Abstract
The establishment of functional retinal circuits in the mammalian retina depends critically on the proper generation and assembly of six classes of neurons, five of which consist of two or more subtypes that differ in morphologies, physiological properties, and/or sublaminar positions. How these diverse neuronal types and subtypes arise during retinogenesis still remains largely to be defined at the molecular level. Here we show that all four family members of the early B-cell factor (Ebf) helix-loop-helix transcription factors are similarly expressed during mouse retinogenesis in several neuronal types and subtypes including ganglion, amacrine, bipolar, and horizontal cells, and that their expression in ganglion cells depends on the ganglion cell specification factor Brn3b. Misexpressed Ebfs bias retinal precursors toward the fates of non-AII glycinergic amacrine, type 2 OFF-cone bipolar and horizontal cells, whereas a dominant-negative Ebf suppresses the differentiation of these cells as well as ganglion cells. Reducing Ebf1 expression by RNA interference (RNAi) leads to an inhibitory effect similar to that of the dominant-negative Ebf, effectively neutralizes the promotive effect of wild-type Ebf1, but has no impact on the promotive effect of an RNAi-resistant Ebf1. These data indicate that Ebfs are both necessary and sufficient for specifying non-AII glycinergic amacrine, type 2 OFF-cone bipolar and horizontal cells, whereas they are only necessary but not sufficient for specifying ganglion cells; and further suggest that Ebfs may coordinate and cooperate with other retinogenic factors to ensure proper specification and differentiation of diverse retinal cell types and subtypes.
Introduction
In the mammalian retina, seven classes of neuronal and glial cells comprise a delicate multilayered sensorineural epithelium. They are the rod and cone photoreceptors; the horizontal, bipolar, and amacrine cell (AC) interneurons; the retinal ganglion cell (RGC) output neurons; and the Müller glial cells. Except for rods, all classes of neurons consist of two or more subtypes that differ in morphologies, physiological properties, and/or sublaminar positions, with ACs and RGCs as the most diversified classes (Wässle and Boycott, 1991; Masland, 2001a,b). For instance, bipolar cells are composed of one type of rod bipolar and nine subtypes of cone bipolar cells, whereas horizontal cells have only two subtypes (Dacey et al., 1996; Hack and Peichl, 1999; Ghosh et al., 2004). RGCs come in 10–15 subtypes, and there are at least 29 subtypes of ACs (Wässle and Boycott, 1991; MacNeil and Masland, 1998; MacNeil et al., 1999; Masland, 2001b). Most ACs contain either glycine or GABA inhibitory neurotransmitters and therefore form two major neurotransmitter subtypes of approximately equal number (Vaney, 1990; Marquardt et al., 2001).
During retinogenesis, the seven classes of cells are generated from multipotent progenitors following a loose temporal order by the coordinated action of various intrinsic and extrinsic factors (Harris, 1997; Cepko, 1999; Livesey and Cepko, 2001). Transcription factors have emerged as the most important intrinsic regulators that control the determination and differentiation of different retinal cell types. In the mouse, for instance, Math5 activates the expression of Brn3b and Isl1, and the three of them constitute a major molecular pathway underlying RGC specification and differentiation (Brown et al., 2001; Wang et al., 2001; Mu et al., 2008; Pan et al., 2008; Qiu et al., 2008; Badea et al., 2009). The specification of ACs involves the concerted activities of Foxn4, Neurod1, Math3, and Ptf1a (Inoue et al., 2002; Li et al., 2004; Fujitani et al., 2006; Nakhai et al., 2007), whereas Bhlhb5, Nr4a2, and Isl1 are involved in specifying GABAergic or cholinergic ACs (Feng et al., 2006; Elshatory et al., 2007; Jiang and Xiang, 2009). Foxn4 also acts upstream of Ptf1a, Prox1, and Lim1 to control the specification and differentiation of horizontal cells (Dyer et al., 2003; Li et al., 2004; Fujitani et al., 2006). Fate determination of bipolar cells relies on the synergistic activities between Chx10 and the basic helix-loop-helix (HLH) factors Mash1, Math3, and Ngn2 (Burmeister et al., 1996; Tomita et al., 2000; Akagi et al., 2004). Bhlhb4 is critically involved in the terminal differentiation of rod bipolar cells, whereas Vsx1, Irx5, and Bhlhb5 are all required for the specification of cone bipolar subtypes (Bramblett et al., 2004; Chow et al., 2004; Ohtoshi et al., 2004; Cheng et al., 2005; Feng et al., 2006).
The vertebrate early B-cell factor (Ebf) proteins (Ebf1-4) belong to a small family of HLH transcription factors that contain a non-basic HLH dimerization domain and an atypical zinc-finger DNA-binding domain (Dubois and Vincent, 2001; Wang et al., 2002). They are expressed in distinct yet overlapping patterns in multiple tissues and cell types, including the B-lymphocytes, adipocytes, osteoblasts, and neuronal cells, and play crucial roles during their development (Lin and Grosschedl, 1995; Akerblad et al., 2002; Wang et al., 2004; Kieslinger et al., 2005; Croci et al., 2006). In the mouse retina, Ebf1 and 3 were previously shown to be expressed by differentiating and mature RGCs and/or ACs (Davis and Reed, 1996; Wang et al., 1997; Trimarchi et al., 2007). Despite this prominent expression, however, the function of Ebfs during retinogenesis is unclear at present. Here we investigated their expression patterns and functions in the mouse retina.
Materials and Methods
Animals.
All experiments with mice were performed in accordance with animal protocols approved by the University of Medicine and Dentistry of New Jersey. The animals were housed and bred at a facility of the University and fed with a standard rodent diet. The C57BL/6J mice were purchased from the Jackson Laboratory, and CD1 mice from the Charles River Laboratories. The Brn3b knock-out mice were generated previously (Gan et al., 1996) and maintained by breeding with C57/BL6J mice. The stage of mouse embryos was determined by taking the morning when the copulation plug was seen as E0.5. An approximately equal number of male and female animals were used, and all genotypes described were confirmed by PCR.
Retrovirus preparation and infection.
The full-length Ebf1, Ebf2, and Ebf3 open reading frames were obtained by reverse transcriptase PCR from C57BL/6J mouse retinal RNA, and verified by DNA sequencing. To construct Ebf-EnR, a DNA fragment corresponding to amino acids 1-430 of Ebf1 was fused with a fragment containing the repressor domain of the Drosophila-engrailed protein (EnR) (Akerblad et al., 2002), followed by a small Myc-tag sequence. These fragments were then ligated into the Control-green fluorescent protein (GFP) retroviral vector (Mo et al., 2004) to yield Ebf1-GFP, Ebf2-GFP, Ebf3-GFP, and Ebf-EnR-GFP viral plasmids. Retroviruses were prepared as described previously (Mo et al., 2004) and stored at −80°C. Before infection, retrovirus mixture was prepared by mixing viruses with polybrene (Sigma) to a final concentration of 8 μg/ml. For in vivo infection, 0.5–1 μl of virus mixture was injected into the subretinal space of postnatal day (P) 0 neonates, and retinal samples were collected at P21 for analysis. For in vitro infection of retinal explants, 20 μl of virus mixture was overlaid onto the explant, and this procedure was repeated once 30 min later. Retinal explant culture was performed as described previously (Tomita et al., 1996).
Small hairpin RNA plasmids and electroporation.
The pU6-Ebf1i5 small hairpin RNA (shRNA) plasmid was generated by inserting the following annealed oligonucleotides into the pBS/U6 (pU6) RNA interference (RNAi) vector (Sui et al., 2002): 5′-GATCCCCAGAGGTTACAGAAGGTCATTTCAAGAGAATGACCTTCTGTAACCTCTGGTTTTTTG-3′ and 5′-AATTCAAAAAACCAGAGGTTACAGAAGGTCATTCTCTTGAAATGACCTTCTGTAACCTCTGGG-3′. pCIG-Ebf1, Ebf2, Ebf3, and Ebf1sm expression plasmids were constructed by inserting the corresponding open reading frames into a modified pCIG vector that contains a cytoplasmic instead of nuclear GFP (Megason and McMahon, 2002). The RNAi-resistant Ebf1sm was obtained by PCR-mediated, site-directed mutagenesis. pBS/U6 and pCIG plasmids were mixed at a ratio of 2:1 μg/μl and 1–2 μl of the mixture were injected into the subretinal space of P0 CD1 mice. Immediately after injection, five square pulses (100 V) of 50 ms duration with 950 ms intervals were applied using the pulse generator ECM 830 (BTX) as described previously (Matsuda and Cepko, 2004). Transfected retinas were collected at P12 for analysis.
Immunostaining and quantification.
Immunostaining of retinal sections and quantification of immunoreactive cells were performed as described previously (Li et al., 2004; Mo et al., 2004). The following primary antibodies were used: rabbit anti-Barhl2 (Mo et al., 2004); mouse anti-bromodeoxyuridine (BrdU)-FITC (BD Biosciences); mouse anti-Brn3a (Millipore); rabbit and goat anti-Brn3b (Santa Cruz Biotechnology); goat anti-Bhlhb5/BETA3 (Santa Cruz Biotechnology); rabbit and mouse anti-calbindin D-28k (Swant); mouse anti-calretinin (Millipore); goat anti-choline acetyltransferase (ChAT) (Millipore); sheep anti-Chx10 (Exalpha); rabbit anti-Dab1 (Santa Cruz Biotechnology); rabbit and goat anti-Ebf (Santa Cruz Biotechnology) (Davis and Reed, 1996); rabbit, mouse, and goat anti-GFP (MBL International, Millipore, and Abcam, respectively); rabbit anti-GABA (Sigma); mouse anti-GAD65 (BD Biosciences); mouse anti-glutamine synthetase (Millipore); rabbit anti-glycine receptor GlyRα1/2 (Abcam); goat anti-GLYT1 (Millipore); mouse anti-Isl1 (DSHB); mouse anti-Ki67 (BD PharMingen); mouse anti-Lim1 (DSHB); rabbit and mouse anti-Pax6 (Millipore and DSHB, respectively); rabbit anti-phosphorylated histone H3 (Millipore); mouse anti-protein kinase C-α (PKCα) (GE Healthcare); rabbit anti-recoverin (Millipore); mouse anti-syntaxin (Sigma); and sheep and rabbit anti-tyrosine hydroxylase (Millipore). Images were captured with a Nikon Eclipse 80i microscope or a laser scanning Leica TCS-SP2 confocal microscope.
To quantify GFP+ cells and GFP+ cells colocalized with cell type-specific markers on retinal sections, positive cells were counted in an optical field using a reticule mounted on the microscope. At least several hundred GFP+ cells (ranging from 201 to 3401 and mostly >1000 depending on frequency of colocalized cells) were scored for each retina, and three or more retinas from different animals were counted for each type. All data were tested for significance using two sample Student's t test with unequal variances.
In situ hybridization and probes.
RNA in situ hybridization was performed as described using digoxigenin-labeled antisense riboprobes (Sciavolino et al., 1997). The DIG RNA Labeling Mix and anti-digoxigenin-AP Fab fragments were obtained from Roche Diagnostics. Riboprobes were prepared following the manufacturer's protocol. The following probes were used: Ebf1 (GenBank accession #NM_007897, 1195–2248 bp); Ebf2 (Mella et al., 2004); Ebf3 (GenBank accession #NM_001113415, 288–1022 bp); and Ebf4 (GenBank accession #NM_001110513, 787–1606 bp).
BrdU labeling and terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling assay.
BrdU labeling and terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) assay were performed as previously described (Li et al., 2004). The mouse FITC-conjugated anti-BrdU antibody was purchased from BD Sciences. TUNEL assays were performed using the In Situ Cell Death Detection Kit TMR Red (Roche Diagnostics). The number of positive cells was scored on retinal sections of the intermediate region in a high-power (600×) optical field using a reticule mounted on the microscope. Six fields were counted on each retina, and at least three retinas were scored for each type.
VISTA analysis.
Ebf noncoding sequences were downloaded from all available vertebrate species using the Non-Coding Sequence Retrieval System (Doh et al., 2007). They were then aligned and compared by the online VISTA tool (http://genome.lbl.gov/vista) (Frazer et al., 2004) to identify conserved regions.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assay was performed with modification according to the protocol by Abcam (http://www.abcam.com/ps/pdf/chromatin/X-ChIP-protocol-card.pdf). Chromatin DNA was prepared from developing mouse retinas pooled from stages E15.5, E17.5, and P0. The anti-Brn3b antibodies Brn3 (C-13) and Brn3b (N-15) are commercially available (Santa Cruz Biotechnology). The PCR primers used to amplify various regions of Ebf3 are as follows: CR, 5′-CTGACAATATCAAATCAGACACCAG-3′ and 5′-CAGAACCAAACAGTACAAACACG-3′; 5′-flanking, 5′-GGACACCTCTGCACTCGAGTCTAG-3′ and 5′-TCTATGGACAACCTCTTGACTCTC-3′; intron, 5′-ACTGAGTGCCTACTATGTGCCAG-3′ and 5′-CGTCAGAACTGACAGGTGTTGTGA-3′; and 3′-flanking, 5′-AGCTGTTCTGTCCAGCACATACCAG-3′ and 5′-ACCCTTCCATACCTGCATCATGTG-3′.
Results
Inactivating Brn3b downregulates expression of Ebf genes in the retina
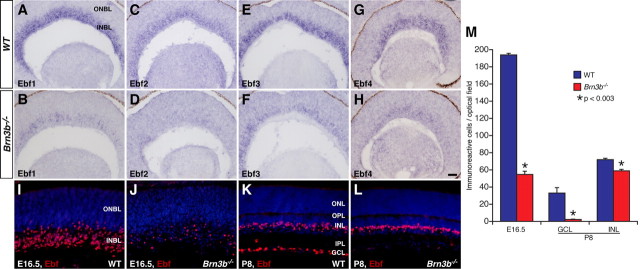
Our previous microarray profiling of gene expression in Brn3b mutant retinas has demonstrated an enrichment of transcription factor genes in the differentially expressed gene set (Qiu et al., 2008). Among them are the Ebf1 and Ebf3 transcription factor genes whose expression is significantly downregulated in Brn3b-null retinas (Qiu et al., 2008). By RNA in situ hybridization, we investigated whether the expression of all members of the Ebf family is downregulated in the Brn3b mutant. In E14.5 wild-type (WT) retinas, all four members of Ebf genes are expressed within the inner neuroblastic layer (INBL) (Fig. 1A,C,E,G). In Brn3b-null retinas, consistent with the microarray result, Ebf1 and Ebf3 transcripts are markedly reduced (Fig. 1A,B,E,F); as well, there is a great reduction of Ebf2 and Ebf4 expression (Fig. 1C,D,G,H). Immunostaining with a pan-Ebf antibody (supplemental Fig. S1, available at www.jneurosci.org as supplemental material) confirmed the dramatic downregulation of Ebf protein expression in Brn3b-null retinas at E16.5 (Fig. 1I,J,M). At P8, however, despite the drastic loss (∼93%; mean ± SD, WT: 33.0 ± 6.5 cells/optical field, n = 4; Brn3b−/−: 2.3 ± 0.9 cells/optical field, n = 4) of Ebf-expressing cells in the ganglion cell layer (GCL), there is only a small decrease (∼18%; mean ± SD, WT: 72.0 ± 6.4 cells/optical field, n = 4; Brn3b−/−: 59.0 ± 2.5 cells/optical field, n = 4) of Ebf-expressing cells within the inner nuclear layer (INL) in the mutant (Fig. 1K–M). This small change is consistent with the previously observed overall reduction (∼15%) of INL nuclei in the Brn3b mutant retina (Gan et al., 1996). Thus, all Ebf genes appear to be expressed in the developing inner retina. Ebf expression in RGCs depends on Brn3b, whereas expression in the INL is most likely independent of Brn3b, suggesting potential roles for Ebfs during RGC and non-RGC development.
Figure 1.
Altered expression of Ebfs in the Brn3b mutant retina. A–H, Sections from E14.5 wild-type and Brn3b−/− retinas were in situ hybridized with the indicated Ebf probes. Compared with the wild-type retina, there is a great decrease of Ebf1-4 expression in the mutant retina. I–L, Retinal sections from wild-type and Brn3b mutant mice at the indicated stages were immunostained with a pan-Ebf antibody and weakly counterstained with DAPI. In the mutant retina, there is a dramatic decrease of Ebf-immunoreactive cells within the INBL and GCL at E16.5 and P8, respectively; whereas, those within the INL at P8 show only a small reduction. M, Quantitation of Ebf-immunoreactive cells in Brn3b wild-type and mutant retinas. Each histogram represents the mean ± SD for 4 retinas. IPL, Inner plexiform layer; ONBL, outer neuroblastic layer; OPL, outer plexiform layer. Scale bar (in H): A–H, 50 μm; I–L, 25 μm.
Ebf3 as a direct Brn3b target
The expression of Ebf genes in RGCs and their downregulation in Brn3b-null retinas suggest that Ebfs may be direct targets of Brn3b. Given that regulatory sequences of a particular gene are usually conserved in the 5′ upstream region among vertebrate paralogs, we took a bioinformatic approach to identify conserved sequences in the 5′ upstream region of Ebf loci as candidate Brn3b cis-regulatory sequences. We used the Non-Coding Sequence Retrieval System (Doh et al., 2007) to download Ebf1-4 noncoding sequences from all available vertebrate species, then applied the VISTA program (Frazer et al., 2004) to identify 5′ conserved regions. We found several small conserved regions in the 5′-flanking sequence of Ebf3, but none were seen in a ∼30 kb 5′-flanking sequence of Ebf1, Ebf2, and Ebf4 (Fig. 2A). Among the conserved mouse Ebf3 sequences, a 149 bp region (CR) is highly conserved from human to zebrafish and contains a core Brn3-binding motif TAATTAAA (Fig. 2A,B) (Gruber et al., 1997).
Figure 2.
In vivo binding of Ebf3 promoter by Brn3b. A, VISTA analysis identifies several regions in the 5′ promoter of Ebf3 conserved among mouse and other vertebrate paralogs. One of them (CR) is ∼5.5 kb upstream and contains a core Brn3-binding motif. The y-axis represents percentage sequence identity. Exons are shown in blue, 5′ untranslated region in cyan, and conserved noncoding sequences in pink. B, Alignment of mouse and zebrafish DNA sequences in the CR region of Ebf3. Identical base pairs are shaded, and the core Brn3-binding motif is outlined. C, ChIP assay showing in vivo occupation by Brn3b of CR but not other Ebf3 sequences in the 5′-flanking, intron, or 3′-flanking regions that do not contain Brn3-binding sites. ChIP assays were performed with chromatin DNA prepared from pooled mouse retinas of stages E15.5, E17.5, and P0. Brn3b(C) and Brn3b(N) refer to antibodies against the C and N termini of Brn3b, respectively. Input lane represents 12% of chromatin DNA used for ChIP assay.
To test the ability for Brn3b to bind and occupy the CR region of Ebf3, we performed ChIP assay using chromatin DNA prepared from developing mouse retinas. Two different anti-Brn3b antibodies specifically immunoprecipitated the CR fragment, whereas the control IgG did not (Fig. 2C). Moreover, neither of the anti-Brn3b antibodies precipitated other Ebf3 sequences in the 5′-flanking, intron, or 3′-flanking regions that do not contain any Brn3-binding sites (Fig. 2C), demonstrating specific in vivo binding of Ebf3 promoter by Brn3b.
Biphasic expression of Ebf factors in developing RGCs and non-RGCs
As a first step to determine all retinal cell types that express Ebf factors and their potential role during retinal development, we performed RNA in situ hybridization and immunostaining analyses to investigate their spatial and temporal expression patterns in the developing mouse retina. Ebf1, Ebf3, and Ebf4 exhibit a nearly identical expression pattern. They all commence their expression around E11.5–E12.5 when their transcripts are seen in cells within the INBL of the central retina (Fig. 3A,C–E,G,H). This expression spreads gradually from the central to the peripheral region, and eventually to the entire retina within the INBL by E14.5 (Fig. 3I,K,L). The onset of Ebf2 expression is delayed until E14.5 when there is a weak Ebf2 expression within the INBL (Fig. 3B,F,J), but from E17.5 onward all Ebf genes exhibit an indistinguishable expression pattern. From E17.5 to P0, their expression maintains in the INBL (Fig. 3M–T). At P10, however, there is a strong expression in the GCL as well as a weak expression in the INL (Fig. 3U–X), suggesting that all Ebf genes are expressed by developing RGCs as well as other cell types located in the INL.
Figure 3.
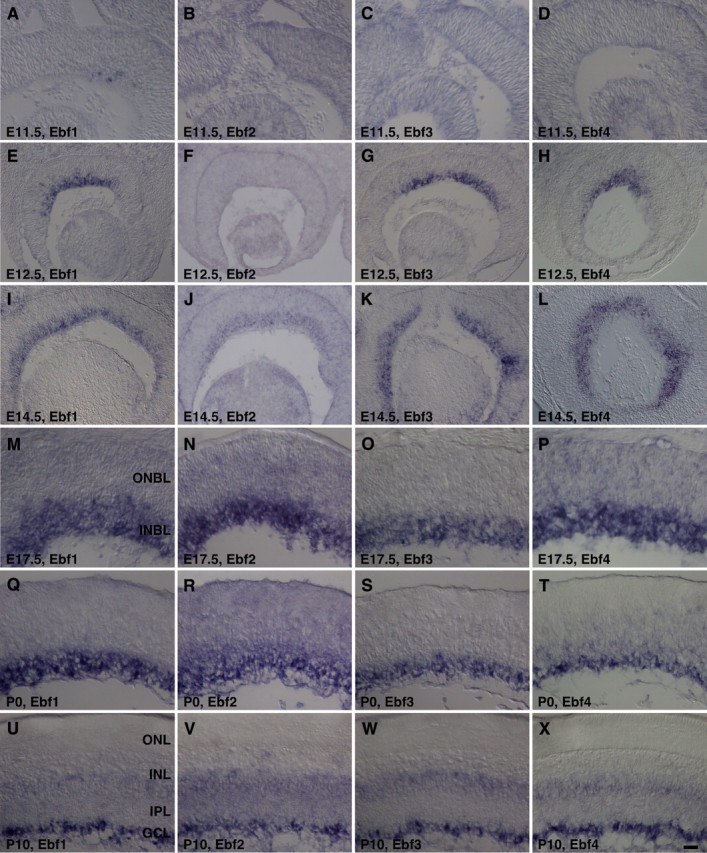
Expression patterns of Ebf genes during mouse retinogenesis. A–X, Retinal sections from the indicated developmental stages were in situ hybridized with Ebf1-4 probes. Ebf1, Ebf3, and Ebf4 have a similar spatiotemporal expression pattern, whereas Ebf2 expression is delayed. They are all expressed in the INBL during embryogenesis and in the GCL and/or INL postnatally. IPL, Inner plexiform layer; ONBL, outer neuroblastic layer. Scale bar (in T): E–L, 50 μm; A–D, M–X, 25 μm.
Pan-Ebf antibodies (supplemental Fig. S1, available at www.jneurosci.org as supplemental material) were used for immunofluorescence to examine the expression patterns of Ebf factors at the cellular level. Consistent with the in situ hybridization pattern, Ebf proteins are initially seen in a few cells in the INBL of the central retina at E11.5 (Fig. 4A). Their expression then gradually expands over time to abundant cells within the INBL by E17.5 (Fig. 4B–D). In postnatal retinas, Ebf factors remain strongly expressed in many cells of the GCL (Fig. 4E–I). Starting from P4, many additional Ebf-expressing cells, initially weakly labeled but later strongly stained, appear in the emerging INL and then maintain in the INL (Fig. 4G–I,P), consistent with the observed Ebf transcript distribution in both the GCL and INL at P10 (Fig. 3U–X). The distribution of Ebf-expressing cells only in the INBL and GCL before P4 implies that Ebf factors may be expressed mostly in RGCs at early stages. This appears to be the case since Ebfs are colocalized with the RGC marker Brn3b in nearly all Ebf-expressing cells at E16.5 (∼98%), but they fail to coexpress in the INL at P5 (Fig. 4M–P). Thus, the expression of Ebf factors is biphasic—embryonic onset of their expression primarily in RGCs and postnatal onset of their expression in non-RGCs.
Figure 4.
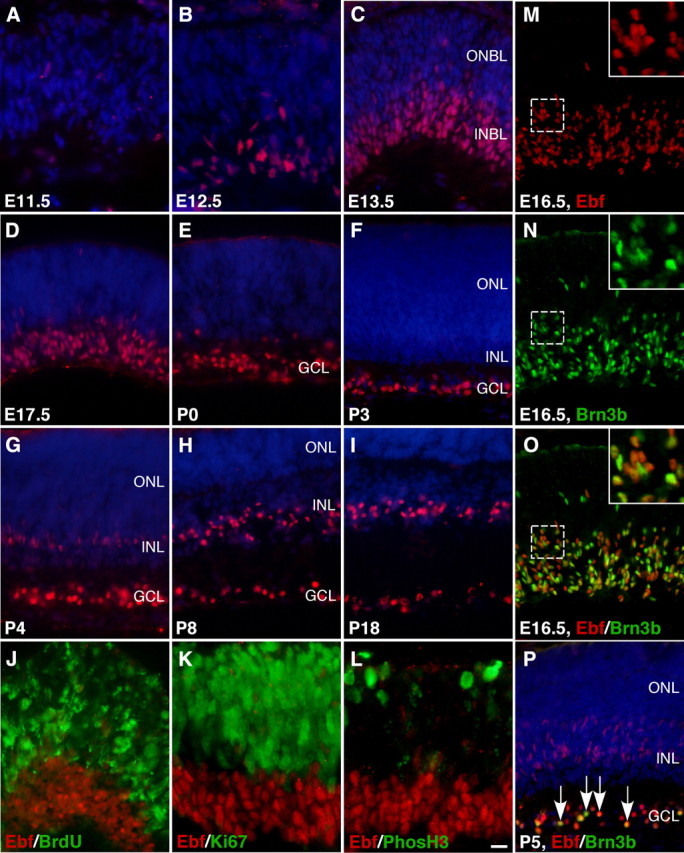
Expression pattern of Ebf proteins during mouse retinal development. A–I, Retinal sections from the indicated developmental stages were immunolabeled with a pan-Ebf antibody and weakly counterstained with nuclear DAPI. Ebf-immunoreactive cells are few at E11.5 but become abundant in the INBL at E13.5–E17.5, and in postnatal retinas are maintained in the GCL and/or INL. J–L, Retinal sections from E14.5 embryos pulse labeled with BrdU were double immunostained with a pan-Ebf antibody and an anti-BrdU antibody (J) or E14.5 retinal sections were double immunostained with a pan-Ebf antibody and antibodies against Ki67 or phosphorylated histone 3 (PhosH3) (K, L). There is no colocalization between Ebfs and BrdU, Ki67, or PhosH3. M–P, Colocalization of Ebfs and Brn3b in RGCs. The great majority of Ebf-expressing cells coexpress Brn3b at E16.5 (M–O); however, Ebfs and Brn3b are coexpressed in the GCL but not in the INL at P5 (P). Insets show corresponding outlined regions at a higher magnification and arrows point to representative colocalized cells. ONBL, Outer neuroblastic layer. Scale bar (in L): C–J, M–P, 16.7 μm; A, B, K, L, 11 μm.
We next investigated whether there is any Ebf expression in dividing retinal progenitors. We found that at E14.5 none of the Ebf-expressing cells could be colabeled by short pulses of BrdU, an S-phase maker, or by an antibody against phosphorylated histone H3, an M-phase marker (Fig. 4J,L). Similarly, they were not coexpressed with Ki67, a protein present during all active phases of the cell cycle (Fig. 4K). Thus, Ebfs appear to be expressed only by postmitotic precursor cells and their derivatives.
Expression of Ebf factors in multiple retinal cell types and subtypes
To identify the types of cells that express Ebfs, we performed a series of double-immunofluorescence labeling in P21 retinal sections using a variety of cell type-specific markers. Consistent with their expression in the GCL and their downregulation in Brn3b mutant retinas (Figs. 1, 3, 4), Ebf proteins are colocalized with Pax6, Barhl2, calretinin, Isl1, and Brn3b, all RGC markers, in many cells of the GCL (Fig. 5A–C,L,M). The proportions of Pax6+ and Brn3b+ cells that coexpress Ebf proteins in the GCL are 53.4 and 97.1%, respectively (supplemental Fig. S2, available at www.jneurosci.org as supplemental material), suggesting that Ebfs are expressed in all or nearly all RGCs.
Figure 5.
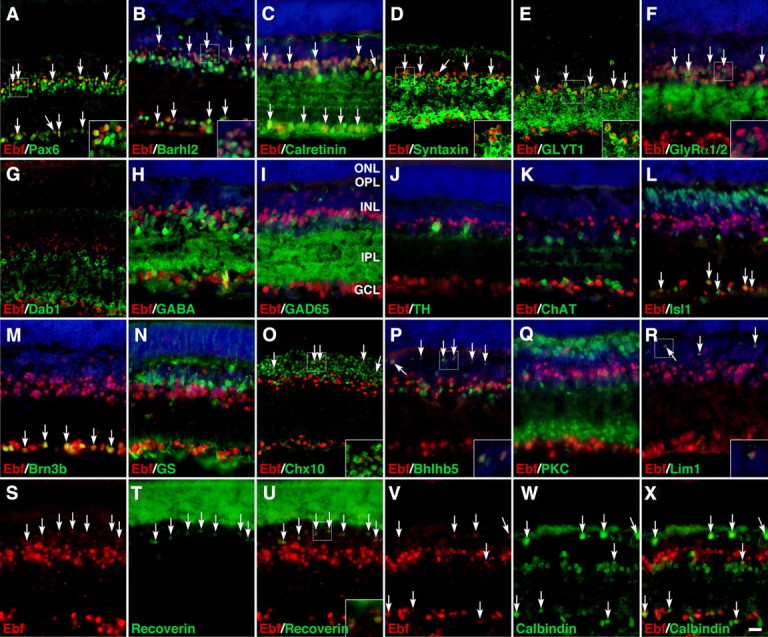
Expression of Ebfs in multiple retinal neuronal types and subtypes. A–X, Sections from P21 mouse retinas were double-immunolabeled with a pan-Ebf antibody and those against the indicated cell type-specific markers. Sections in (B, C, F, H–N, P–R) were also weakly counterstained with DAPI. There is colocalization between Ebfs and Pax6, Barhl2, calretinin, syntaxin, GLYT1, or GlyRa1/2 in ACs and/or RGCs (A–F) but no colocalization with Dab1, GABA, GAD65, TH, ChAT, or Isl1 in ACs (G–L). Ebfs are coexpressed with Isl1 or Brn3b in RGCs (L, M), with Chx10, Bhlhb5, or recoverin but not with PKC in bipolar cells (O–Q, S–U), with Lim1 or calbindin in horizontal cells (R, V–X); however, they are not coexpressed with GS in Müller cells (N). Arrows point to representative colocalized cells, and insets show corresponding outlined regions at a higher magnification. IPL, Inner plexiform layer; OPL, outer plexiform layer. Scale bar (in X): A, D, E, G, O, 19 μm; B, C, F, H–N, P–X, 16.7 μm.
Within the inner half of the INL, there is colocalization in many cells between Ebfs and AC markers Pax6, Barhl2, calretinin, and syntaxin, with 30.7% of Pax6+ and 36.3% of syntaxin+ cells coexpressing Ebf proteins (Fig. 5A–D; supplemental Fig. S2, available at www.jneurosci.org as supplemental material), suggesting that Ebfs are expressed in a subpopulation of ACs. We next determined the AC subgroup that expresses Ebfs by glycinergic and GABAergic AC markers. Ebfs are expressed by many glycinergic ACs immunoreactive for the glycine transporter GLYT1 (54.8%) or glycine receptor GlyRα1/2 but not in cells labeled by Dab1 (Fig. 5E–G; supplemental Fig. S2, available at www.jneurosci.org as supplemental material), a marker for the AII cells that represent a glycinergic AC subtype bridging the rod pathway and the cone circuitry (Kolb and Famiglietti, 1974; Strettoi et al., 1992; Rice and Curran, 2000). Thus, Ebfs are expressed in a subset of glycinergic ACs distinct from AII cells. There is no expression of Ebfs in GABAergic ACs immunoreactive for GABA, GAD65, or Bhlhb5, in dopaminergic ACs immunoreactive for tyrosine hydroxylase (TH), or in cholinergic ACs immunoreactive for ChAT or Isl1 (Elshatory et al., 2007) (Figs. 5H–L,P; supplemental Fig. S2, available at www.jneurosci.org as supplemental material).
In the outer half of the INL, Ebfs are seen in a small fraction (3.7%) of Chx10-positive bipolar cells, although there is no coexpression between them and bipolar cell makers PKCα (for rod bipolar cells) and Isl1 (Figs. 5L,O,Q; supplemental Fig. S2, available at www.jneurosci.org as supplemental material). This small number of double-labeled bipolar cells appear to be the type 2 OFF-cone bipolar cells because Ebfs are found in most cells (∼83%) that express Bhlhb5 or recoverin (Fig. 5P,S–U; supplemental Fig. S2, available at www.jneurosci.org as supplemental material), both of which are protein markers for this particular cone subtype (Feng et al., 2006). At the outer edge of the INL, Ebfs are coexpressed with Lim1 or calbindin in most horizontal cells (∼82%) (Fig. 5R,V–X; supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Compared with their strong expression in ganglion and amacrine cells, Ebf proteins appear to be much more weakly expressed in bipolar and horizontal cells (Fig. 5S,V). Ebfs do not colocalize with glutamine synthetase (GS), a Müller cell marker, or with recoverin in the outer nuclear layer (ONL) where rod and cone cells reside (Fig. 5N,S–U), thereby demonstrating the lack of Ebf expression in glial and photoreceptor cells. Thus, these colocalization experiments together suggest that Ebf factors may be expressed in most ganglion and horizontal cells as well as in a subset of glycinergic AC and cone bipolar cells.
Ebf factors promote the differentiation of glycinergic amacrine and cone bipolar cells
Given the expression of Ebfs in retinal precursors and multiple cell types, we investigated whether they are involved in retinal cell differentiation by a gain-of-function approach using a replication-incompetent murine retroviral vector that carries a GFP reporter (supplemental Fig. S3A, available at www.jneurosci.org as supplemental material) (Mo et al., 2004). Retinal progenitors were infected at P0 by subretinal injection of Ebf1-GFP or Control-GFP viruses, and we analyzed the laminar position and morphology of GFP+ cells in infected retinas at P21. While the great majority of GFP+ cells in control retinas differentiate as photoreceptors randomly distributed within the entire ONL, fewer GFP+ cells are present in the ONL of retinas infected with Ebf1-GFP viruses, and these are mostly located at the outer edge of the ONL without the characteristic inner and outer segments of photoreceptors (supplemental Fig. S3B–D, available at www.jneurosci.org as supplemental material). In the INL, by contrast, the fraction of GFP+ cells doubled in retinas infected with Ebf1-GFP viruses (supplemental Fig. S3B–D, available at www.jneurosci.org as supplemental material). Thus, Ebf1 misexpression in progenitors significantly alters the morphology and proportion of progeny distributed in different retinal cellular layers.
The elevated GFP+ cells in the INL of retinas infected with Ebf1-GFP viruses could represent more amacrine, bipolar, horizontal, and/or Müller cells. To distinguish these possibilities, we used various cell type-specific markers to analyze the types of cells that were differentiated from the virus-transduced progenitors. First, misexpressed Ebf1 increased fourfold to sevenfold the number of GFP+ ACs that are immunoreactive for Pax6 or GLYT1 while completely inhibiting the formation of Dab1-positive AII cells (Fig. 6A,B,D,E,Y), demonstrating a potent activity for Ebf1 to promote non-AII glycinergic AC differentiation. However, it had no effect on the number of GABAergic ACs immunoreactive for GABA, Gad65, or Bhlhb5 (Fig. 6J,K, M,N,Y). Second, misexpressed Ebf1 also increased up to threefold the number of GFP+ cells that are immunoreactive for Bhlhb5 within the outer half of the INL, which represent type 2 OFF-cone bipolar cells (Fig. 6M,N,Y). Finally, it reduced the proportion of Chx10+ bipolar cells from 5.5 to 2.7% (mean ± SD, control: 5.5 ± 0.7%, n = 3; Ebf1: 2.7 ± 0.9%, n = 3); recoverin+ photoreceptor cells from 84.5 to 0.21% (mean ± SD, control: 84.5 ± 2.5%, n = 3; Ebf1: 0.21 ± 0.07%, n = 3); and GS+ Müller cells from 2.54 to 0.21% (mean ± SD, control: 2.54 ± 0.7%, n = 3; Ebf1: 0.21 ± 0.12%, n = 4), but had no effect on horizontal cells (Fig. 6P,Q,S,T,V,W,Y). The inhibitory effect of Ebf1 on selective cell types does not appear to be caused by increased cell death since misexpressed Ebf1 exerted no effect on apoptosis (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). Thus, when misexpressed in P0 retinal progenitors, Ebf1 can bias them toward the fate of non-AII glycinergic ACs and type 2 OFF-cone bipolar cells while suppressing the differentiation of other bipolar cells and photoreceptor and Müller cells.
Figure 6.
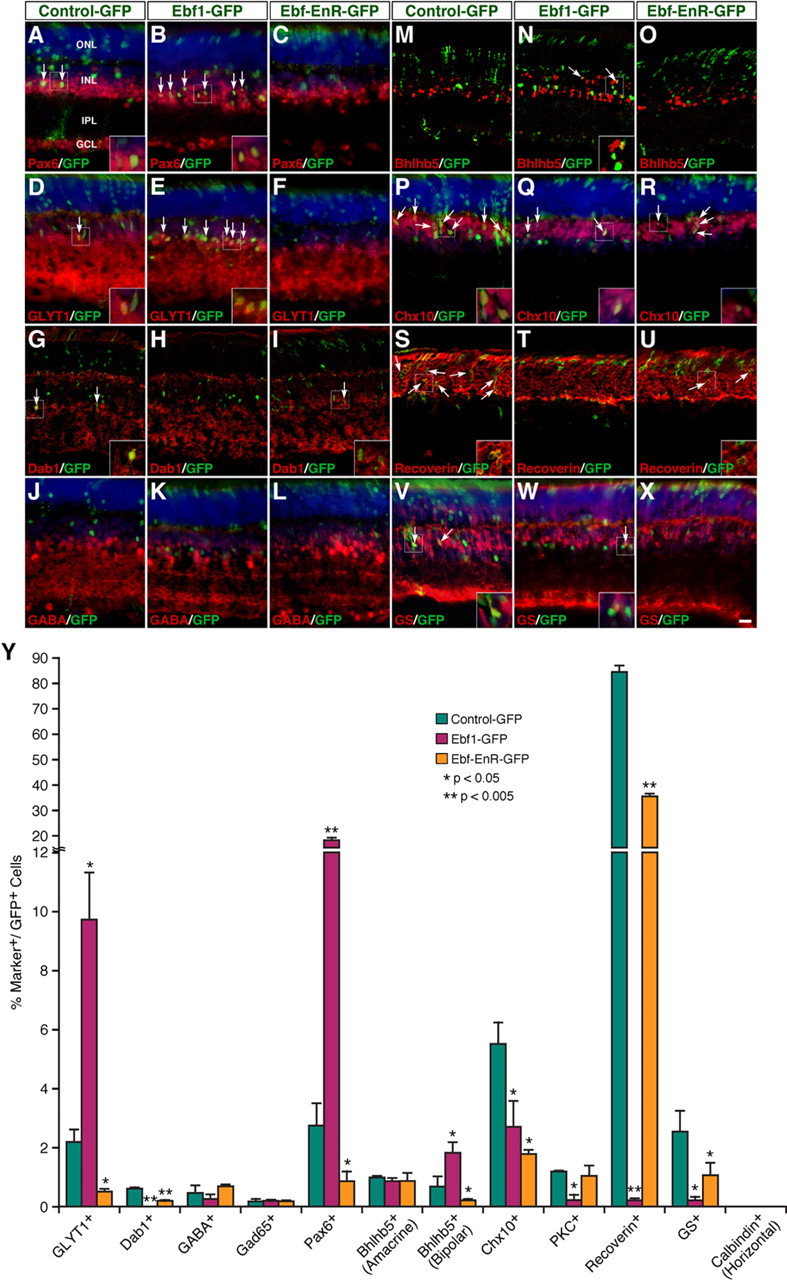
Effect of Ebf1 and a dominant-negative Ebf misexpressed at P0 on the formation of different retinal cell types. A–X, Sections from retinas infected with control-GFP, Ebf1-GFP, or Ebf-EnR-GFP viruses were double immunostained with an anti-GFP antibody and antibodies against Pax6, GLYT1, Dab1, GABA, Bhlhb5, Chx10, recoverin, or GS. Sections in (A–F, J–L, P–R, V–X) were also weakly counterstained with DAPI. Misexpression of wild-type Ebf1 results in a significant increase of cells immunoreactive for Pax6, GLYT1, or Bhlhb5, whereas the dominant-negative form has the opposite effect (A–F, M–O); however, both the wild-type and dominant-negative forms of Ebf1 suppress the formation of AII cells immunoreactive for Dab1 (G–I), bipolar cells immunoreactive for Chx10 (P–R), photoreceptor cells immunoreactive for recoverin (S–U), and Müller cells immunoreactive for GS (V–X). They have no effect on GABAergic ACs immunoreactive for GABA (J–L). Arrows point to representative colocalized cells, and insets show corresponding outlined regions at a higher magnification. Images in G–I, M–O, and S–U represent confocal images with an optical thickness of 7 μm. Y, Quantitation of virus-transduced retinal cells that become immunoreactive for various cell type-specific markers. Each histogram represents the mean ± SD for 3–5 retinas. More than 520 GFP+ cells were scored in each retina. IPL, Inner plexiform layer. Scale bar (in X): G–I, M–O, 19 μm; A–F, J–L, P–R, V–X, 16.7 μm; S–U, 12 μm.
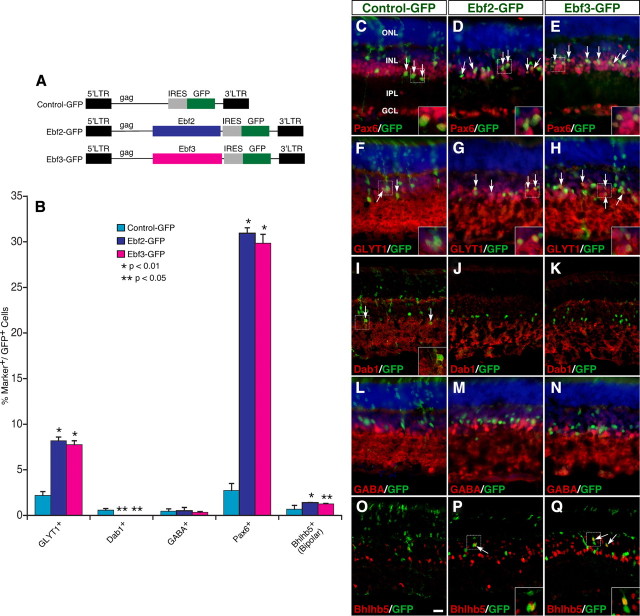
Given their similar expression pattern during retinogenesis and their high sequence homology in the DNA-binding domain (Fig. 3) (Wang et al., 2002), we asked whether all Ebfs could similarly affect retinal cell differentiation. When misexpressed in P0 retinal progenitors, similar to Ebf1, both Ebf2 and Ebf3 increased GFP+ cells in the INL but decreased them in the ONL (supplemental Fig. S3D, available at www.jneurosci.org as supplemental material). They increased GFP+ ACs that are immunoreactive for Pax6 and GLYT1 by ∼11-fold and 4-fold, respectively, and completely suppressed AII cell differentiation (Fig. 7A–K); whereas, neither had any effect on the number of GABAergic ACs (Fig. 7B,L–N). In addition, retinas infected with Ebf2-GFP and Ebf3-GFP viruses exhibited approximately a twofold increase in the number of GFP+ bipolar cells that are immunoreactive for Bhlhb5 (Fig. 7B,O–Q). Therefore, Ebf2 and Ebf3, similar to Ebf1, have the ability to specifically promote differentiation of non-AII glycinergic ACs and type 2 OFF-cone bipolar cells, suggesting that different members of the Ebf family are likely to play similar roles during retinal development.
Figure 7.
Effect of Ebf2 and Ebf3 misexpressed at P0 on the formation of amacrine and bipolar subtypes. A, Schematics of Control-GFP, Ebf2-GFP, and Ebf3-GFP retroviral constructs. The internal ribosomal entry site (IRES) allows for expression of both Ebfs and GFP. B, Quantitation of virus-transduced retinal cells that become immunoreactive for several cell type-specific markers. Each histogram represents the mean ± SD for 3–5 retinas. More than 650 GFP+ cells were scored in each retina. C–Q, Sections from retinas infected with Control-GFP, Ebf2-GFP, or Ebf3-GFP viruses were double immunostained with an anti-GFP antibody and antibodies against Pax6, GLYT1, Dab1, GABA, or Bhlhb5. Sections in C–H and L–N were also weakly counterstained with DAPI. Misexpressed Ebf2 and Ebf3 significantly increase cells immunoreactive for Pax6, GLYT1, or Bhlhb5 (C–H, O–Q), reduce Dab1+ cells (I–K), but have no effect on GABAergic ACs (L–N). Arrows point to representative colocalized cells, and insets show corresponding outlined regions at a higher magnification. Images in I–K and O–Q represent confocal images with an optical thickness of 7 μm. IPL, Inner plexiform layer. Scale bar (in O): I–K, O–Q, 19 μm; C–H, L–N, 16.7 μm.
Inhibition of differentiation of glycinergic amacrine and cone bipolar cells by a dominant-negative form of Ebf factors
Given the sufficiency for Ebf factors to promote differentiation of amacrine and cone bipolar subtypes, we next determined whether they are also necessary and act as transcriptional activators for their differentiation. A dominant-negative form of Ebf, Ebf-EnR, was constructed by fusing the repressor domain of the Drosophila-engrailed protein to the Ebf1 N terminus containing the DNA-binding and HLH domains (supplemental Fig. S3A, available at www.jneurosci.org as supplemental material). Because the four Ebf proteins share >90% of amino acid sequence identity in the DNA-binding domain and all of them act as activators in transcriptional assays (Wang et al., 1997, 2002), it is expected that Ebf-EnR would function as a dominant-negative form for all Ebf factors (Akerblad et al., 2002). In retinas transfected with Ebf-EnR-GFP viruses, there was a significant reduction of GFP+ cells in the INL with a simultaneous increase in the ONL (supplemental Fig. S3D, available at www.jneurosci.org as supplemental material). The proportions of GFP+ cells immunoreactive for Pax6 or GLYT1 and fraction of Bhlhb5+ bipolar cells were all reduced by threefold to fourfold compared with control retinas (Fig. 6A,C,D,F,M,O,Y), suggesting that Ebf factors are both necessary and sufficient for differentiation of glycinergic ACs and type 2 OFF-cone bipolar cells. While having no effect on GABAergic ACs immunoreactive for GABA and Bhlhb5 (Fig. 6J,L,M,O,Y), forced Ebf-EnR expression significantly diminished the number of GFP+ cells that were positive for Dab1, Chx10, recoverin, or GS (Fig. 6G,I,P,R,S,U,V,X,Y), consistent with the notion that Ebfs act as repressors for the differentiation of AII, non-type 2 OFF-cone bipolar, photoreceptor, and Müller cells. However, Ebf-EnR appears not to be as effective an inhibitor as wild-type Ebf1 since it has a weaker activity in suppressing AII, photoreceptor, and Müller cell differentiation (Fig. 6Y). Similar to Ebf1, misexpressed Ebf-EnR did not cause increased apoptotic cell death (supplemental Fig. S4, available at www.jneurosci.org as supplemental material).
Ebf1 knockdown suppresses differentiation of glycinergic and cone bipolar cells
To ask whether there is a specific requirement for individual Ebf factors in amacrine and bipolar subtype specification, we knocked down Ebf1 expression by shRNA-mediated interference. An Ebf1 shRNA, Ebf1i5, when expressed from the RNAi vector pU6 (supplemental Fig. S5A, available at www.jneurosci.org as supplemental material) (Sui et al., 2002), was found to dramatically knock down GFP expression in 293T cells cotransfected with the pCIG-Ebf1 expression plasmid but not in cells cotransfected with a modified pCIG GFP expression vector (supplemental Fig. S5B–E, available at www.jneurosci.org as supplemental material) (Megason and McMahon, 2002). To rule out any nonspecific effects of Ebf1 RNAi, an Ebf1i5-resistant Ebf1 form (Ebf1sm) was designed that contains mutations only at the third positions of amino acid codons in the target region so that the protein sequence remains unchanged (supplemental Fig. S5A, available at www.jneurosci.org as supplemental material). Ebf1sm expression was not affected by Ebf1i5 in 293T cells, nor was Ebf2 or Ebf3 expression (supplemental Fig. S5F–K, available at www.jneurosci.org as supplemental material), demonstrating the specificity and efficiency of Ebf1i5 in knocking down Ebf1 expression.
To investigate the effect of Ebf1i5 on retinal cell differentiation, we coelectroporated pU6-Ebf1i5 or pU6 plasmids with the pCIG vector into P0 mouse retinas and collected them at P12 for cell type analysis. In retinas transfected with pU6-Ebf1i5, GFP+ cells significantly reduced in the INL but increased in the ONL (supplemental Fig. S6, available at www.jneurosci.org as supplemental material). There was an approximately twofold decrease in the percentage of Pax6+ or GLYT1+ ACs and Bhlhb5+ bipolar cells compared with pU6-transfected retinas (Fig. 8A–D,I,J,Q), indicating a requirement for Ebf1 in differentiation of glycinergic ACs and type 2 OFF-cone bipolar cells. In contrast, Ebf1i5 increased Dab1+ AII and GS+ Müller cells by 57% (mean ± SD, control: 0.56 ± 0.09%, n = 3; Ebf1i5: 0.88 ± 0.11%, n = 3) and 21% (mean ± SD, control: 2.25 ± 0.20%, n = 6; Ebf1: 2.73 ± 0.14%, n = 5), respectively, and exerted no effect on the number of GABAergic ACs, Chx10+ bipolar cells, and recoverin+ photoreceptors (Fig. 8E–H,K–Q). The increase of AII cells is consistent with Ebf1 being a suppressor of AII AC fate, and the increased Müller cells may reflect a general repressor activity by proneural HLH factors on glial cell specification (Cai et al., 2000).
Figure 8.
Effect of knocking down Ebf1 expression on the formation of different retinal cell types. A–P, Sections from retinas coelectroporated with pCIG and RNAi vectors pU6 or pU6-Ebf1i5 were double immunostained with an anti-GFP antibody and antibodies against Pax6, GLYT1, GABA, Dab1, Bhlhb5, Chx10, GS, or recoverin. Knockdown of Ebf1 expression leads to a significant decrease of ACs immunoreactive for Pax6 or GLYT1 and bipolar cells immunoreactive for Bhlhb5 (A–D, I, J), a significant increase of AII cells immunoreactive for Dab1 (G, H) and Müller cells immunoreactive for GS (M, N), but no change in the number of GABAergic ACs (E, F), bipolar cells immunoreactive for Chx10 (K, L), and photoreceptor cells immunoreactive for recoverin (O, P). Arrows point to representative colocalized cells, and insets show corresponding outlined regions at a higher magnification. All images are confocal with an optical thickness of 7 μm. Q, Quantitation of GPF+ cells that are immunoreactive for various cell type-specific markers. Each histogram represents the mean ± SD for 3–6 retinas. More than 200 GFP+ cells (ranging from 201 to 2022 depending on frequency of colocalized cells) were scored in each retina. IPL, Inner plexiform layer. Scale bar: J (for A–P), 47.6 μm.
Consistent with the results obtained by retrovirus-mediated misexpression (Fig. 6), misexpression of Ebf1 and its silent mutant Ebf1sm by electroporation similarly promoted the formation of Pax6+ and GLYT1+ ACs and Bhlhb5+ bipolar cells, but had no effect on GABAergic AC differentiation (supplemental Fig. S7, available at www.jneurosci.org as supplemental material). Cotransfection of pU6-Ebf1i5 with pCIG-Ebf1 could effectively bring down the number of Pax6+ and GLYT1+ ACs and Bhlhb5+ bipolar cells to the level in control retinas transfected with pU6 and pCIG, whereas cotransfection of pU6-Ebf1i5 with pCIG-Ebf1sm did not have any effect (supplemental Fig. S7, available at www.jneurosci.org as supplemental material). Thus, the repressive effect of Ebf1i5 on AC and bipolar subtype specification is specific to the wild-type and endogenous Ebf1.
Requirement for Ebf factors in ganglion and horizontal cell differentiation
The expression of Ebf factors in ganglion and horizontal cells suggests a role for them in the development of these two cell types. The prenatal birth of nearly all ganglion and horizontal cells, however, prevented us from analyzing in postnatal retinas the effect of misexpressed Ebf factors on their differentiation. Thus, to investigate a possible role for Ebfs during ganglion and horizontal cell development, we infected retinal explants with Control-GFP, Ebf1-GFP, or Ebf-EnR-GFP viruses at E13.5 when progenitors are still competent for producing all early-born cell types. The infected retinas were harvested after 4 d in vitro (4 DIV) to analyze ganglion and horizontal cells or collected after 18 DIV for the analysis of other cell types. We found that forced Ebf-EnR expression reduced the proportion of Brn3a- and Brn3b-immunoreactive RGCs by 46% (mean ± SD, control: 2.95 ± 0.23%, n = 3; Ebf-EnR: 1.59 ± 0.27%, n = 3) and 39% (mean ± SD, control: 1.96 ± 0.33%, n = 3; Ebf-EnR: 1.20 ± 0.08%, n = 3), respectively, whereas Ebf1 exerted no effect on them (Fig. 9A–F,J). Therefore, Ebfs appear to be necessary but not sufficient to promote RGC differentiation. For Lim1-immunoreactive horizontal cells, however, misexpressed Ebf1 increased their number by more than twofold, while Ebf-EnR decreased them by ∼40% (mean ± SD, control: 0.24 ± 0.03%, n = 3; Ebf-EnR: 0.15 ± 0.01%, n = 3) (Fig. 9G–J), suggesting that Ebfs are both necessary and sufficient to promote horizontal cell differentiation. Similar to its effect on retinas infected at P0 (Fig. 6), misexpressed Ebf1 also prominently promoted Pax6+ and GLYT+ ACs, while having no effect on GABA+ ACs and reducing Chx10+ bipolar, recoverin+ photoreceptor, and GS+ Müller cells (supplemental Fig. S8, available at www.jneurosci.org as supplemental material). By contrast, Ebf-EnR significantly reduced Pax6+ and GLYT+ ACs (supplemental Fig. S8, available at www.jneurosci.org as supplemental material).
Figure 9.

Effect of Ebf1 and a dominant-negative Ebf misexpressed at E13.5 on the formation of ganglion and horizontal cells. A–I, Sections from retinas infected with Control-GFP, Ebf1-GFP, or Ebf-EnR-GFP viruses were double immunostained with an anti-GFP antibody and antibodies against Brn3a, Brn3b, or Lim1. Misexpressed wild-type Ebf1 significantly increases horizontal cells immunoreactive for Lim1 but does not change the number of RGCs immunoreactive for Brn3a or Brn3b; whereas, the dominant-negative form diminishes cells immunoreactive for any of the three markers. Arrows point to representative colocalized cells, and insets show corresponding outlined regions at a higher magnification. All images are confocal with an optical thickness of 7 μm. J, Quantitation of virus-transduced retinal cells that become immunoreactive for Brn3a, Brn3b, or Lim1. Each histogram represents the mean ± SD for three retinas. More than 1000 GFP+ cells were scored in each retina. Scale bar: I (for A–I), 34 μm.
Discussion
Multiple roles for Ebf factors in specifying retinal cell types and subtypes
Despite the previous demonstration of Ebf1 and Ebf3 expression in the retina (Davis and Reed, 1996; Wang et al., 1997; Trimarchi et al., 2007), it is unclear whether all Ebfs are expressed during retinogenesis. In this work, we systematically analyzed the expression of all Ebf members during mouse retinogenesis and determined the types and subtypes of retinal cells that express them. The finding that all Ebfs are expressed during embryogenesis and exhibit a similar spatial pattern of expression suggests that they may share common functions during retinal development. However, the temporal delay in the onset of Ebf2 expression may hint at a differential temporal requirement for individual Ebfs. For instance, we have shown previously that in the Brn3 family, it is the member with the earliest onset of expression in a given tissue that plays the leading role in the differentiation of RGCs, inner ear sensory neurons, and trigeminal ganglion neurons (Gan et al., 1996; Xiang, 1998; Huang et al., 1999, 2001). The observed expression of Ebfs only in postmitotic precursors and differentiated neurons during retinal development is consistent with their expression in other areas of the CNS (Garel et al., 1997) and suggests that Ebfs are unlikely to have a role in retinal progenitor proliferation.
The use of pan-Ebf antibodies has allowed us to identify all retinal cell types and subtypes that express Ebf proteins. This analysis has confirmed their strong expression in RGCs and ACs as previously observed for Ebf1 (Davis and Reed, 1996) as well as revealed additional positive cell types—mostly horizontal cells and type 2 OFF-cone bipolar cells. It appears that Ebfs are expressed in all RGCs but only in a subset of ACs. Based on their colocalization with GLYT1 and GlyRα1/2 but not with the AII cell marker Dab1 (Rice and Curran, 2000), we are able to identify Ebf-expressing ACs as a subset of glycinergic ACs distinct from AII cells. Similarly, we have identified all Ebf-expressing bipolar cells as type 2 OFF-cone bipolar cells because of the colocalization between Ebfs and recoverin and Bhlhb5, two markers for type 2 OFF-cone bipolar cells (Feng et al., 2006).
Several lines of evidence suggest that Ebfs are involved in specifying amacrine and bipolar cell subtypes during retinogenesis (supplemental Fig. S9, available at www.jneurosci.org as supplemental material). First, as mentioned above, Ebfs are expressed only in non-AII glycinergic ACs and type 2 OFF-cone bipolar cells among the numerous AC and bipolar cell subtypes. Second, misexpressed Ebf1, Ebf2, and Ebf3 are able to bias retinal precursors toward the fates of non-AII glycinergic ACs and type 2 OFF-cone bipolar cells. Third, the differentiation of these two subtypes can be significantly suppressed by a dominant-negative Ebf form, Ebf-EnR, which is supposed to inhibit transcription through the formation of heterodimers with all Ebfs (Wang et al., 2002). Finally, knockdown of Ebf1 expression by RNAi leads to an inhibitory effect similar to that of Ebf-EnR as well as increased AII cells. Moreover, this knockdown can effectively neutralize the promotive effect of Ebf1 on glycinergic ACs and type 2 OFF-cone bipolar cells but has no effect on the promotive effect of the RNAi-resistant form Ebf1sm. Thus, our data from gain- and loss-of-function experiments together suggest that Ebfs are both necessary and sufficient to promote the identity of non-AII glycinergic ACs and type 2 OFF-cone bipolar cells.
We investigated the function of Ebfs during development of RGCs and horizontal cells in E13.5 retinal explants due to the fact that nearly all of them are born before birth and postnatal retinas are not conducive to their differentiation. In agreement with their functions in AC and bipolar cell development, Ebfs appear to be both necessary and sufficient for specifying horizontal cells since their differentiation is promoted by misexpressed Ebf1 but inhibited by Ebf-EnR. In contrast, misexpressed Ebf1 fails to promote the RGC fate from retinal precursors even though Ebf-EnR suppresses RGC differentiation, indicating that Ebfs are necessary but not sufficient for RGC differentiation. This contrasts also with the role of the Ebf upstream regulator Brn3b, which has been shown to be both necessary and sufficient for the specification of RGCs (Gan et al., 1996; Liu et al., 2000; Qiu et al., 2008). Thus, there appears to be a differential requirement for Ebf function during the development of different retinal cell types and subtypes.
Our finding that Ebf1–3 have a similar capacity in specifying glycinergic ACs and type 2 OFF-cone bipolar cells indicates that Ebfs exhibit functional redundancy during retinal development. This is in keeping with the postulated functional redundancy between Ebfs in other neural tissues. For instance, eliminating Ebf1 does not appear to cause any phenotype in the olfactory epithelium where all Ebfs are expressed but results in a neuronal differentiation defect in the striatum where only Ebf1 is expressed (Lin and Grosschedl, 1995; Garel et al., 1999). However, more recent studies have indicated that the functions of Ebfs may be only partially redundant and/or dependent on dosage. Thus, Ebf2 and Ebf3 single mutants and their double heterozygotes display a similar defect in the projection of olfactory axons (Wang et al., 2004), and knocking down Ebf1 or Ebf2 expression independently blocks adipogenesis by preadipocytes (Jimenez et al., 2007). We have found that knocking down Ebf1 expression in postnatal retinal precursors inhibits the differentiation of non-AII glycinergic ACs and type 2 OFF-cone bipolar cells, albeit much less potently than Ebf-EnR (Figs. 6, 8). This may also result from a partial functional nonredundancy among Ebf family members during retinogenesis or from a dosage effect due to the reduction of overall Ebf protein levels. A careful examination of all Ebf single and compound mutants is required to distinguish these possibilities in future experiments.
Ebfs participate in the regulatory gene network of retinal cell development
Similar to many other retinogenic factors, Ebfs function not only as positive regulators of cell development but negative regulators as well. In addition to the requirement for them in specifying RGCs, glycinergic ACs, type 2 OFF-cone bipolar cells, and horizontal cells, Ebfs appear to suppress the formation of AII, Müller, photoreceptor and other bipolar cells (supplemental Fig. S9, available at www.jneurosci.org as supplemental material). The downregulation of Ebfs in Brn3b mutant retinas implicates them as Brn3b downstream genes during RGC development, and our ChIP assay indicates Ebf3 as a direct target. We have shown previously that Brn3b directly or indirectly represses a network of retinogenic factor genes to prevent non-RGC fates while promoting the RGC fate from competent precursors (Qiu et al., 2008). Conceivably, Ebfs may act downstream of Brn3b to mediate part of its function by inhibiting alternative non-RGC differentiation pathways to AII, Müller, photoreceptor, and bipolar cells. Our data thus place Ebfs downstream of the Math5-Brn3b pathway of RGC differentiation (Gan et al., 1996; Wang et al., 2001; Qiu et al., 2008).
ACs are the most diversified cell type in the mammalian retina. The determination and specification of ACs and horizontal cells have been shown to critically depend on the activities of Foxn4 and Ptf1a (Li et al., 2004; Fujitani et al., 2006; Nakhai et al., 2007). In addition, Math3 and Neurod1 are redundantly required for specifying ACs and Prox1 for horizontal cells (Inoue et al., 2002; Dyer et al., 2003). The relatively late onset of Ebf expression in ACs suggests that Ebfs may act genetically downstream of these specifier genes to control subtype specification of ACs (supplemental Fig. S9, available at www.jneurosci.org as supplemental material). They appear to be the first transcription factors found to be specifically expressed in glycinergic versus GABAergic ACs and be required for their differentiation. By contrast, Barhl2 is expressed in both glycinergic and GABAergic ACs and is involved in specifying a subset of each neurotransmitter subtype (Mo et al., 2004; Ding et al., 2009). Bhlhb5 is crucial for specifying a subset of GABAergic ACs (Feng et al., 2006), and so is the orphan nuclear receptor Nr4a2 for a different GABAergic AC subset (Jiang and Xiang, 2009). Isl1 plays an essential role in specifying cholinergic ACs (Elshatory et al., 2007). Our data thus suggest that Ebfs join these transcriptional regulators to coordinately specify appropriate AC subtypes during retinogenesis.
Previous studies have demonstrated that commitment to a bipolar cell fate requires the combinatorial action of Chx10, Mash1, Math3, and Ngn2 in progenitor cells (Burmeister et al., 1996; Tomita et al., 2000; Akagi et al., 2004). In the mouse retina, there exist one type of rod bipolar cells and at least nine subtypes of morphologically and physiologically distinct cone bipolar cells (Ghosh et al., 2004). At present, it is unclear how each of these subtype identities is specified, although strides have been made to identify some of the intrinsic factors involved in their differentiation. For instance, Bhlhb4 is required for terminal differentiation and survival of rod bipolar cells (Bramblett et al., 2004). Vsx1, Irx5, and Bhlhb5 are all involved in the specification of cone bipolar subtypes including the type 2 OFF-cone bipolar cells (Chow et al., 2004; Ohtoshi et al., 2004; Cheng et al., 2005; Feng et al., 2006). Given our demonstration of the requirement for Ebfs also in specifying type 2 OFF-cone bipolar cells (supplemental Fig. S9, available at www.jneurosci.org as supplemental material), it will be interesting to determine the regulatory relationship between Ebfs and Vsx1, Irx5, and Bhlhb5, and how they act in concert to specify the type 2 OFF-cone bipolar subtype.
In the olfactory system, Ebf1 is implicated to control olfactory neuron differentiation by interacting with Roaz (Tsai and Reed, 1997). During B-lymphocyte development, Ebf1 specifies the B-cell fate by activating the expression of a set of genes that define the B-cell lineage through multiple mechanisms. Ebf1 appears to enhance its own DNA-binding and transcriptional activities by direct association with Pax5 (Merluzzi et al., 2004). In particular, it binds collaboratively with E2A and Runx1 to promoters of B cell-specific genes to increase promoter accessibility by inducing DNA demethylation and chromatin remodeling (Lukin et al., 2010; Sigvardsson et al., 1997; O'Riordan and Grosschedl, 1999; Maier et al., 2004). Interestingly, E2A is expressed in retinal precursors and Runx1 in a subset of ACs (Stewart et al., 2005; Yang et al., 2009). Conceivably, Ebfs may also cooperate with these and other retinogenic factors to specify multiple retinal cell types and subtypes.
Footnotes
This work was supported by National Institutes of Health Grants EY12020 and EY015777 (to M.X.). We thank Drs. Andrew McMahon (Harvard University, Cambridge, MA) and Yang Shi (Harvard Medical School, Boston, MA) for the pCIG and pBS/U6 vectors, respectively; Dr. Randall Reed (Johns Hopkins University School of Medicine, Baltimore, MD) for the rabbit anti-Ebf antibody; Dr. Alain Vincent (Centre National de la Recherche Scientifique/Université Paul Sabatier, Toulouse, France) for the Ebf2 plasmid; and members of the Xiang laboratory for valuable discussion of the project and thoughtful comments on the manuscript.
References
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol. 2002;22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors Brn3a and Brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009;61:852–864. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43:779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, Roderick TH, Taylor BA, Hankin MH, McInnes RR. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Cai L, Morrow EM, Cepko CL. Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development. 2000;127:3021–3030. doi: 10.1242/dev.127.14.3021. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Chow RL, Lebel M, Sakuma R, Cheung HO, Thanabalasingham V, Zhang X, Bruneau BG, Birch DG, Hui CC, McInnes RR, Cheng SH. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev Biol. 2005;287:48–60. doi: 10.1016/j.ydbio.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Chow RL, Volgyi B, Szilard RK, Ng D, McKerlie C, Bloomfield SA, Birch DG, McInnes RR. Control of late OFF-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci U S A. 2004;101:1754–1759. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci L, Chung SH, Masserdotti G, Gianola S, Bizzoca A, Gennarini G, Corradi A, Rossi F, Hawkes R, Consalez GG. A key role for the HLH transcription factor EBF2COE2,O/E-3 in Purkinje neuron migration and cerebellar cortical topography. Development. 2006;133:2719–2729. doi: 10.1242/dev.02437. [DOI] [PubMed] [Google Scholar]
- Dacey DM, Lee BB, Stafford DK, Pokorny J, Smith VC. Horizontal cells of the primate retina: cone specificity without spectral opponency. Science. 1996;271:656–659. doi: 10.1126/science.271.5249.656. [DOI] [PubMed] [Google Scholar]
- Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Chen H, Xie X, Libby RT, Tian N, Gan L. BARHL2 differentially regulates the development of retinal amacrine and ganglion neurons. J Neurosci. 2009;29:3992–4003. doi: 10.1523/JNEUROSCI.5237-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh ST, Zhang Y, Temple MH, Cai L. Non-coding sequence retrieval system for comparative genomic analysis of gene regulatory elements. BMC Bioinformatics. 2007;8:94. doi: 10.1186/1471-2105-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois L, Vincent A. The COE-Collier/Olf1/EBF-transcription factors: structural conservation and diversity of developmental functions. Mech Dev. 2001;108:3–12. doi: 10.1016/s0925-4773(01)00486-5. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007;27:12707–12720. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Xie X, Joshi PS, Yang Z, Shibasaki K, Chow RL, Gan L. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;133:4815–4825. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Marín F, Mattéi MG, Vesque C, Vincent A, Charnay P. Family of Ebf/Olf-1-related genes potentially involved in neuronal differentiation and regional specification in the central nervous system. Dev Dyn. 1997;210:191–205. doi: 10.1002/(SICI)1097-0177(199711)210:3<191::AID-AJA1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Garel S, Marín F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wässle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- Gruber CA, Rhee JM, Gleiberman A, Turner EE. POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol. 1997;17:2391–2400. doi: 10.1128/mcb.17.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack I, Peichl L. Horizontal cells of the rabbit retina are non-selectively connected to the cones. Eur J Neurosci. 1999;11:2261–2274. doi: 10.1046/j.1460-9568.1999.00647.x. [DOI] [PubMed] [Google Scholar]
- Harris WA. Cellular diversification in the vertebrate retina. Curr Opin Genet Dev. 1997;7:651–658. doi: 10.1016/s0959-437x(97)80013-5. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Zang K, Schmidt A, Saulys A, Xiang M, Reichardt LF. POU domain factor Brn-3a controls the differentiation and survival of trigeminal neurons by regulating Trk receptor expression. Development. 1999;126:2869–2882. doi: 10.1242/dev.126.13.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Liu W, Fritzsch B, Bianchi LM, Reichardt LF, Xiang M. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development. 2001;128:2421–2432. doi: 10.1242/dev.128.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Jiang H, Xiang M. Subtype specification of GABAergic amacrine cells by the orphan nuclear receptor Nr4a2/Nurr1. J Neurosci. 2009;29:10449–10459. doi: 10.1523/JNEUROSCI.3048-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieslinger M, Folberth S, Dobreva G, Dorn T, Croci L, Erben R, Consalez GG, Grosschedl R. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev Cell. 2005;9:757–767. doi: 10.1016/j.devcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Kolb H, Famiglietti EV. Rod and cone pathways in the inner plexiform layer of cat retina. Science. 1974;186:47–49. doi: 10.1126/science.186.4158.47. [DOI] [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Liu W, Khare SL, Liang X, Peters MA, Liu X, Cepko CL, Xiang M. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development. 2000;127:3237–3247. doi: 10.1242/dev.127.15.3237. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Lukin K, Fields S, Lopez D, Cherrier M, Ternyak K, Ramirez J, Feeney AJ, Hagman J. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc Natl Acad Sci U S A. 2010;107:7869–7874. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998;20:971–982. doi: 10.1016/s0896-6273(00)80478-x. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH. The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J Comp Neurol. 1999;413:305–326. [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001a;11:431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001b;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Mella S, Soula C, Morello D, Crozatier M, Vincent A. Expression patterns of the coe/ebf transcription factor genes during chicken and mouse limb development. Gene Expr Patterns. 2004;4:537–542. doi: 10.1016/j.modgep.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Merluzzi S, Moretti M, Altamura S, Zwollo P, Sigvardsson M, Vitale G, Pucillo C. CD40 stimulation induces Pax5/BSAP and EBF activation through a APE/Ref-1-dependent redox mechanism. J Biol Chem. 2004;279:1777–1786. doi: 10.1074/jbc.M305418200. [DOI] [PubMed] [Google Scholar]
- Mo Z, Li S, Yang X, Xiang M. Role of the Barhl2 homeobox gene in the specification of glycinergic amacrine cells. Development. 2004;131:1607–1618. doi: 10.1242/dev.01071. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, Wang SW, Maeda H, Saszik SM, Frishman LJ, Klein WH, Behringer RR. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–536. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F, Jiang H, Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J Neurosci. 2008;28:3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DS, Curran T. Disabled-1 is expressed in type AII amacrine cells in the mouse retina. J Comp Neurol. 2000;424:327–338. doi: 10.1002/1096-9861(20000821)424:2<327::aid-cne10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- Stewart L, Potok MA, Camper SA, Stifani S. Runx1 expression defines a subpopulation of displaced amacrine cells in the developing mouse retina. J Neurochem. 2005;94:1739–1745. doi: 10.1111/j.1471-4159.2005.03336.x. [DOI] [PubMed] [Google Scholar]
- Strettoi E, Raviola E, Dacheux RF. Synaptic connections of the narrow-field, bistratified rod amacrine cell (AII) in the rabbit retina. J Comp Neurol. 1992;325:152–168. doi: 10.1002/cne.903250203. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007;502:1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaney DI. The mosaic of amacrine cells in the mammalian retina. Prog Retinal Res. 1990;9:49–100. [Google Scholar]
- Wang SS, Tsai RY, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Betz AG, Reed RR. Cloning of a novel Olf-1/EBF-like gene, O/E-4, by degenerate oligo-based direct selection. Mol Cell Neurosci. 2002;20:404–414. doi: 10.1006/mcne.2002.1138. [DOI] [PubMed] [Google Scholar]
- Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wässle H, Boycott BB. Functional architecture of the mammalian retina. Physiol Rev. 1991;71:447–480. doi: 10.1152/physrev.1991.71.2.447. [DOI] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998;197:155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Silva AO, Koyano-Nakagawa N, McLoon SC. Progenitor cell maturation in the developing vertebrate retina. Dev Dyn. 2009;238:2823–2836. doi: 10.1002/dvdy.22116. [DOI] [PubMed] [Google Scholar]