Abstract
Nitrogen (N) is an essential macronutrient for plants. In nature, N cycles between different inorganic and organic forms some of which can serve as nutrients for plants. The inorganic N forms nitrate and ammonium are the most important sources of N for plants. However, plants can also uptake and use organic N forms such as amino acids and urea. Beside their nutritional role, nitrate and other forms of N can also act as signals that regulate the expression of hundreds of genes causing modulation of plant metabolism, physiology, growth and development. Although many genes and processes affected by changes in external or internal N have been identified, the molecular mechanisms involved in N sensing and signaling are still poorly understood. Classic reverse and forward genetics and more recently the advent of genomic and systems approaches have helped to characterize some of the components of the signaling pathways directing Arabidopsis responses to N. Here we provide an update on recent advances to identify components involved in N sensing and signaling in Arabidopsis and their importance for the plant response to N.
N nutrients and metabolites can act as potent signals to regulate genome-wide gene expression in plants. Microarray analyses performed using nitrate, nitrite, glutamic acid and other forms of N revealed a large set of genes participating in many different plant processes (reviewed in [1, 2]), however the sensing and signaling pathways underlying N responses have yet to be fully identified. Work in bacterial and fungal systems has shown that these organisms posses sophisticated sensing mechanisms to monitor and adjust to N status. Although there might be some conserved features between plants and other organisms, thus far the mechanisms of N sensing in other organisms have not directly extrapolated to plants [3]. This review will focus on the regulatory networks for N sensing and signaling in the model plant Arabidopsis thaliana.
Keywords: Arabidopsis thaliana, Nitrogen, N-sensing and signaling, Gene regulatory networks, Systems biology
Regulatory mechanisms of the N response
Nitrate transporters
Most of the recent advances in deciphering the regulatory mechanisms underlying N responses have come from phenotypic analysis of root responses to N availability. N effects on root system architecture (RSA) depend on the N source and concentration available to plants and require the coordination of many downstream signaling pathways (Fig.1). In Arabidopsis, N effects over RSA include changes in primary root growth [4], lateral root initiation [5-7] and lateral root elongation [8, 9]. In the last years, the dual-affinity nitrate transporter NRT1.1 and the high-affinity transporter NRT2.1 have been implicated in nitrate signaling leading to changes in RSA. The NRT2.1 transporter has been involved in lateral root (LR) repression in media that contain a high C/N ratio [7] and in LR initiation control in response to a low nitrate supply [5]. Both effects have been shown to be independent of nitrate transport by NRT2.1 [5, 7]. NRT1.1 signaling function was first suggested by SAGE analysis of the transcriptome of the NRT1.1 mutant chl1-5 [10]. Among the differentially expressed genes, the NRT2.1 transcript showed a higher expression in the mutant when compared with the wild-type plant, and this was correlated with an induction in high-affinity nitrate transport [10]. NRT1.1 was later shown to be involved in NRT2.1 repression by high-N, regulating nitrate transport depending on external/internal N-availability [11]. Recent transcriptomics analysis using the ATH1 chip have shown that NRT1.1 mutants are impaired in nitrate regulation of nitrate regulated genes [12, 13]. This misregulation is independent of nitrate transport [12, 13], consistent with NRT1.1 role as a dual-function transporter/sensor. NRT1.1 role in RSA regulation has also been shown to be independent of nitrate transport. Mutation of NRT1.1 has been shown to affect lateral root primordia maturation or elongation at low nitrate concentrations and the first stages of primary root growth of plants growing at different nitrate concentrations [14]. More recently, this transporter has been involved in root colonization of nitrate-rich patches by promoting LR elongation in response to a localized high nitrate supply [15]. This effect has been shown to depend on a local NO -3 signaling and not on a nutritional effect of N [8]. In roots, NRT1.1 is expressed in the apex and base of LRs, young emergent LRs, LR primordia and the apex of the primary root [14, 15]. NRT1.1 expression was shown to co-express in roots with ANR1 expression [15]. ANR1 is a MADS-box transcription factor whose accumulation in roots is required for LR elongation in response to a localized N supply [15-17]. This observation, and the fact that the ANR1 RNA levels are diminished in the chl1-5 mutant, suggests that ANR1 works downstream of NRT1.1 to promote LR growth in response to localized N. NRT1.1 is also involved in nitrate-mediated repression of primary root growth inhibition by glutamate (Fig.1A, Fig. 2). Similar to nitrate, exogenous L-Glu elicits complex changes in RSA [18-20]. L-Glu is sensed stereospecifically at the root tip, inhibits primary root growth and increases branching in the apical zone [19]. Interestingly, nitrate antagonizes the L-Glu effect on primary root elongation and this effect is abolished in the chl1-5 mutant [20]. The nitrate antagonism of the Glu effect is rescued when chl1-5 constitutively expresses a normal NRT1.1 but not when it constitutively expresses a mutated version of NRT1.1 that harbors a Thr101Ala substitution. Given that 35S::CHL1Thr101Ala presents a normal low-affinity nitrate transport and is more sensitive to nitrate repression of the Glu effect than wild-type plants, the authors concluded that the phosphorylated NRT1.1 acts as a nitrate sensor [20]. The phosphorylated NRT1.1 would activate a signaling cascade leading to the activation or repression of Glu signaling to adjust primary root growth to the available N sources in soils [20]. Further evidence has recently shown that in fact NRT1.1 acts as a nitrate sensor [21]. Low nitrate availability triggers NRT1.1 phosphorylation in Thr101, which in turn switches between the low and high transporter affinity [22]. Phosphorylation of this Thr101 by the calcineurin B-like protein-interacting kinase 23 (CIPK23) was shown to reduce nitrate primary response to low levels in low nitrate concentrations whereas in high nitrate concentrations high expression of the primary response genes is correlated with a low phosphorylation status of the transporter [21]. Thus, NRT1.1 phosphorylation generates different levels of expression of primary nitrate response genes according to nitrate availability.
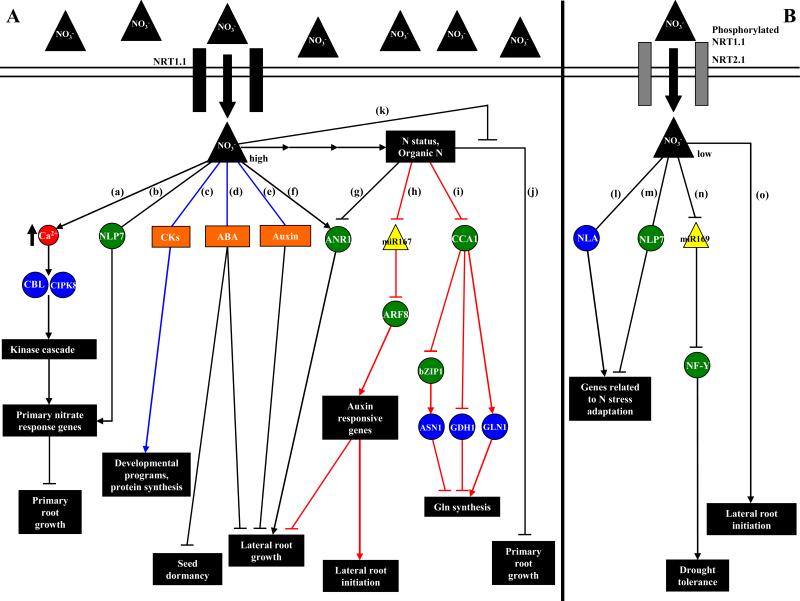
Figure 1. Recent advances in identifying the molecular pathways controlling Arabidopsis responses to N.
Green circles represent transcription factors, blue circles represent enzymes and yellow triangles represent miRNAs. Black lines represent relationships inferred by genetic approaches, red lines represent relationships inferred by systems biology approaches and blue lines represent relationships inferred by both genetic and systems biology approaches. (A) High nitrate situation: nitrate is transported and probably sensed by the NRT1.1 transporter. Inside the cell, nitrate can regulate nitrate primary response genes such as NIA, NiR and NRT2.1 and primary root growth by a pathway involving calcium accumulation, the CBL proteins and the CIPK8 protein kinase [12] or the NLP7 transcription factor [35] (a,b). Nitrate can also interact with the cytokinin, ABA and/or auxin signaling pathways to regulate developmental programs and protein synthesis [59-63] (c), repress seed dormancy [57] (d) and repress lateral root growth [52-54] (e). Localized high nitrate supply is also known to induce the MADS-box transcription factor ANR1 to promote lateral root elongation [15, 17] (f). N metabolites produced by nitrate reduction and assimilation also have signaling roles repressing lateral root growth by a pathway involving ANR1 [16] (g), determining the ratio between initiating and elongating lateral roots by regulation of the miR167/ARF8 module in the pericycle [6] (h), regulating Gln biosynthesis by regulation of the ASN1, GDH1 and GLN1.3 genes by the master clock gene CCA1 [74] (i) and repressing primary root growth [18-20] (j). Nitrate is also able to alleviate primary root growth inhibition by glutamate (k). (B) Low nitrate situation: nitrate can be transported and probably sensed by the high-affinity NRT2.1 transporter. Limiting nitrate conditions can regulate drought tolerance by repressing miR169 and inducing the A5 subunit of the NF-Y transcription factor [47] (l), regulate the expression of genes related to N stress adaptation by pathways involving NLP7 and NLA [30-32, 35] (m,n) and induce lateral root initiation when plants are transferred from high to low nitrate conditions [5] (o). See the text for more details.
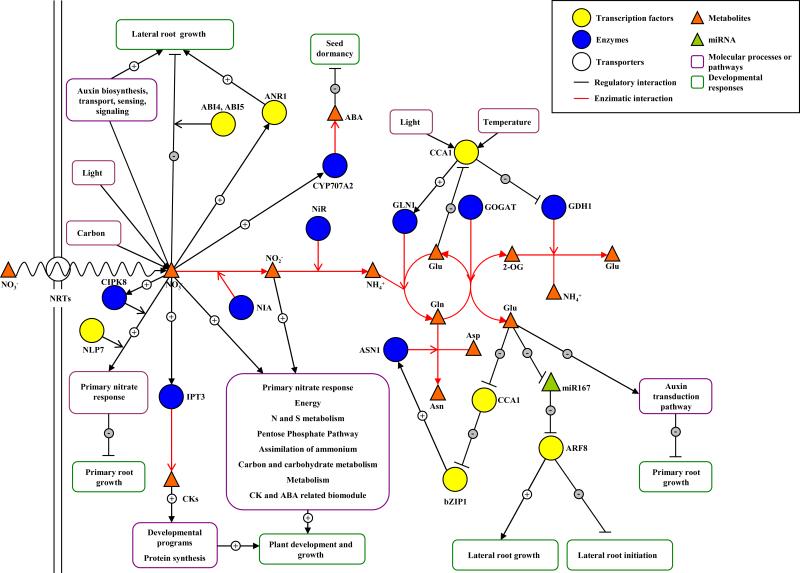
Figure 2. Regulatory networks for N responses in Arabidopsis.
Nitrate and other N sources produced by nitrate reduction by the nitrate assimilatory pathway are able to act as signals that regulate molecular processes and pathways (purple rounded rectangles) leading to regulation of developmental processes (green rounded rectangles). The arrow heads or lines at the end of the edges indicate the direction of information or metabolic flow. 2-OG: 2-oxoglutarate; ASN1: Asparragine synthetase 1; CCA1: Circadian-clock associated 1; CKs: Cytokinins; GDH1: Glutamate dehydrogenase 1; GLN1: Glutamine synthetase 1; GOGAT: Glutamate synthase; IPT3: Isopentenyl transferase 3; NRTs : Nitrate transporters; NIA : Nitrate reductase; NiR: Nitrite reductase. See the text for more details.
Nitrogen regulatory networks downstream of the N nutrient/metabolite receptors
A search for possible signaling mechanisms downstream of NRT1.1 signaling identified a calcineurin B-like interacting protein kinase gene, CIPK8, whose nitrate response was altered in the chl1-5 mutant [12]. The calcineurin B-like proteins (CBLs) are plant-specific proteins that act as Ca2+ sensors and activate CIPKs upon Ca2+ binding to trigger downstream signaling cascades [23]. CBL-CIPKs have been involved in stress responses and nutrient signaling (reviewed in [24]). A link between Ca2+ signaling and NO3- signaling had been suggested from studies with compounds that reduce Ca2+ availability [25]. Plants treated with EGTA, a specific Ca2+ chelator, or La3+ , a Ca2+ channel blocker, show a diminished induction of key genes of the nitrate assimilatory pathway such as nitrate reductase and nitrite reductase [25]. The use of protein kinase inhibitors also affected the nitrate response of primary response genes [26]. CIPK8 appears to act upstream of nitrate primary response genes since there is a 40% reduction in the plant response to nitrate treatments in cipk8 mutants [12] (Fig. 1A, Fig.2). CIPK8 can also be involved in a nitrate signaling pathway controlling primary root growth (Fig. 1A, Fig. 2) as cipk8 mutants show an altered high-nitrate inhibition of primary root growth [12]. Interestingly, CIPK8 appears to be involved only in the response to high concentrations of nitrate. These results suggest there are at least two signal transduction pathways for nitrate, one acting at low and another at high nitrate concentrations and that the low-affinity pathway might depend in part on Ca2+ and a kinase cascade [12] (Fig. 1A).
N-limitation is also known to cause global changes in RNA levels [27]. These conditions are known to cause changes in plant phenotype such as reduced size and accelerated senescence [27-29]. Reverse genetic approaches have identified an E3 ubiquitin ligase NLA that is involved in plant adaptation responses to N-limitation [30] (Fig. 1B). Genomic analysis of the nla mutant showed that these plants have an altered expression of genes involved in the synthesis of nitrogenous compounds, amino acid degradation, starch accumulation, anthocyanin and phenylpropanoid biosynthesis and senescence-related genes under N-limitation when compared with wild-type plants [31, 32]. These results indicate that protein degradation by the proteasomal pathway can be an important mechanism of plant adaptation to N stress.
In legume species, nodule development allows a symbiotic interaction between the plant and nitrogen-fixing bacteria and this process depends on nodule inception protein (NIN) genes, a family of transcription factors [33]. NIN-like proteins are found among plant species including plants that do not form symbiotic interactions to fix N such as Arabidopsis [34]. The Nodule Inception Like 7 protein (NLP7) of Arabidopsis has recently been shown to act in nitrate signaling (Fig. 1). nlp7 mutants show a constitutive response to N starvation, suggesting that NLP7 action inhibits the plant adaptation to N-starvation as opposed to the role of NLA [35] (Fig. 1B). These plants also show a reduced NR and glutamine syntethase (GS) activity and a reduced response of the nitrate-responsive genes NIA1, NIA2 and NRT2.1 [35], suggesting a general defect in N signal transmission (Fig. 1A). Consistent with its role in N sensing, NLP7 is highly expressed in root hairs, emerging lateral roots, vascular tissue of stems, leaf parenchyma and stomata, tissues that are involved in transport and sensing of nutrients and assimilates [35]. Although NLP7 has been shown to be located in the nucleus, its direct action on the transcription of NR, GS or other genes involved in N metabolism has not yet been confirmed [35].
Post-transcriptional regulation by N-responsive miRNAs
Over the last years, microRNAs (miRNAs) have arisen as key regulators of gene expression in animals and plants [36-38]. In plants, these non-coding small RNAs have been involved in several aspects of plant growth and development and other processes such as hormone responses and biotic and abiotic stress responses [39, 40]. For example both miR395 and miR399 have been characterized as important players in sulfur and phosphorus-limitation responses respectively [41-45]. In their 2007 publication, Gutierrez et al. reported that 27 miRNA target transcripts were regulated by nitrate and/or sucrose treatments in Arabidopsis roots, based on network analysis of microarray data [46]. This result suggested that miRNA mediated N-modulation of target mRNA levels is an important mechanism for the control of gene expression in plants. Later work showed that miR167 and its target the auxin-responsive factor ARF8 participates in an organic-N responsive module in pericycle cells that regulates the ratio of initiating and emerging lateral roots to adjust lateral root growth to N availability (Fig. 1A, Fig. 2)(see below)[6]. More recently, Pant et al. developed a qPCR-based platform for the analysis of 177 known Arabidopsis primary miRNAs (pri-miRNAs) and used this approach to discover N- and P-limitation responsive pri-miRNAs [47]. They found that specific family members were differentially expressed under a full-nutrition condition or an N- or P- limited condition. For example, pri-miR169h through pri-miR169l were repressed by N-limitation while pri-miR169m and pri-miR169n were induced by P-limitation. Pri-miRNA analysis identified N-limitation responsive pri-miRNAs corresponding to 5 different miRNA families (miR169, miR398, miR447, miR156 and miR157). However, analysis of mature miRNA expression showed that only mature miR398 and miR169 were differentially expressed under N-limiting conditions. miR398 was repressed by N-, P- and C- limitation, indicating that this miRNA is responding to general nutrient stresses. This is in agreement with the function of the miR398 targets, copper superoxide dismutases, which are involved in detoxification of reactive oxygen species [42, 48, 49]. Mature miR169 levels are specifically repressed by N-limitation [47]. miR169 repression by N-limitation can lead to induction of its target HAP2-1. The homolog of HAP2-1 in Medicago truncatula has been described as a key regulator of nodulation [50]. However, the possible role of this miRNA:target module in Arabidopsis is unknown. The authors discuss that miR169 repression by N-limitation can also lead to drought tolerance by regulating the A5 subunit of NF-Y, a transcription factor expressed in guard cells, crucial for expression of drought resistance genes [51] (Fig. 1B). miR169 was also found repressed by N-limitation in rapeseed phloem sap, suggesting that this miRNA can act as a signal from shoot to root communicating N status of the plant [47].
Nitrogen:hormone interactions
Nitrate regulation of lateral root growth has been shown to depend in part on the hormone auxin (Fig. 1A, Fig. 2). Auxin has been proposed as the long-range signal from shoot to root mediating the release of systemic inhibition of LR development of plants growing in high nitrate concentrations [19, 52]. Auxin concentration increases in Arabidopsis seedlings transferred from high to low nitrate concentrations and decreases in roots of maize and soybean exposed to high nitrate supply [53, 54], indicating that high nitrate can inhibit auxin biosynthesis or its translocation from shoots to roots [19, 52, 54]. Besides auxin concentration, nitrate regulates the expression of genes involved in the auxin signaling pathway such as specific auxin receptors [46](Vidal and Gutierrez unpublished results), ARF transcription factors and Aux/IAA genes [6, 46]. Auxin is also known to induce the NRT1.1 transporter [55], whose role in RSA modulation has already been discussed (see above). These data suggest a regulatory role of nitrate on auxin transport, biosynthesis and/or signaling and a reciprocal regulation of nitrate transport and/or signaling by auxin.
There is also evidence that another plant hormone, abscisic acid (ABA) may participate in regulating LR growth in response to endogenous nitrate pools (Fig. 1A, Fig. 2). ABA signaling mutants abi4 and abi5 are insensitive to nitrate repression of LR growth and ABA biosynthesis mutants show a reduced sensitivity to nitrate repression [56]. ABA also participates in a nitrate mediated signaling pathway controlling seed dormancy. Nitrate releases seed dormancy [57] by inducing the catabolic ABA gene CYP707A2 and decreasing ABA levels in seeds [58] (Fig. 1A, Fig. 2).
Nitrate is also known to interact with the cytokinin signaling pathway. The expression of the Arabidopsis isopentenyl transferase 3 (IPT3) gene, encoding a key enzyme for cytokinin biosynthesis, is induced by nitrate supplementation and is essential for nitrate induced cytokinin synthesis [59, 60]. Cytokinins translocate into the xylem vessels and communicate N availability from roots to shoots [61]. Cytokinin serves as a feedback signal to maintain a balance in nitrate acquisition, as it downregulates the NRT2.1 transporter [62]. Two main signaling pathways of nitrate responses have been proposed: a nitrate-specific pathway, involved in nitrate assimilation and synthesis of amino acids and nucleotides and a cytokinin-dependent pathway, involved in the control of developmental programs and protein synthesis [63] (Fig. 1A, Fig. 2).
N-responsive subnetworks acting at the cell-specific level
It is known that organ development in multicellular organisms requires coordination of growth and developmental processes between different types of cells. Over the last years, tools have been developed in Arabidopsis to study transcriptional profiles of individual cell types. One such technique takes advantage of plants expressing the green fluorescent protein in specific root cell types. The roots of the plants are dissociated into single cells by enzymatic digestion of their cell walls (protoplasting) and the GFP-tagged cells are then isolated by fluorescence activated cell sorting (FACS) for posterior genomic analyses of the cell transcriptome [64, 65]. This technique has been used to address how nitrate regulates global gene expression at the cell type-specific level in Arabidopsis roots [6]. In this analysis, an N responsive module that included miR167 and one of its targets the auxin responsive factor ARF8 was identified in the pericycle [6] (Fig. 1A, Fig. 2). The authors found that repression of miR167 by organic N leads to an induction of ARF8 levels in pericycle cells (which are precursors of lateral roots). Induction of ARF8 leads to an increase in the ratio of initiating versus emerging lateral roots (Fig. 1A, Fig. 2). 126 potential ARF8 targets in pericycle were also identified based on their induction by N in pericycle cells, the presence of an ARF-binding site on their promoters and their co-expression with ARF8 in microarray experiments deposited in the NASC database [6, 66]. Transcriptomic analyses showed that miR167, ARF8 and its 126 targets defined an organic-N gene subnetwork that might participate in adjusting lateral root outgrowth in response to external and internal N availability [6].
Systems biology approaches to map N-regulatory networks
Genetic and genomic approaches have provided us with regulatory factors involved in plant responses to N. However these approaches do not present us with an integrated vision of the plant response to N that includes the relationships between the many N responsive processes or components of the system. In the last years, plant biologists have adopted systems biology approaches to integrate data obtained from diverse high-throughput genome-scale techniques to identify transcriptional networks underlying plant growth, development and plant responses to environmental cues [67, 68]. A systems approach to study biological processes is an iterative process that includes 1. identification of the components of the molecular network and their relationships, 2. modeling the network, 3. perturbing the network and monitoring the network response at a global scale and 4. generating new hypotheses for experimental validation [68]. The global view of the plan circuitry provided by integrative network biology approaches has been valuable to generate new hypotheses and discovering new regulatory networks mediating plant N responses.
Qualitative model of the Arabidopsis molecular networks
The first qualitative network model of Arabidopsis molecular interactions [46] integrated information from metabolic pathways and known and predicted protein-DNA, protein-protein and miRNA:RNA interactions obtained from publicly available databases such as KEGG [69], ARACYC [70], AGRIS [71] and miRBase [72] and from literature based interactions obtained from data mining of published papers using Geneways [73]. This multinetwork model is currently available through VirtualPlant (http://www.virtualplant.org), a software platform that offers multiple tools to analyze and visualize genomic data from a systems perspective. This first network model of Arabidopsis molecular interactions was used to understand the effect of nitrate and/or sucrose treatments in Arabidopsis roots [46]. The network was queried with quantitative microarray data generated from Arabidopsis roots exposed to nitrate, sucrose or nitrate and sucrose treatments to dissect the specific gene networks modulated by nitrate and sucrose in roots [46]. Analysis of the CN-regulated network identified gene subnetworks that include components of important plant processes such as metabolism, protein synthesis and degradation and signaling transduction pathways including an auxin regulatory subnetwork. This analysis suggested that plant roots are able to modulate multiple interconnected molecular networks to adjust their growth and development according to external C and N nutrient supply. Moreover, this analysis highlighted the role of hormones such as auxin (Fig. 1A, Fig.2) in the plant response to changes in CN and proposed miRNAs as important regulatory factors in the CN response of Arabidopsis roots [46].
Systems approaches using network analysis were also used to analyze the plant response to organic forms of N [74]. Using an updated multinetwork model that included predicted protein:DNA interactions, an organic-N responsive subnetwork was identified [74]. The protein:DNA interactions were predicted based on overrepresentation of regulatory motifs for a given transcription factor on a gene promoter region and correlation of gene expression between the transcription factor and its putative target in microarray data [74]. Putative “master regulators” of the organic-N response were identified based on their high degree of connectivity (high number of connections) in the organic-N subnetwork. Two transcription factors were identified: the master clock control gene Circadian Clock Associated-1 (CCA1), and the golden 2-like transcription factor 1 (GLK1). Both transcription factors were repressed by organic N and were predicted to positively affect gene expression of Glutamine synthetase 1.3 (GLN1.3) and to negatively affect the expression of Glutamate dehydrogenase 1 (GDH1) and bZIP1 transcription factor. bZIP1 was predicted to positively affect the expression of Asparagine synthetase 1 (ASN1) ([74] and Fig. 1A, Fig. 2). The regulatory predictions in this subnetwork were validated by analyzing the expression levels of GLN1.3, GDH1, bZIP1 and ASN1 in CCA1 overexpressor lines. All four genes showed altered expression in the CCA1 overexpressor as predicted by the model. Predicted binding of CCA1 to the promoter regions of GLN1.3, GDH1 and bZIP1 was validated using chromatin immunoprecipitation assays. These results verified the subnetwork predictions and proposed CCA1 as a key regulator of N assimilation. In turn, these experiments lead to the discovery that N metabolites modulate CCA1 expression allowing N-assimilation to feedback on the Arabidopsis circadian clock [74]. This study highlights the power of network biology and systems approaches to identify previously unknown regulatory mechanisms that integrate different processes such as circadian rhythms and N-assimilation in plants.
Interaction of N signaling pathways and other pathways
Besides network representations, other bioinformatics tools have been developed to derive new biological hypotheses from genome wide approaches. The Sungear tool (also available through VirtualPlant) is a visualization tool that generates multidimensional Venn diagrams to analyze data from multiple origins [2]. Sungear analysis of microarray data from different N-treatments has shown that only a small subset of genes (including genes involved in nitrate assimilation, glycolysis, amino acid and organic acid metabolism and energy production) was regulated across all the experiments tested [2]. The majority of the genes respond to nitrogen treatments depending on the specific experimental conditions [2]. The context-dependent nature of N regulation of gene expression has also been highlighted in two studies of the analysis of the crosstalk between N and C signaling [46] and N, C and light (L) signaling in roots and shoots [75]. It was shown that there are few genes that respond to N independent from C or L signals [46, 75]. Moreover, modeling of the N, C and L gene expression responses in shoot and root organs revealed that interactions between N, C and L are most prominent in the root organ [75]. A surprising finding in this study was the observation that Arabidopsis utilize a very small subset from all theoretically possible regulatory models to change gene expression in response to N, C or L signals. This revealed a major constraining structure for plant N, C, and L signaling networks and suggested the existence of a code of signal integration in Arabidopsis [75].
As previously stated, N:hormone interactions can translate N signals into developmental processes to adjust plant growth to nutrient availability. Recently, an analysis of N co-regulated genes using biclustering provided a potential mechanistic explanation for the link between N and hormones. Biclustering is a tool that can cluster both the genes and the treatments to identify genes co-regulated by N only in a subset of the N-conditions analyzed [76]. Biclustering identified an N-responsive bicluster that contained genes from all the steps of N-uptake and assimilation and whose functions were biologically relevant [76]. The genes on this bicluster were queried on the Arabidopsis multinetwork model, resulting in a highly connected subnetwork termed “biomodule” that was directly induced by nitrate as a signal [76]. The genes in the biomodule were further biclustered with microarray data obtained from hormone-related experimental conditions from NASC [66], finding co-regulation mainly with cytokinin and ABA treatments, suggesting these genes can be regulated by an interaction between the nitrate signaling pathway and cytokinin and ABA signaling pathways [76]. Authors further discuss that nitrate/hormone responsive genes are more strongly induced or repressed than genes responding just to nitrate indicating that hormones can enhance the nitrate response. This N:hormone interaction would allow the plant to fine-tune and coordinate the response levels of genes in a biomodule allowing metabolic processes to be regulated according to the growth rate of the plant.
Final remarks
Given the importance of N for plant growth and development, intense fertilization programs are in use today to ensure agricultural productivity throughout the world. The large amount of N in soils has clear negative consequences for the environment, affecting the function and composition of natural ecosystems and climate. It is therefore crucial to elucidate how plants sense and respond to different N forms to regulate growth and development. Understanding these molecular mechanisms is the first step to develop biotechnological solutions for sustainable agricultural practices.
Here we have briefly discussed some of the recent advances in elucidating N sensing and signaling mechanisms in Arabidopsis. The study of RSA modulation by N availability and the use of mutants in known key components of N metabolism have been important sources of knowledge on how the N signal is sensed and transmitted. The recent application of systems biology to the study of plant N responses has shown the power of this approach to discover new molecular mechanisms that link N nutrition to other plant processes. Figure 2 represents a network view of the nitrate assimilatory pathway and how nitrate and downstream metabolites generated by nitrate reduction and assimilation into amino acids can interact with the various signaling pathways discussed in this review. Most of the edges on the network shown in Fig. 2 represent connections between N metabolites and components of hormonal pathways, indicating that hormones are crucial players in plant developmental responses to N. This interaction between N and hormones is generated at different levels, such as hormone metabolism, hormone transport, hormone sensing and hormone signaling (Fig. 2). Given that hormones such as cytokinin and auxin are known to directly regulate components of the cell cycle [77, 78], hormones provide a natural link between N nutrition and plant growth and development.
Nitrate interaction with other important regulatory networks such as those controlled by C, L and the circadian cycle (Fig. 2) has also been uncovered by the use of systems approaches, highlighting the highly integrated nature of plant N responses. Organic N metabolites directly regulate the master clock gene CCA1 and can cause a shift in the phase of the circadian clock [74], potentially leading to a change in the expression of many circadian-regulated genes.
Undoubtedly this is just the beginning. Systems approaches are a powerful way to uncover new connections between plant nutrition and other processes that are not easily revealed by classical genetic, molecular or biochemical approaches. A systems view of N nutrient and metabolite responses will continue to be essential to construct an integrated view of plant N nutrition that connects N to C, L, hormonal signaling pathways and other plant processes with the underlying regulatory mechanisms (Fig. 1, Fig. 2). However, for widespread use of systems approaches we still need to develop new user-friendly bioinformatics tools to visualize and integrate genomics data from a systems view (such as the VirtualPlant platform). In addition, we need to continue promoting the training of young researchers in biology and informatics, biology and statistics, and other interdisciplinary programs. These are essential factors that will permit the routine use of systems biology in research programs throughout the world and enable the construction of the holistic view of N nutrition in plants.
Acknowledgements
Research in R.A.G.'s laboratory in this area is supported by grants from FONDECYT (1060457), ICGEB (CRPCHI0501), NIH-FIRCA (1R03TW007823-01A1) and ICM-MIDEPLAN (MN-PFG P06-009-F). We apologize to the authors of many publications which we were unable to cite due to space constraints.
Contributor Information
Elena A Vidal, Departamento de Genética Molecular y Microbiología, Pontificia Universidad Católica de Chile eavidal@uc.cl.
Karem P Tamayo, Departamento de Genética Molecular y Microbiología, Pontificia Universidad Católica de Chile kptamayo@uc.cl.
Rodrigo A Gutierrez, Departamento de Genética Molecular y Microbiología, Pontificia Universidad Católica de Chile rgutierrez@uc.cl.
References
- 1.Vidal EA, Gutiérrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol. 2008;11:521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez RA, Gifford ML, Poultney C, et al. Insights into the genomic nitrate response using genetics and the Sungear Software System. J Exp Bot. 2007;58:2359–2367. doi: 10.1093/jxb/erm079. [DOI] [PubMed] [Google Scholar]
- 3.Miller AJ, Fan X, Orsel M, et al. Nitrate transport and signalling. J Exp Bot. 2007;58:2297–2306. doi: 10.1093/jxb/erm066. [DOI] [PubMed] [Google Scholar]
- 4.Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008;54:820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- 5.Remans T, Nacry P, Pervent M, et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006;140:909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gifford ML, Dean A, Gutierrez RA, et al. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Little DY, Rao H, Oliva S, et al. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci U S A. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Jennings A, Barlow PW, et al. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci U S A. 1999;96:6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 10.Muños S, Cazettes C, Fizames C, et al. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krouk G, Tillard P, Gojon A. Regulation of the high-affinity NO3- uptake system by NRT1.1-mediated NO3- demand signaling in Arabidopsis. Plant Physiol. 2006;142:1075–1086. doi: 10.1104/pp.106.087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu H, Wang Y, Tsay Y. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009;57:264–278. doi: 10.1111/j.1365-313X.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang R, Xing X, Wang Y, et al. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 2009;151:472–478. doi: 10.1104/pp.109.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo FQ, Wang R, Chen M, et al. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell. 2001;13:1761–1777. doi: 10.11054/TPC.010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remans T, Nacry P, Pervent M, et al. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci U S A. 2006;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan Y, Filleur S, Rahman A, et al. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta. 2005;222:730–742. doi: 10.1007/s00425-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 18.Walch-Liu P, Liu L-H, Remans T, et al. Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:1045–1057. doi: 10.1093/pcp/pcj075. [DOI] [PubMed] [Google Scholar]
- 19.Walch-Liu P, Ivanov II, Filleur S, et al. Nitrogen regulation of root branching. Ann Bot (Lond) 2006;97:875–881. doi: 10.1093/aob/mcj601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008;54:820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- 21.Ho C-H, Lin S-H, Hu H-C, et al. CHL1 functions as a nitrate sensor in plants. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Liu KH, Tsay YF. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albrecht V, Ritz O, Linder S, et al. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001;20:1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Sakakibara H, Takei K, Sugiyama T. Isolation and characterization of a cDNA that encodes maize uroporphyrinogen III methyltransferase, an enzyme involved in the synthesis of siroheme, which is a prosthetic group of nitrite reductase. Plant J. 1996;10:883–892. doi: 10.1046/j.1365-313x.1996.10050883.x. [DOI] [PubMed] [Google Scholar]
- 26.Sakakibara H, Kobayashi K, Deji A, et al. Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 1997;38:837–843. [Google Scholar]
- 27.Bi Y-M, Wang R-L, Zhu T, et al. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics. 2007;8:281. doi: 10.1186/1471-2164-8-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz C, Saliba-Colombani V, Loudet O, et al. Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:74–83. doi: 10.1093/pcp/pci225. [DOI] [PubMed] [Google Scholar]
- 29.Tschoep H, Gibon Y, Carillo P, et al. Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ. 2009;32:300–318. doi: 10.1111/j.1365-3040.2008.01921.x. [DOI] [PubMed] [Google Scholar]
- 30.Peng M, Hannam C, Gu H, et al. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2007;50:320–337. doi: 10.1111/j.1365-313X.2007.03050.x. [DOI] [PubMed] [Google Scholar]
- 31.Peng M, Hudson D, Schofield A, et al. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J Exp Bot. 2008;59:2933–2944. doi: 10.1093/jxb/ern148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng M, Bi Y-M, Zhu T, et al. Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol. 2007;65:775–797. doi: 10.1007/s11103-007-9241-0. [DOI] [PubMed] [Google Scholar]
- 33.Schauser L, Roussis A, Stiller J, et al. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 34.Schauser L, Wieloch W, Stougaard J. Evolution of NIN-Like Proteins in Arabidopsis, Rice, and Lotus japonicus. J. Mol. Evol. 2005;60:229–237. doi: 10.1007/s00239-004-0144-2. [DOI] [PubMed] [Google Scholar]
- 35.Castaings L, Camargo A, Pocholle D, et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2008;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- 36.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 37.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Ann Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 38.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 39.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Sunkar R, Chinnusamy V, Zhu J, et al. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Bari R, Datt Pant B, Stitt M, et al. PHO2, microRNA399, and PHR1 define a phosphate-ssignaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 43.Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, et al. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57:313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- 44.Fujii H, Chiou TJ, Lin SI, et al. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Pant BD, Buhtz A, Kehr J, et al. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2007;53(5):731–738. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez RA, Lejay LV, Dean A, et al. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pant BD, Musialak-Lange M, Nuc P, et al. Identification of nutrient-responsive Arabidopsis and Rapeseed MicroRNAs by comprehensive Real-Time Polymerase Chain Reaction profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamasaki H, Abdel-Ghany SE, Cohu CM, et al. Regulation of copper homeostasis by microRNA in Arabidopsis. J Biol Chem. 2007;282(22):16369–16378. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- 49.Dugas DV, Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol. 2008;67(4):403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]
- 50.Combier J-P, Frugier F, de Billy F, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W-X, Oono Y, Zhu J, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forde BG. Local and long-range signaling pathways regulating plant responses to nitrate. Ann Rev Plant Biol. 2002;53:203–224. doi: 10.1146/annurev.arplant.53.100301.135256. [DOI] [PubMed] [Google Scholar]
- 53.Caba JM, Centeno ML, Fernández B, et al. Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta. 2000;211:98–104. doi: 10.1007/s004250000265. [DOI] [PubMed] [Google Scholar]
- 54.Tian Q, Chen F, Liu J, et al. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol. 2008;165:942–951. doi: 10.1016/j.jplph.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Guo FQ, Wang R, Crawford NM. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J Exp Bot. 2002;53:835–844. doi: 10.1093/jexbot/53.370.835. [DOI] [PubMed] [Google Scholar]
- 56.Signora L, De Smet I, Foyer CH, et al. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001;28:655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- 57.Alboresi A, Gestin C, Leydecker MT, et al. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005;28:500–512. doi: 10.1111/j.1365-3040.2005.01292.x. [DOI] [PubMed] [Google Scholar]
- 58.Matakiadis T, Alboresi A, Jikumaru Y, et al. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009;149:949–960. doi: 10.1104/pp.108.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004;37:128–138. doi: 10.1046/j.1365-313x.2003.01945.x. [DOI] [PubMed] [Google Scholar]
- 60.Takei K, Ueda N, Aoki K, et al. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 2004;45:1053–1062. doi: 10.1093/pcp/pch119. [DOI] [PubMed] [Google Scholar]
- 61.Takei K, Takahashi T, Sugiyama T, et al. Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot. 2002;53:971–977. doi: 10.1093/jexbot/53.370.971. [DOI] [PubMed] [Google Scholar]
- 62.Brenner WG, Romanov GA, Kollmer I, et al. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 2005;44:314–333. doi: 10.1111/j.1365-313X.2005.02530.x. [DOI] [PubMed] [Google Scholar]
- 63.Sakakibara H, Takei K, Hirose N. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 2006;11:440–448. doi: 10.1016/j.tplants.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Birnbaum K, Shasha DE, Wang JY, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 65.Birnbaum K, Jung JW, Wang JY, et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- 66.Craigon DJ, James N, Okyere J, et al. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Long TA, Brady SM, Benfey PN. Systems approaches to identifying gene regulatory networks in plants. Ann Rev Cell Dev Biol. 2008;24:81–103. doi: 10.1146/annurev.cellbio.24.110707.175408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutierrez RA. Shasha, DE and Coruzzi, GM. Systems biology for the virtual plant. Plant Physiol. 2005;138:550–554. doi: 10.1104/pp.104.900150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Yeast. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davuluri RV, Sun H, Palaniswamy SK, et al. AGRIS: Arabidopsis Gene Regulatory InformationServer, an information resource of Arabidopsis cis-regulatory elements and transcription factors. Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griffiths-Jones S, GR, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rzhetsky A, Iossifov I, Koike T, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 74.Gutierrez RA, Stokes TL, Thum K, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci U S A. 2008;105:4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krouk G, Tranchina D, Lejay L, et al. A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLoS Comput Biol. 2009;5:e1000326. doi: 10.1371/journal.pcbi.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nero D, Krouk G, Tranchina D, et al. A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule”. BMC Syst Biol. 2009;3:59. doi: 10.1186/1752-0509-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Himanen K, Boucheron E, Vanneste S, et al. Auxin-mediated cell cycle activation during early lateral root Initiation. Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riou-Khamlichi C, Huntley R, Jacqmard A, et al. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]