Abstract
Objective
To address mechanisms that control the activity of human peptidyl arginine deiminase type 4 (PAD-4).
Methods
PAD-4 autocitrullination was determined by anti–modified citrulline immunoblotting, using purified recombinant and endogenous PAD-4 from activated human primary neutrophils and cell lines expressing PAD-4. The citrullination sites in PAD-4 were determined by mass spectrometry. Mechanisms of autocitrullination-induced inactivation and the functional consequences of autocitrullination in PAD-4 polymorphic variants were addressed using purified components and cell lines expressing PAD-4 wild-type, PAD-4 mutant, and PAD-4 polymorphic variants relevant to rheumatoid arthritis (RA).
Results
PAD-4 is autocitrullinated in vitro and during activation of primary cells and cell lines expressing PAD-4. Interestingly, this modification inactivated the function of the enzyme. The efficiency of inactivation differed among genetically defined PAD-4 variants relevant to RA. PAD-4 was citrullinated at 10 sites, which are clustered into 3 distinct regions, including a cluster of arginines around the active site cleft where Arg-372 and -374 were identified as the potential autocitrullination targets that inactivate the enzyme. Autocitrullination also modified the structure of PAD-4, abrogating its recognition by multiple rabbit antibodies, but augmenting its recognition by human anti–PAD-4 autoantibodies.
Conclusion
Our findings suggest that autocitrullination regulates the production of citrullinated proteins during cell activation, and that this is affected by structural polymorphisms in PAD-4. Autocitrullination also influences PAD-4 structure and immune response.
Introduction
Posttranslational modifications of proteins greatly diversify the functional repertoire of these molecules, rapidly shaping cell functions to accommodate changes in the extracellular environment. These covalent modifications produce important effects on the structure, function, and likely the immunogenicity of the target protein (1–4). Although the discovery of nonribosomally encoded citrulline in proteins was first reported >50 years ago (5, 6), the importance of citrullination remained unclear until the last 10 years, when 2 major discoveries brought attention to this modification. The first finding was that patients with rheumatoid arthritis (RA) produce autoantibodies that recognize epitopes containing peptidylcitrulline, and that these autoantibodies are both highly specific for diagnosis and predictive of disease severity (7, 8). The second discovery was that histones become citrullinated (9), raising the possibility that, like other posttranslational histone modifications (i.e., phosphorylation, acetylation, and methylation), histone citrullination may regulate chromatin-templated nuclear events, including transcription (10, 11). The functional role of histone citrullination remains unclear (12).
The peptidyl arginine deiminase (PAD) enzymes hydrolyze guanidinium side chains in peptidyl arginine to yield peptidylcitrulline and ammonia, and belong to a larger group of guanidino-modifying enzymes called the amidinotransferase superfamily (13). To date, 5 human PAD isoenzymes have been identified (14). For historical reasons, these enzymes are designated PAD-1–PAD-4 and PAD-6 (14). PAD-4 is a homodimer that is distinguished by the insertion of a nuclear localization sequence and is the only PAD localized to the cell nucleus (15, 16). Among the PAD enzymes, PAD-4 has gained special attention as a potential candidate that may drive citrullination of self antigens in RA (8). The specific immune response to citrullinated proteins, the presence of increased levels of citrullinated proteins in synovial tissue and fluid from RA patients (17–19), and the genetic association of PADI4 polymorphisms with RA in some populations (20–23) strongly suggest that pathways which promote and/or restrain protein citrullination may be altered in this disease. Understanding the mechanisms that regulate PAD activity under physiologic or pathologic conditions is therefore a high priority.
In this study, we show that autocitrullination of PAD-4 inactivates its function and that the efficiency of this process (i.e., citrullination-induced inactivation of PAD-4) is distinct in the different PAD-4 variants relevant to RA. We identified multiple citrullination sites in PAD-4, and further defined Arg-372 and -374 as the potential autocitrullination targets that inactivate the enzyme. Finally, autocitrullination also modified the structure of PAD-4, augmenting its recognition by human autoantibodies. Taken together, these findings suggest that the extent of citrullination during cell activation represents an integrated function regulated by PAD-4 activation and by the efficiency of autocitrullination-induced inactivation of PAD-4, and that this process is influenced by known PAD-4 polymorphisms associated with RA. In addition, PAD-4 autocitrullination is a potential mechanism to explain its targeting by RA autoantibodies. Autocitrullination, which influences PAD-4, both enzymatically and immunologically, may play an important role in RA pathogenesis.
Materials and Methods
Human PAD-4 cloning, expression vectors, and recombinant human PAD-4 (rhPAD-4) purification
Total RNA was purified from ATRA-differentiated HL-60 cells and reverse-transcribed to generate complementary DNA (cDNA). PAD-4 cDNA was amplified by polymerase chain reaction and cloned into the Gateway (Invitrogen) vector pDEST-51 for mammalian expression and the pDEST-17 prokaryotic expression vector to generate an N-terminal His6-tagged fusion protein that was further purified by ÄKTA Prime Chromatography system (Amersham Biosciences). The purified protein was dialyzed against 10 mM Tris (pH 7.4), 300 mM NaCl, 200 µM dithiothreitol, 1 mM EDTA, and 10% glycerol. The purity of the preparation was >98% as assessed by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and Coomassie blue staining (data not shown). To generate plasmids encoding PAD-4 containing 3 nonsynonymous polymorphisms associated with RA susceptibility (PAD-4snp [Gly55→Ser, Val82→Ala, and Gly112→Ala]), PAD-4R372K and PAD-4R374K, PAD-4 cDNA into pDEST-51 was used as template for site-directed mutagenesis (Stratagene). The cDNA encoding PAD-4snp was also cloned into pDEST-17 for protein purification as described above.
Cell culture and cell transfection
HL-60 and 293T cells were cultured using standard procedures. Cell lines were transfected using Lipofectamine 2000 (Invitrogen).
In vitro citrullination assays
Using siliconized tubes (Sigma), 700 nM human rhPAD-4 was incubated alone or 0–700 nM rhPAD-4 was coincubated with 700 nM human recombinant histone H3.1 (New England Biolabs) in buffer A (i.e., 100 mM Tris [pH 7.6]) in the absence or presence of 10 mM CaCl2 or 5 mM EDTA as indicated in the figure legends. After 0–120 minutes at 37°C, reactions were stopped by adding SDS sample buffer and boiling. In some experiments, following rhPAD-4 incubation (i.e., after 0, 30, 60, or 120 minutes) in the absence or presence of 10 mM CaCl2, 200 nM noncitrullinated or citrullinated rhPAD-4 was further incubated in the presence of HL-60 cell lysate (7 × 106 cells/ml) generated in buffer B (100 mM Tris [pH 7.6], 150 mM NaCl, 1% Nonidet P40 [NP40], and protease inhibitors), plus 10 mM CaCl2. After 15 or 60 minutes at 37°C, reactions were stopped by adding SDS sample buffer and boiling. Protein citrullination was determined by anti–modified citrulline immunoblotting, according to the recommendations of the manufacturer (Millipore). Additionally, noncitrullinated and citrullinated rhPAD-4 were also used for mass spectrometry or immunoprecipitation assays. For these studies, 700 nM rhPAD-4 was incubated for 2 hours, which corresponds to the time at which PAD-4 autocitrullination reaches its plateau.
Neutrophil isolation
After institutional review board approval and informed consent were obtained, neutrophils were isolated from heparinized venous blood from healthy volunteers. Briefly, after Ficoll-Hypaque isolation of mononuclear cells, neutrophils were isolated by 2 cycles of red blood cell lysis using ACK lysing buffer (Quality Biological). The purified neutrophils were washed and resuspended at 3 × 106 cells/ml in Hanks' balanced salt solution without calcium/magnesium. At this point, neutrophil viability was typically >98%, as assessed by trypan blue dye exclusion.
Ca2+/ionomycin-induced cell activation
Neutrophils or 293T cells were stimulated with or without 1 µM ionomycin (Sigma) for 1–3 hours at 37°C as indicated in the figure legends. Following the incubation, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors, sonicated, and cleared by centrifugation. The supernatants were further used for immunoprecipitation and/or immunoblotting. In some experiments, following the incubation, the cells were directly lysed and boiled in SDS sample buffer for further analysis by immunoblotting. Protein citrullination was determined by anti–modified citrulline immunoblotting.
Antibodies and immunoprecipitation assays
Anti-human PAD-4 antibodies recognizing the C-terminal region (amino acids 519–528) were generated in rabbit (Covance) and affinity-purified using PAD-4 peptide 519–528. The anti-human PAD-4 N-terminal (amino acids 1–15) antibody was from Sigma, and the anti–green fluorescent protein (anti-GFP) antibody was from Invitrogen. Human anti–PAD-4 antisera have been described previously (24). Four anti–PAD-4 sera confirmed by Western blotting were randomly selected for the immunoprecipitation assays. Immunoprecipitations were performed using neutrophil or 293T cell lysate in RIPA buffer, and noncitrullinated or citrullinated rhPAD-4 (10 µg/ml) in buffer C (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP40 [pH 7.4]), as indicated in the figure legends.
Mass spectrometry analysis
Native or in vitro autocitrullinated PAD-4 were reduced, alkylated, and digested with trypsin (sequencing grade; Promega). Peptides were analyzed by liquid chromatography/tandem mass spectrometry on an LTQ ion trap mass spectrometer (Thermo Fisher Scientific) or a QSTAR/Pulsar mass spectrometer (Applied Biosystems/MDX Sciex) interfaced with a 2-dimensional nanoLC system (Eksigent). Peptides were fractionated in a reverse-phase C18 (5 µm, 120Å, YMC ODS-AQ; Waters) 75 µm × 100 mm column with a 10-µm emitter using 0–60% acetonitrile/0.5% formic acid gradient over 30 minutes at 300 nl/minute. For iTRAQ experiments, the iTRAQ reagents were dissolved in isopropanol, added to the digests, and incubated at room temperature for 2 hours. After labeling, the combined peptide mixture was fractionated by strong cation exchange chromatography on an Agilent high-performance liquid chromatography system using a PolySulfoethyl A column (2.1 × 100 mm, 5 µm, 300Å) (PolyLC). The absorbance at 214 nm was monitored, and 5 strong cation exchange fractions were collected along the gradient. Peptide sequences were identified using Mascot (www.matrixscience.com) or ProteinPilot (Applied Biosystems) software to search the National Center for Biotechnology Information nonredundant database with acquired fragmentation data using human as species, trypsin as enzyme (one missed cleavage allowed), Arg deamination as a variable modification, and cysteine static modification with methylmethanethiosulfate or amines with iTRAQ 8. The peptide confidence threshold cutoff for this study was at least 95% confidence using Mascot and 90% confidence using ProteinPilot.
Results
Self-citrullination of rhPAD-4 in vitro
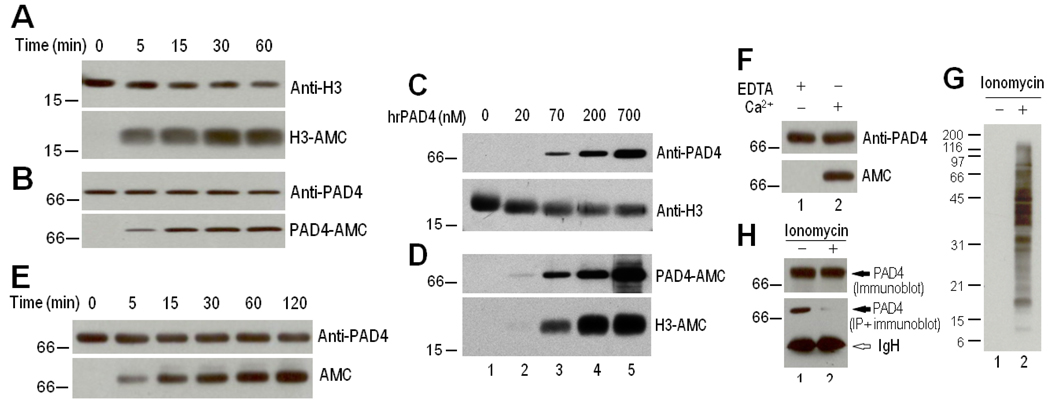
In preliminary in vitro studies of histone citrullination by PAD-4, we observed that rhPAD-4 is autocitrullinated. When recombinant histone H3.1 was coincubated with rhPAD-4, both proteins became citrullinated in a time-dependent manner, as determined by anti–modified citrulline immunoblotting (Figures 1A and B). Interestingly, citrullination of rhPAD-4 is as efficient as histone H3.1 citrullination even in the presence of excess amounts of substrate (Figures 1C and D), suggesting that citrullination of rhPAD-4 likely involves intramolecular and/or intermolecular events within the PAD-4 dimer complex. Citrullination of rhPAD-4 is therefore not affected by enzyme concentration. Furthermore, the incubation of rhPAD-4 alone in the absence of any substrate induced a similar pattern of autocitrullination of the enzyme (Figure 1E). Autocitrullination was confirmed to be calcium dependent (Figure 1F).
Figure 1.
Autocitrullination of recombinant human peptidyl arginine deiminase type 4 (rhPAD-4) in vitro. A–E, Recombinant human PAD-4 (700 nM in A, B, and E or 0–700 nM in C and D) was coincubated with 700 nM human recombinant histone H3.1 or incubated alone (E) in the presence of 10 mM CaCl2 at 37°C for 0–60 minutes (A and B), for 60 minutes (C and D), or for 0–120 minutes (E). F, Recombinant human PAD-4 (700 nM) was incubated in the presence of 5 mM EDTA or 10 mM CaCl2 for 120 minutes at 37°C. After termination of the reactions, the samples were electrophoresed and noncitrullinated and citrullinated proteins were detected by immunoblotting with anti–histone H3, anti–PAD-4 C-terminal antibody, and anti–modified citrulline (AMC). G and H, Immunoprecipitation (IP) of PAD-4 from control and activated neutrophils. Primary human neutrophils in Hanks' balanced salt solution containing 2 mM CaCl2 were incubated in the absence (lane 1) or presence (lane 2) of 1 µM ionomycin for 2 hours at 37°C. Following the incubation, the cells were lysed in radioimmunoprecipitation assay buffer. The samples were directly electrophoresed (G and top panel of H) or used to immunoprecipitate endogenous PAD-4 using a rabbit anti-human PAD-4 C-terminal antibody (bottom panel of H). Protein citrullination was detected by anti–modified citrulline immunoblotting (G), and endogenous PAD-4 (top panel of H) or immunoprecipitated PAD-4 (bottom panel of H) was detected by immunoblotting using the rabbit anti–PAD-4 C-terminal antibody.
Modification of the native structure of PAD-4 by citrullination
To further address whether PAD-4 is self-citrullinated in vivo, we initially attempted to isolate endogenous PAD-4 by immunoprecipitation using a rabbit anti-human PAD-4 antibody that recognizes an epitope in the C-terminal region of PAD-4 (amino acids 519–528). Lysates were generated from control neutrophils or activated neutrophils in which citrullination was induced by Ca2+/ionomycin (Figure 1G). Surprisingly, although PAD-4 was detected by immunoblotting using the anti–PAD-4 C-terminal antibody in both control and ionomycin-activated neutrophils, the antibody precipitated PAD-4 from control but not activated neutrophil lysates (Figure 1H). Thus, although this antibody recognizes denatured PAD-4 equally in control and ionomycin-activated neutrophils, the same antibody is unable to immunoprecipitate native PAD-4 exclusively from ionomycin-activated neutrophils. Since protein citrullination produces changes in protein structure (3), it is likely that autocitrullination of PAD-4 alters its structure and recognition by antibodies generated against noncitrullinated PAD-4. To further investigate this, we took a similar approach using PAD-4 that was citrullinated in vitro. Two different rabbit anti–PAD-4 antibodies (recognizing C-terminal epitopes [amino acids 519–528] and N-terminal epitopes [amino acids 1–15; Sigma] of human PAD-4) efficiently immunoprecipitated control but not citrullinated rhPAD-4 (Figure 2A, lanes 1–4). Because these rabbit antibodies were generated against small peptides derived from the PAD-4 molecule, they have a very restricted recognition and, therefore, they may lose reactivity against the citrullinated form of the protein. Since RA autoantibodies preferentially recognize citrullinated autoantigens, and PAD-4 is an important antibody target in RA (24, 25), we investigated whether human anti–PAD-4 sera might recognize citrullinated PAD-4. Notably, in contrast to the rabbit anti–PAD-4 antibodies, human anti–PAD-4 RA sera immunoprecipitated both noncitrullinated and citrullinated rhPAD-4, with a general preference for the citrullinated form (Figure 2A). In one case (serum 2489), the RA serum recognized citrullinated PAD-4 alone by immunoprecipitation. Taken together, these data demonstrate that citrullination of PAD-4 strikingly modifies the structure of the protein, decreasing recognition by antipeptide rabbit antibodies, and maintaining and enhancing its recognition by human RA autoantibodies.
Figure 2.
Autocitrullination of PAD-4 during cell activation. A, Immunoprecipitation of control and citrullinated rhPAD-4 by rabbit antibodies and by human rheumatoid arthritis (RA) sera. Noncitrullinated (native) rhPAD-4 and citrullinated (cit) rhPAD-4 were immunoprecipitated using rabbit anti-human PAD-4 antibodies against the C-terminal (Cterm) or N-terminal (Nterm) domains, or using human anti–PAD-4 RA sera (i.e., 2454, 1067, 2489, and 2314). Purified immune complexes were electrophoresed, and PAD-4 was detected by immunoblotting using anti–PAD-4 C-terminal antibody. B, Immunoprecipitation of PAD-4 from control and activated neutrophil lysates. Cell lysates were generated in radioimmunoprecipitation assay (RIPA) buffer from control neutrophils (lanes 1 and 3) and ionomycin-activated neutrophils (lanes 2 and 4), and endogenous PAD-4 was immunoprecipitated using human anti–PAD-4 serum. Then, control and ionomycin-activated samples were divided in two and immunoblotted using anti–PAD-4 C-terminal antibody (lanes 1 and 2) or anti–modified citrulline (lanes 3 and 4). C, Protein citrullination in 293T cells that were transiently transfected to express green fluorescent protein (GFP)–PAD-4. After 48 hours, the cells were incubated in the absence (lane 1) or presence (lane 2) of 1 µM ionomycin for 2 hours at 37°C. Following the incubation, the cells were lysed in RIPA buffer and electrophoresed to detect protein citrullination by anti–modified citrulline. D, Immunoprecipitation of GFP–PAD-4 from control and activated 293T-transfected cells. Purified immune complexes were electrophoresed, and equal protein loading was visualized by ponceau S staining (top) prior to anti–modified citrulline immunoblotting (bottom). See Figure 1 for other definitions.
Citrullination of PAD-4 during cell activation
Using human anti–PAD-4, we evaluated whether PAD-4 is autocitrullinated in vivo. Endogenous PAD-4 was immunoprecipitated from lysates generated from control or ionomycin-activated neutrophils using human anti–PAD-4 sera, and PAD-4 citrullination was evaluated by anti–modified citrulline immunoblotting. Notably, although RA sera immunoprecipitated PAD-4 from both control and activated neutrophil lysates (Figure 2B, lanes 1 and 2), a prominent citrullinated protein with molecular weight identical to that of PAD-4 was detected exclusively in lysates from ionomycin-activated neutrophils (Figure 2B, lane 4). These data strongly suggest that PAD-4 is indeed citrullinated in activated primary neutrophils. Since RA sera may contain antibodies that could precipitate other citrullinated proteins with a molecular weight similar to that of PAD-4, we generated a mammalian expression vector in which PAD-4 was N-terminally tagged with GFP. GFP–PAD-4 was expressed by transient transfection in 293T cells, and the GFP tag was used to immunoprecipitate PAD-4. Prominent protein citrullination was induced when GFP–PAD-4–expressing 293T cells were incubated in the presence of Ca2+/ionomycin (Figure 2C), demonstrating that GFP–PAD-4 is functional. This was similar to 293T cells expressing the untagged PAD-4 construct (as discussed below). GFP–PAD-4 was efficiently immunoprecipitated from both control and ionomycin 293T-transfected cells (Figure 2D, top panel). However, citrullination of GFP–PAD-4 was only detected in cells activated with the calcium ionophore (Figure 2D, bottom panel). Taken together, these data demonstrate that upon PAD-4 activation in cells, PAD-4 itself is a genuine target for citrullination.
Self-citrullination of PAD-4 inhibits its function
To determine the functional consequences of PAD-4 citrullination, we directly measured the function of rhPAD-4 (control and citrullinated) against macromolecular substrates. Thus, rhPAD-4 was incubated for 2 hours in the absence or presence of Ca2+ to generate noncitrullinated and citrullinated rhPAD-4 (Figure 3A). These were further incubated for 15 minutes and 60 minutes in the presence of HL-60 cell lysate as a source of macromolecular targets for citrullination (Figure 3B). As a negative control, HL-60 cell lysate was incubated alone. In the absence of added PAD-4, no citrullination was detected in the lysates by anti–modified citrulline immunoblotting at either 15 minutes or 60 minutes of incubation (Figure 3B, top panel, lanes 1 and 4). When noncitrullinated rhPAD-4 was coincubated with the cell lysate, time-dependent protein citrullination was observed (Figure 3B, top panel, lanes 2 and 5). In contrast, citrullinated PAD-4 had minimal citrullination activity (Figure 3B, top panel, lanes 3 and 6). No differences were found in the levels of control and citrullinated rhPAD-4 by immunoblotting, either at 15 minutes or 60 minutes of incubation in the lysates (Figure 3B, middle panel), confirming that these differences in PAD-4 activity did not result from rhPAD-4 degradation. These data demonstrate that autocitrullination of PAD-4 inactivates the function of the enzyme.
Figure 3.
Autocitrullination of PAD-4 inhibits its function. A, Generation of noncitrullinated rhPAD-4 (lane 1) and citrullinated rhPAD-4 (lane 2). Recombinant human PAD-4 was incubated in the absence or presence of 10 mM CaCl2 for 120 minutes at 37°C. Samples were immunoblotted with rabbit anti–PAD-4 (top) and anti–modified citrulline (bottom). B, Citrullination activity of control and citrullinated rhPAD-4. HL-60 cell lysates were incubated with buffer alone (lanes 1 and 4), noncitrullinated rhPAD-4 (lanes 2 and 5), or citrullinated (cit) rhPAD-4 (lanes 3 and 6) for 15 minutes (lanes 1–3) or 60 minutes (lanes 4–6) at 37°C. After termination of the reactions, the samples were electrophoresed and immunoblotted using anti–modified citrulline, anti–PAD-4 C-terminal antibody, and anti–β-actin (loading control). See Figure 1 for other definitions.
Potential citrullination sites in PAD-4
Citrullination is a posttranslational modification on arginine and is manifested as a loss of 0.98 daltons as compared with the mass of the “original” arginine. We used 2 different methods to identify the possible autocitrullination sites on PAD-4. The first method consisted of in solution digestion of citrullinated PAD-4 with trypsin and subsequent analysis on an LTQ ion trap mass spectrometer. The raw data file was searched with Mascot, using Deamidation (R) as a variable modification (deamidation has the same mass change as deimination). (See Supplementary Figure 1A, available on the Arthritis & Rheumatism Web site at http://www3.interscience.wiley.com/journal/76509746/home.) Using this method, 5 possible sites were identified as Arg-383, Arg-394, Arg-495, Arg-536, and Arg-544. The second approach was using iTRAQ technology and analysis on a QSTAR mass spectrometer. The iTRAQ technology enables relative quantitation of possible citrullination sites as compared with the unoccupied sites. Thus, native and citrullinated PAD-4 were independently digested and labeled, mixed together, and analyzed on the QSTAR. The raw files were searched with ProteinPilot software (see Supplementary Figure 1B, available on the Arthritis & Rheumatism website at http://www3.interscience.wiley.com/journal/76509746/home). The advantage of this approach is that it gives ratios between citrullinated peptide in nonmodified and modified samples. Citrullinated sites obtained by this method were Arg-205, Arg-212, Arg-218, Arg-372, Arg-374, and Arg-383. The complementary results probably arise from differences between the duty cycles and mass analyzers in the different mass spectrometers, as well as the possibility that iTRAQ labeling in the second approach may change the ionization efficiency of peptides and enable one to see peptides that might not be identified otherwise.
Interestingly, the 10 citrullination sites are clustered in 3 regions within the PAD-4 molecule: one N-terminal region and 2 C-terminal regions (Figure 4). The N-terminal cluster is located in the PAD-4 subdomain 2 (based on the crystal structure of PAD-4 [16,26]) and includes Arg-205, -212, and -218. In the C-terminal region, one cluster is located at the surface of -helix 11 and -helix 12 (Arg-495, -536, and -544), and the last cluster spans the active site cleft of PAD-4 (Arg-372, -374, -383, and -394). In this regard, Arg-372 and -374 are known to be directly involved in the recognition of substrate by the enzyme (26), and Arg-374 is absolutely required for citrullination of the target (26).
Figure 4.
Clustering of the potential citrullination sites in peptidyl arginine deiminase type 4 (PAD-4) into 3 distinct regions. A, Schematic representation of the secondary structure of PAD-4 (adapted from ref.24). The secondary structure elements in the N-terminal and C-terminal domains are shown in blue and yellow, respectively. Bars show -helices; arrows show β strands; and broken lines show disordered regions. The potential arginine targets for citrullination are shown in red. B, Tertiary structure of PAD-4, revealing clustering of citrullination sites into 3 distinct regions. The cluster comprising Arg-372, -374, -383, and -394 spans the active site cleft. The N-terminal and C-terminal domains are shown in blue and yellow, respectively, while the sites of arginine deimination are indicated in red. The model shown was generated from the Molecular Modeling Database (National Center for Biotechnology Information), according to coordinates generated by Arita et al (16). Adapted by permission from Macmillan Publishers Ltd: Nat Struct Mol Biol 2004;11(8):777–83. Copyright 2004.
Mechanisms of autocitrullination-induced inactivation of PAD-4
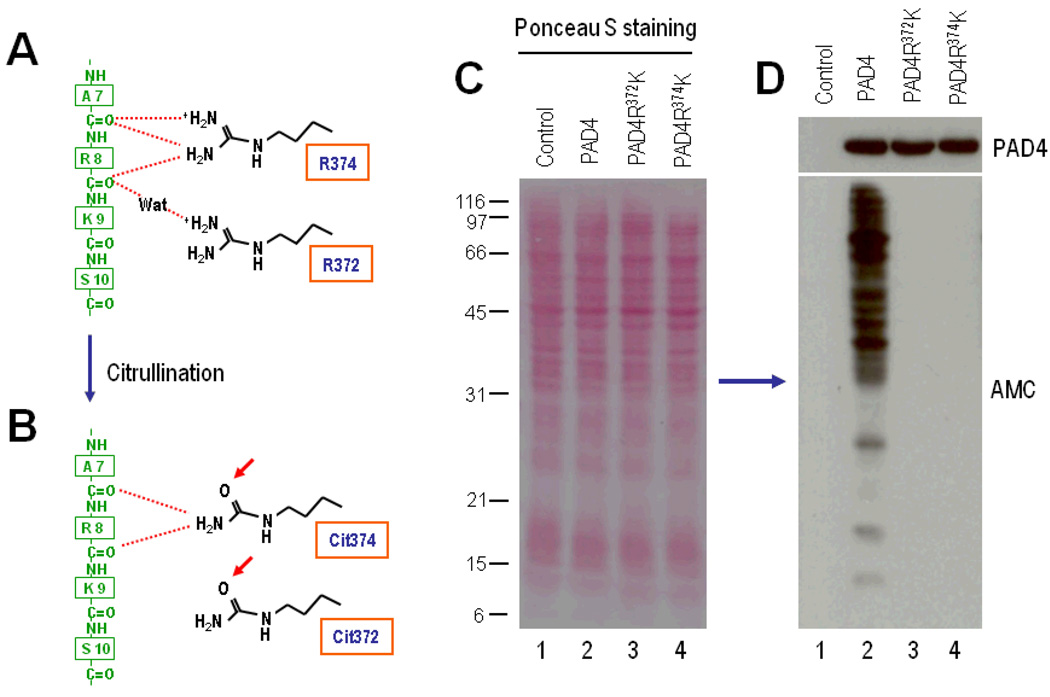
To further define the functional consequences of PAD-4 autocitrullination during cell activation, we focused on those citrullination sites that surround the active site cleft in PAD-4 (26). In this regard, the study of the tertiary structure of PAD-4 in complex with histone N-terminal peptides (26) has shown that the citrullination targets Arg-372 and Arg-374 are directly involved in substrate recognition (Figure 5A) and therefore, by affecting the enzyme–substrate interactions (Figure 5B), their citrullination may explain the inactivation of PAD-4. Arg-372 and Arg-374 were changed to the conserved amino acid lysine (i.e., PAD-4R372K and PAD-4R374K, respectively) to preserve charge but to lack the guanidino group removed during citrullination. Wild-type and mutant PAD-4 were expressed by transient transfection in 293T cells, and their citrullination activity was assessed by anti–modified citrulline immunoblotting in ionomycin-activated cells. Similar PAD-4 expression was observed among the wild-type and mutant variants (Figure 5D, top panel), and no citrullination activity was found in activated mock-transfected cells (Figure 5D, bottom panel, lane 1). Although protein citrullination was prominent in cells expressing wild-type PAD-4 (Figure 5D, bottom panel, lane 2), no citrullination activity was observed upon cell activation in cells expressing the PAD-4R372K and PAD-4R374K mutants (Figure 5D, bottom panel, lanes 3 and 4), highlighting the absolute requirement of the guanidino group of arginine at those sites in PAD-4 function, and strongly indicating that the modification of this group during citrullination events around the active site cleft (Figures 5A and B) may be responsible for inhibition of enzyme function.
Figure 5.
The PAD-4 citrullination sites Arg-372 and Arg-374 regulate PAD-4 activity during cell activation. A, Schematic diagram of the interactions between Arg-374 and Arg-372 at the active site cleft of PAD-4 and N-terminal residues in histone H3-1 (adapted from ref.26). Arg-374 makes multiple hydrogen bonds with backbone carbonyl oxygens of the Ala-7 (A7) and Arg-8 (R8) residues in histone H3.1 (green), and Arg-372 recognizes the carbonyl oxygen of the Arg-8 residue by means of water (Wat)–mediated hydrogen bonds. B, Conversion of Arg-372 and Arg-374 to citrullines (Cit) during PAD-4 autocitrullination, potentially distressing the interactions shown in A. C and D, The PAD4 mutants PAD4R372K and PAD4R374K lack citrullination activity. The 293T cells were mock transfected (lane 1) or transiently transfected to express PAD-4 (lane 2), PAD-4R372K (lane 3), or PAD-4R374K (lane 4). At 48 hours posttransfection, the cells were stimulated with 1 εM ionomycin for 1 hour. After termination of the reactions, the samples were electrophoresed, and equal protein loading was visualized by ponceau S staining (C) prior to anti–modified citrulline immunoblotting (bottom panel of D). PAD-4 expression was visualized by immunoblotting (top panel of D). See Figure 1 for other definitions.
Functional consequences of autocitrullination in polymorphic PAD-4 variants
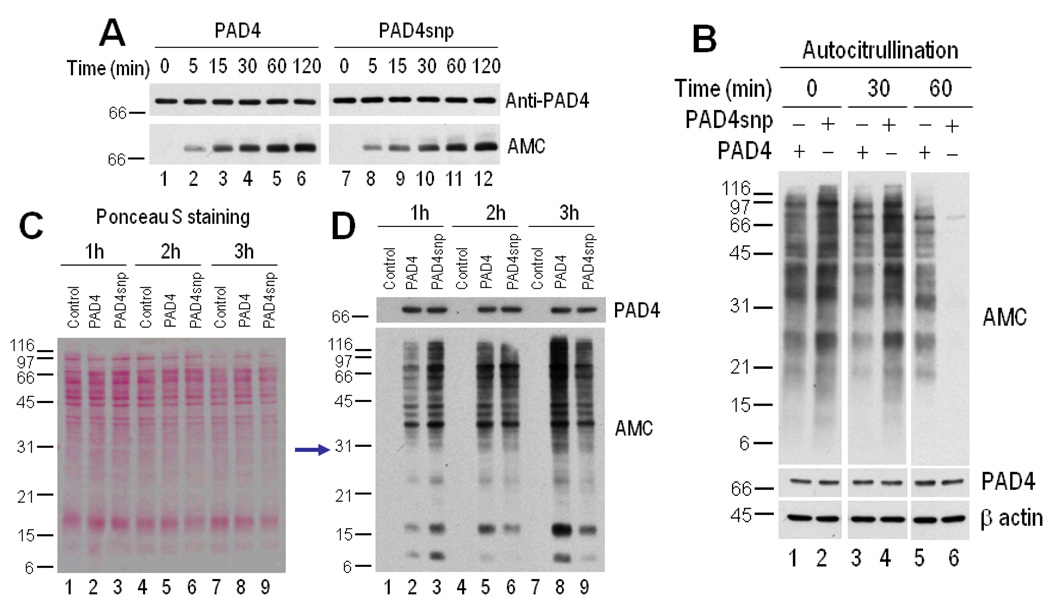
To gain insight into potential differences among PAD-4 structural variants relevant to RA, we addressed whether the efficiency of autocitrullination and/or citrullination-induced inactivation differed between the “nonsusceptible” variant of PAD-4 (used in the experiments described above) and the PAD-4 variant that has been associated with RA (referred to as PAD-4snp in this article) (20–23). Interestingly, although the kinetics of autocitrullination of rhPAD-4 and rhPAD-4snp were similar (Figure 6A), we found a striking difference in their pattern of citrullination-induced inactivation. Recombinant PAD-4 and rhPAD-4snp underwent autocitrullination for increasing time periods (i.e., 0, 30, and 60 minutes), and their residual activity was determined by further incubation for 15 minutes in the presence of HL-60 cell lysate (as a source of citrullination targets). This time of incubation was selected after preliminary experiments showed that protein citrullination by PAD-4 falls within the linear range of the assay at this time point. Interestingly, the rhPAD-4snp variant showed enhanced activity compared with rhPAD-4 when the enzymes were noncitrullinated (i.e., time 0), as well as early after autocitrullination (i.e., 30 minutes of autocitrullination before adding the substrate) (anti–modified citrulline in Figure 6B). However, as the process of autocitrullination progressed (i.e., 60 minutes postcitrullination), rhPAD-4 clearly retained better enzyme activity than rhPAD-4snp (anti–modified citrulline in Figure 6B). To further address whether the functional differences observed using purified PAD-4 variants also occur during cell activation, we defined the patterns of protein citrullination during activation of 293T cells that express PAD-4 or PAD-4snp by transient transfection. The data were fully consistent in the 2 systems. Thus, cells expressing PAD-4 showed prominent accumulation of citrullinated proteins over time, reaching a maximum at 3 hours after activation (anti–modified citrulline in Figure 6D). This time point may represent the point at which the enzyme is largely inactivated. In contrast, although cells expressing PAD-4snp showed a slight increase in protein citrullination compared with those expressing PAD-4 during early activation (anti–modified citrulline at 1 and 2 hours) (Figure 6D), the PAD-4snp cells showed decreased citrullination at later time points (anti–modified citrulline at 3 hours) (Figure 6D). Taken together, these data suggest that PAD-4snp is more efficiently inactivated than PAD-4.
Figure 6.
PAD-4 variants have distinct patterns of inactivation induced by autocitrullination. A, Autocitrullination of rhPAD-4 and rhPAD-4snp variants. Recombinant human PAD-4 and rhPAD-4snp were incubated in the presence of 10 mM CaCl2 at 37°C for 0–120 minutes. Noncitrullinated and citrullinated proteins were detected by immunoblotting with anti–PAD-4 (top) and anti–modified citrulline (bottom), respectively. B, Citrullination activity of control and citrullinated rhPAD-4 and rhPAD-4snp. Recombinant human PAD-4 (lanes 1, 3, and 5) or rhPAD-4snp (lanes 2, 4, and 6) were incubated in the presence of 10 mM CaCl2 for 0, 30, or 60 minutes at 37°C, and further incubated with HL-60 cell lysate for an additional 15 minutes at 37°C. After termination of the reactions, the samples were electrophoresed and immunoblotted using anti–modified citrulline, anti–PAD-4, and anti–β-actin (loading control). C and D, Citrullination activity of PAD-4 and PAD-4snp during cell activation. The 293T cells were mock transfected (lanes 1, 4, and 7) or transiently transfected to express PAD-4 (lanes 2, 5, and 8) or PAD-4snp (lanes 3, 6, and 9). At 48 hours posttransfection, the cells were stimulated with 1 µM ionomycin for 1 hour (lanes 1–3), 2 hours (lanes 4–6), 3 hours (lanes 7–9), and 5–6 hours (results not shown). After termination of the reactions, the samples were electrophoresed, and equal protein loading was visualized by ponceau S staining (C) prior to anti–modified citrulline immunoblotting (bottom panel of D). PAD-4 expression was visualized by immunoblotting (top panel of D). See Figure 1 for other definitions.
Discussion
Protein citrullination is emerging as an important posttranslational modification in both human disease and gene regulation. Among the PAD enzymes, PAD-4 has recently become a target of significant interest because of its suspected role in the pathogenesis of RA, and also because of its potential role in gene regulation. However, significant questions about PAD-4 regulation remain unanswered. Using biochemical and cellular approaches in primary human cells and cell lines, we demonstrated that PAD-4 is autocitrullinated in vitro and during cell activation, and that autocitrullination of PAD-4 has important effects on both the structure and the function of the enzyme. Autocitrullination thus provides an efficient mechanism of autoregulation that limits PAD-4–mediated protein citrullination.
Arginine deimination in PAD-4 occurs at 10 sites, corresponding to 37% of the arginine content in the enzyme and introducing a net of 1.5% citrullines in PAD-4. This magnitude of citrulline content can begin to induce disorder in organized protein structures (3). Indeed, striking changes in PAD-4 structure upon autocitrullination are clearly evident in the loss of citrullinated PAD-4 recognition by immunoprecipitation when using multiple different anti–PAD-4 antibodies. In contrast, human RA sera containing anti–PAD-4 antibodies recognize both noncitrullinated and citrullinated PAD-4 by immunoprecipitation, with a preference for the citrullinated form. Although it is not yet clear whether the same human antibodies recognize both noncitrullinated and citrullinated PAD-4, the recognition of citrullinated PAD-4 by RA sera is very clear, and quite distinct from the 2 rabbit anti–PAD-4 antibodies. The data suggest that these autoantibodies either recognize a part of PAD-4 whose access does not change significantly after citrullination or potentially that they recognize the modified molecule.
Based on the tertiary structure of PAD-4 in complex with histone N-terminal peptides (26), we focused the study of autocitrullination-induced inactivation of PAD-4 on those arginine residues located around the active site cleft in PAD-4, which we demonstrated to undergo citrullination. Among these sites, Arg-372 and Arg-374 are especially interesting because they are directly involved in substrate recognition (26) and, therefore, their citrullination may have critical consequences for the function of the enzyme. Indeed, our findings demonstrated that the guanidino group of the arginines at positions 372 and 374 is absolutely required for efficient PAD-4–mediated citrullination, as evidenced by the lack of citrullination activity of PAD-4 mutants in which these residues were changed to lysine. These results are consistent with the observation that the guanidino groups of these arginines make critical interactions with backbone carbonyl oxygens in the histone peptides (Figure 5A), stabilizing the substrate structure within the active site cleft of the enzyme (26). The lysine substitution used in this study is more conservative than the alanine replacement that has been used previously (26) and provides stronger evidence of a critical role of the guanidino group, which is converted to a ureido group during citrullination. We propose that citrullination at these sites, by affecting substrate recognition by the enzyme, plays an essential role in autocitrullination-induced inactivation of PAD-4. However, further studies are required to determine whether other citrullination sites may also affect the function of PAD-4 and whether arginines 372 and 374 are the only citrullination targets for PAD-4 inactivation. Regardless, our data suggest that the inactivation of PAD-4 through autocitrullination may play a role in limiting the production of citrullinated proteins, ensuring that protein citrullination is appropriate to physiologic state.
Despite the genetic association of PADI4 polymorphisms with RA in some populations (20–23), the role (if any) of the PAD-4 structural variant that is associated with RA susceptibility (PAD-4snp) remains unclear. When we explored whether the process of citrullination-induced inactivation of PAD-4 differed among the “nonsusceptible” and the “susceptible” PAD-4 variants, we found that while PAD-4snp was a little more active initially, it lost function more rapidly and more completely after autocitrullination. This was a highly reproducible observation, both in assays using purified components as well as in intact cell systems. Since the overall kinetics of autocitrullination appear to be similar in PAD-4 and PAD-4snp, this enhanced inhibition of PAD-4snp may reflect more efficient deimination of “critical” activity-related arginines (i.e., 372 and/or 374) in PAD-4snp. In support of this idea, the internal location of these arginines into the catalytic site of PAD-4 strongly supports that deimination at these sites is mediated through intramolecular events. Thus, taking into account that PAD-4snp appears to be more active than PAD-4, the initial high rate of PAD-4snp citrullination may also increase autocitrullination at the active site cleft and, consequently, induce a more rapid inactivation of the enzyme. To evaluate this hypothesis, further studies will require the development of novel tools to directly define the arginine-specific kinetics of autocitrullination among PAD-4 and PAD-4snp during cell activation.
Our findings appear to be inconsistent with current models that posit that the effect of PAD-4snp in RA is due to enhanced accumulation of citrullination in target antigens. Several possibilities are relevant in this regard. First, the activity of PAD-4snp appears to be higher than that of the nonsusceptible variant initially, but is later obscured by more efficient inactivation. It is possible that early citrullination activity is particularly important in the pathogenesis of RA. Second, PAD-4 (specifically PAD-4snp) may exert its effects on RA disease susceptibility as a consequence of its immunogenicity, rather than its enzyme activity. In support of this hypothesis is the observation that the production of anti–PAD-4 antibodies is strongly associated with PAD-4snp (24), and that anti–PAD-4 antibodies are associated with more severe, erosive disease in RA (24, 27, 28). Thus, it is possible that PAD-4snp structure may change more dramatically upon autocitrullination or that autocitrullination may occur at different and/or additional sites compared with PAD-4, allowing a more efficient inactivation of the enzyme, and modifying immunogenicity. Other PADs (e.g., PAD-2) may have more critical roles in the production of citrullinated proteins targeted in RA. Regardless of the mechanism, our findings suggest that the distinct inhibition pattern of PAD-4snp by autocitrullination reflects an important difference, which should be further evaluated. Demonstrating the presence of citrullinated PAD-4 in vivo in RA remains an important priority.
Supplementary Material
Acknowledgments
F.A. is supported a Dana Foundation Scholars Program in Human Immunology, The Donald B. and Dorothy L. Stabler Foundation and NIH grant P30 AR053503. A.R. is supported by NIH Grant R37 DE-12354 and ACR-REF Within our Reach grant. F.A is a Lowe Family Scholar in the Johns Hopkins Bayview Center for Innovative Medicine. A.R. is a Hugh and Renna Cosner Scholar in Translational Research.
REREFENCES
- 1.Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biol Chem. 2008;389(4):353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q, Wang Y. High mobility group proteins and their post-translational modifications. Biochim Biophys Acta. 2008;1784(9):1159–1166. doi: 10.1016/j.bbapap.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem. 1996;271(48):30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- 4.Doyle HA, Mamula MJ. Posttranslational modifications of self-antigens. Ann N Y Acad Sci. 2005;1050:1–9. doi: 10.1196/annals.1313.001. [DOI] [PubMed] [Google Scholar]
- 5.Rogers GE. Occurrence of citrulline in proteins. Nature. 1962;194:1149–1151. doi: 10.1038/1941149a0. [DOI] [PubMed] [Google Scholar]
- 6.Rogers GE, Simmonds DH. Content of citrulline and other amino-acids in a protein of hair follicles. Nature. 1958;182(4629):186–187. doi: 10.1038/182186a0. [DOI] [PubMed] [Google Scholar]
- 7.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101(1):273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klareskog L, Ronnelid J, Lundberg K, Padyukov L, Alfredsson L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol. 2008;26:651–675. doi: 10.1146/annurev.immunol.26.021607.090244. [DOI] [PubMed] [Google Scholar]
- 9.Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem Biophys Res Commun. 2002;290(3):979–983. doi: 10.1006/bbrc.2001.6303. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 11.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1(7):433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 13.Shirai H, Blundell TL, Mizuguchi K. A novel superfamily of enzymes that catalyze the modification of guanidino groups. Trends Biochem Sci. 2001;26(8):465–468. doi: 10.1016/s0968-0004(01)01906-5. [DOI] [PubMed] [Google Scholar]
- 14.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25(11):1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 15.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277(51):49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 16.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11(8):777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 17.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, et al. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum. 2008;58(8):2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 18.Matsuo K, Xiang Y, Nakamura H, Masuko K, Yudoh K, Noyori K, et al. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res Ther. 2006;8(6):R175. doi: 10.1186/ar2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S, et al. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 2005;44(1):40–50. doi: 10.1093/rheumatology/keh414. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 21.Kang CP, Lee HS, Ju H, Cho H, Kang C, Bae SC. A functional haplotype of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Koreans. Arthritis Rheum. 2006;54(1):90–96. doi: 10.1002/art.21536. [DOI] [PubMed] [Google Scholar]
- 22.Hoppe B, Haupl T, Gruber R, Kiesewetter H, Burmester GR, Salama A, et al. Detailed analysis of the variability of peptidylarginine deiminase type 4 in German patients with rheumatoid arthritis: a case-control study. Arthritis Res Ther. 2006;8(2):R34. doi: 10.1186/ar1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77(6):1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris ML, Darrah E, Lam GK, Bartlett SJ, Giles JT, Grant AV, et al. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum. 2008;58(7):1958–1967. doi: 10.1002/art.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takizawa Y, Sawada T, Suzuki A, Yamada R, Inoue T, Yamamoto K. Peptidylarginine deiminase 4 (PADI4) identified as a conformation-dependent autoantigen in rheumatoid arthritis. Scand J Rheumatol. 2005;34(3):212–215. doi: 10.1080/03009740510026346-1. [DOI] [PubMed] [Google Scholar]
- 26.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci U S A. 2006;103(14):5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halvorsen EH, Pollmann S, Gilboe IM, van der HD, Landewe R, Odegard S, et al. Serum IgG antibodies to peptidylarginine deiminase 4 in rheumatoid arthritis and associations with disease severity. Ann Rheum Dis. 2008;67(3):414–417. doi: 10.1136/ard.2007.080267. [DOI] [PubMed] [Google Scholar]
- 28.Halvorsen EH, Haavardsholm EA, Pollmann S, Boonen A, van der HD, Kvien TK, et al. Serum IgG antibodies to peptidylarginine deiminase 4 predict radiographic progression in patients with rheumatoid arthritis treated with tumour necrosis factor-alpha blocking agents. Ann Rheum Dis. 2009;68(2):249–252. doi: 10.1136/ard.2008.094490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.