appropriately validated) reference gene.
In addition to exhibiting stable expression across all experimental conditions, an ideal reference gene should have comparable transcript abundance to a target gene of interest [9]. For this reason, we intentionally excluded genes with extremely high or low mean fluorescent intensities (upper or lower vigintiles) across arrays and focused our qRT-PCR validation experiments on candidates in the mid-90% of the dataset. Additionally, the three candidate genes selected from our array dataset represented low (Ablim1), midrange (Sptbn1) and high (Wrnip1) transcript abundance within the mid-90%. This strategy improved the likelihood of validating stably expressed reference genes with comparable transcript abundance to a wide range of possible target genes; however, it is important to note that a number of genes with high transcript abundance (upper vigintile) demonstrated even greater stability than our candidate selections; many of these genes encode ribosomal proteins (e.g., Rps3a, Rps4×, Rpl9, Rps6, Rpl3). This observation is consistent with a number of recent microarray-based publications that have identified ribosomal protein genes as stably expressed reference gene candidates across a range of experimental systems [25, 26, 29]. Hence, in the vocal fold mucosal injury system reported here, a ribosomal protein gene may be a suitable choice in experiments requiring normalization of a target gene with comparable high transcript abundance.
Acknowledgments
This study was supported by grants R01 DC004428 and R01 DC004428-S1 from the National Institute on Deafness and Other Communication Disorders. We gratefully acknowledge microarray expertise provided by the Gene Expression Center at the University of Wisconsin Biotechnology Center, and statistical consultation provided by Alejandro Muñoz del Río and Glen Leverson.
Abbreviations
- ANOVA
analysis of variance
- CT
cycle threshold
- CV
coefficient of variation
- qRT-PCR
quantitative real-time polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–84. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 3.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–9. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 4.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 5.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 6.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 7.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters IR, Helps CR, Hall EJ, Day MJ. Real-time RT-PCR: considerations for efficient and sensitive assay design. J Immunol Methods. 2004;286:203–17. doi: 10.1016/j.jim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–7. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 10.Foldager CB, Munir S, Ulrik-Vinther M, Soballe K, Bunger C, Lind M. Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol. 2009;10:94. doi: 10.1186/1471-2199-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–4. 116, 118–9. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- 12.Bemeur C, Ste-Marie L, Desjardins P, Hazell AS, Vachon L, Butterworth R, Montgomery J. Decreased beta-actin mRNA expression in hyperglycemic focal cerebral ischemia in the rat. Neurosci Lett. 2004;357:211–4. doi: 10.1016/j.neulet.2003.12.081. [DOI] [PubMed] [Google Scholar]
- 13.Schulz WA, Eickelmann P, Hallbrucker C, Sies H, Haussinger D. Increase of beta-actin mRNA upon hypotonic perfusion of perfused rat liver. FEBS Lett. 1991;292:264–6. doi: 10.1016/0014-5793(91)80880-c. [DOI] [PubMed] [Google Scholar]
- 14.Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59:566–73. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 15.Mansur NR, Meyer-Siegler K, Wurzer JC, Sirover MA. Cell cycle regulation of the glyceraldehyde-3-phosphate dehydrogenase/uracil DNA glycosylase gene in normal human cells. Nucleic Acids Res. 1993;21:993–8. doi: 10.1093/nar/21.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–5. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee PD, Sladek R, Greenwood CM, Hudson TJ. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002;12:292–7. doi: 10.1101/gr.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 21.Akilesh S, Shaffer DJ, Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res. 2003;13:1719–27. doi: 10.1101/gr.533003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller F, Kulle B, Schwager S, Gunawan B, von Heydebreck A, Sultmann H, Fuzesi L. Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalization. Anal Biochem. 2004;335:1–9. doi: 10.1016/j.ab.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Gubern C, Hurtado O, Rodriguez R, Morales JR, Romera VG, Moro MA, Lizasoain I, Serena J, Mallolas J. Validation of housekeeping genes for quantitative real-time PCR in in-vivo and in-vitro models of cerebral ischaemia. BMC Mol Biol. 2009;10:57. doi: 10.1186/1471-2199-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zampieri M, Ciccarone F, Guastafierro T, Bacalini MG, Calabrese R, Moreno-Villanueva M, Reale A, Chevanne M, Burkle A, Caiafa P. Validation of suitable internal control genes for expression studies in aging. Mech Ageing Dev. 2010;131:89–95. doi: 10.1016/j.mad.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, Lim QE, Wan G, Too HP. Normalization with genes encoding ribosomal proteins but not GAPDH provides an accurate quantification of gene expressions in neuronal differentiation of PC12 cells. BMC Genomics. 2010;11:75. doi: 10.1186/1471-2164-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popovici V, Goldstein DR, Antonov J, Jaggi R, Delorenzi M, Wirapati P. Selecting control genes for RT-QPCR using public microarray data. BMC Bioinformatics. 2009;10:42. doi: 10.1186/1471-2105-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Jo M, Lee J, Koh SS, Kim S. Identification of novel universal housekeeping genes by statistical analysis of microarray data. J Biochem Mol Biol. 2007;40:226–31. doi: 10.5483/bmbrep.2007.40.2.226. [DOI] [PubMed] [Google Scholar]
- 28.Kwon MJ, Oh E, Lee S, Roh MR, Kim SE, Lee Y, Choi YL, In YH, Park T, Koh SS, Shin YK. Identification of novel reference genes using multiplatform expression data and their validation for quantitative gene expression analysis. PLoS One. 2009;4:e6162. doi: 10.1371/journal.pone.0006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benninger MS, Alessi D, Archer S, Bastian R, Ford C, Koufman J, Sataloff RT, Spiegel JR, Woo P. Vocal fold scarring: current concepts and management. Otolaryngol Head Neck Surg. 1996;115:474–82. doi: 10.1177/019459989611500521. [DOI] [PubMed] [Google Scholar]
- 31.Rosen CA, Lee AS, Osborne J, Zullo T, Murry T. Development and validation of the voice handicap index-10. Laryngoscope. 2004;114:1549–56. doi: 10.1097/00005537-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 32.Lim X, Tateya I, Tateya T, Munoz-Del-Rio A, Bless DM. Immediate inflammatory response and scar formation in wounded vocal folds. Ann Otol Rhinol Laryngol. 2006;115:921–9. doi: 10.1177/000348940611501212. [DOI] [PubMed] [Google Scholar]
- 33.Ohno T, French LC, Hirano S, Ossoff RH, Rousseau B. Effect of hepatocyte growth factor on gene expression of extracellular matrix during wound healing of the injured rat vocal fold. Ann Otol Rhinol Laryngol. 2008;117:696–702. doi: 10.1177/000348940811700912. [DOI] [PubMed] [Google Scholar]
- 34.Rousseau B, Ge PJ, Ohno T, French LC, Thibeault SL. Extracellular matrix gene expression after vocal fold injury in a rabbit model. Ann Otol Rhinol Laryngol. 2008;117:598–603. doi: 10.1177/000348940811700809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thibeault SL, Duflo S. Inflammatory cytokine responses to synthetic extracellular matrix injection to the vocal fold lamina propria. Ann Otol Rhinol Laryngol. 2008;117:221–6. doi: 10.1177/000348940811700310. [DOI] [PubMed] [Google Scholar]
- 36.Welham NV, Lim X, Tateya I, Bless DM. Inflammatory factor profiles one hour following vocal fold injury. Ann Otol Rhinol Laryngol. 2008;117:145–52. doi: 10.1177/000348940811700213. [DOI] [PubMed] [Google Scholar]
- 37.Ohno T, Hirano S, Rousseau B. Extracellular matrix gene expression during wound healing of the injured rat vocal fold. Otolaryngol Head Neck Surg. 2009;140:757–61. doi: 10.1016/j.otohns.2008.12.065. [DOI] [PubMed] [Google Scholar]
- 38.Ohno T, Hirano S, Rousseau B. Gene expression of transforming growth factor-beta1 and hepatocyte growth factor during wound healing of injured rat vocal fold. Laryngoscope. 2009;119:806–10. doi: 10.1002/lary.20174. [DOI] [PubMed] [Google Scholar]
- 39.Ohno T, Yoo MJ, Swanson ER, Hirano S, Ossoff RH, Rousseau B. Regeneration of aged rat vocal folds using hepatocyte growth factor therapy. Laryngoscope. 2009;119:1424–30. doi: 10.1002/lary.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y, Yamashita M, Zhang J, Ling C, Welham NV. Pulsed dye laser-induced inflammatory response and extracellular matrix turnover in rat vocal folds and vocal fold fibroblasts. Lasers Surg Med. 2009;41:585–94. doi: 10.1002/lsm.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling C, Yamashita M, Waselchuk EA, Raasch JL, Bless DM, Welham NV. Alteration in cellular morphology, density and distribution in rat vocal fold mucosa following injury. Wound Repair Regen. 2010;18:89–97. doi: 10.1111/j.1524-475X.2009.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Branski RC, Rosen CA, Verdolini K, Hebda PA. Acute vocal fold wound healing in a rabbit model. Ann Otol Rhinol Laryngol. 2005;114:19–24. doi: 10.1177/000348940511400105. [DOI] [PubMed] [Google Scholar]
- 43.Tateya T, Tateya I, Sohn JH, Bless DM. Histologic characterization of rat vocal fold scarring. Ann Otol Rhinol Laryngol. 2005;114:183–91. doi: 10.1177/000348940511400303. [DOI] [PubMed] [Google Scholar]
- 44.Tateya T, Tateya I, Sohn JH, Bless DM. Histological study of acute vocal fold injury in a rat model. Ann Otol Rhinol Laryngol. 2006;115:285–92. doi: 10.1177/000348940611500406. [DOI] [PubMed] [Google Scholar]
- 45.Welham NV, Montequin DW, Tateya I, Tateya T, Choi SH, Bless DM. A rat excised larynx model of vocal fold scar. J Speech Lang Hear Res. 2009;52:1008–20. doi: 10.1044/1092-4388(2009/08-0049). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, Vailaya A, Kincaid R, Tsalenko A, Deng DX, Connolly A, Zhang P, Yang E, Watt C, Yakhini Z, Ben-Dor A, Adler A, Bruhn L, Tsao P, Quertermous T, Ashley EA. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–18. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 47.Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem. 2008;283:35173–85. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roof DJ, Hayes A, Adamian M, Chishti AH, Li T. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol. 1997;138:575–88. doi: 10.1083/jcb.138.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis L, Abdi K, Machius M, Brautigam C, Tomchick DR, Bennett V, Michaely P. Localization and structure of the ankyrin-binding site on beta2-spectrin. J Biol Chem. 2009;284:6982–7. doi: 10.1074/jbc.M809245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank S, Madlener M, Werner S. Transforming growth factors beta1, beta2, and beta3 and their receptors are differentially regulated during normal and impaired wound healing. J Biol Chem. 1996;271:10188–93. doi: 10.1074/jbc.271.17.10188. [DOI] [PubMed] [Google Scholar]
- 51.Eslami A, Gallant-Behm CL, Hart DA, Wiebe C, Honardoust D, Gardner H, Hakkinen L, Larjava HS. Expression of integrin alphavbeta6 and TGF-beta in scarless vs scar-forming wound healing. J Histochem Cytochem. 2009;7:543–57. doi: 10.1369/jhc.2009.952572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szpaderska AM, Zuckerman JD, DiPietro LA. Differential injury responses in oral mucosal and cutaneous wounds. J Dent Res. 2003;82:621–6. doi: 10.1177/154405910308200810. [DOI] [PubMed] [Google Scholar]