Abstract
Listeria monocytogenes is an intracellular pathogen responsible for severe foodborne infections. It can replicate in both phagocytic and nonphagocytic mammalian cells. The infectious process at the cellular level has been studied extensively, but how the bacterium overcomes early host innate immune responses remains largely unknown. Here we show that InlC, a member of the internalin family, is secreted intracellularly and directly interacts with IKKα, a subunit of the IκB kinase complex critical for the phosphorylation of IκB and activation of NF-κB, the major regulator of innate immune responses. Infection experiments with WT Listeria or the inlC-deletion mutant and transfection of cells with InlC reveal that InlC expression impairs phosphorylation and consequently delays IκB degradation normally induced by TNF-α, a classical NF-κB stimulator. Moreover, infection of RAW 264.7 macrophages by the inlC mutant leads to increased production of proinflammatory cytokines compared with that obtained with the WT. Finally, in a peritonitis mouse model, we show that infection with the inlC mutant induces increased production of chemokines and increased recruitment of neutrophils in the peritoneal cavity compared with infection with WT. Together, these results demonstrate that InlC, by interacting with IKKα, dampens the host innate response induced by Listeria during the infection process.
Keywords: anti-inflammation, NF-κB, virulence, cytokines, internalin
The Gram-positive bacterium Listeria monocytogenes infects human and animal hosts and causes foodborne infections that can lead to bacteremia and meningitis. It mainly affects immunocompromised patients, pregnant women, and newborns. Once inside the host, L. monocytogenes can invade both phagocytic and nonphagocytic cell types, replicate intracellularly, and spread directly from cell to cell, thereby escaping the humoral immune response. The successive steps of this intracellular parasitism are dependent on various virulence factors, including the surface proteins InlA and InlB, required for entry into cells; secreted proteins listeriolysin O (LLO) and phospholipases, involved in escape from the primary and secondary vacuoles; and ActA, responsible for actin-based intracellular and intercellular movements. These virulence factors are positively controlled by the transcriptional activator PrfA (1–3).
The complete genome sequence of L. monocytogenes strain EGD-e has revealed the presence of 25 genes encoding proteins of the internalin family (4–6). Proteins of this family are characterized by the presence of a leucine-rich repeat (LRR) domain. Most of these are surface proteins attached to the bacterial surface by different anchoring motifs, in particular the LPXTG motif, which mediates covalent binding to the peptidoglycan. Some of these internalin proteins are well-characterized virulence factors, including internalin A (InlA, the prototype of the family) and InlB, which are involved in the crossing of intestinal and placental barriers (7, 8). Only four proteins of the internalin family are predicted to be secreted proteins (5), and among these, only InlC has received attention. This protein, whose gene is present in the pathogenic L. monocytogenes and L. ivanovii species but absent in nonpathogenic Listeria species, has been identified by searching PrfA-regulated genes in strains overexpressing prfA (9, 10). inlC encodes a small protein of 297 aa that displays a signal peptide of 34 aa, no known anchoring motif, and six LRRs of 22 aa, followed by an Ig-like domain (Fig. 1A). This latter domain has been shown in other internalins to stabilize the LRR domain and favor protein–protein interactions. The InlC 3D structure has been determined (11). The LRR domain is similar to that of other internalins. The Ig-like domain displays a high concentration of aromatic residues, suggesting a self-association of InlC molecules or the possibility of InlC association with partner proteins through this domain as well as with the LRR domain.
Fig. 1.
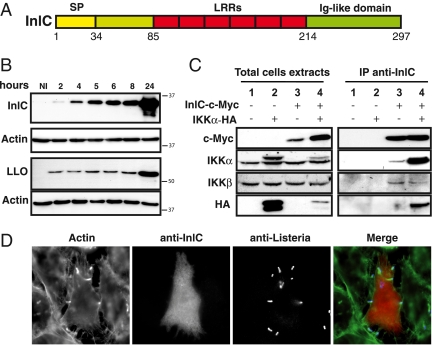
InlC binds IKKα. (A) Schematic representation of InlC, a protein containing a signal peptide (SP), six LRRs, and an Ig-like domain. (B) Kinetics of InlC and LLO expression in infected HeLa cells. Total cell extracts at different time points after infection were analyzed by Western-blot using anti-InlC, anti-LLO, and anti-actin antibodies. Actin is used as loading control of protein. (C) Interaction of IKKα with InlC in transfected HEK-293 T-REx cells. Whole cell lysates from HEK-293 T-REx cells, nontransfected or transfected with the indicated plasmids (InlC c-Myc, IKKα-HA) were immunoprecipited with anti-InlC antibodies, followed by immunoblotting with anti-c-Myc and anti-HA antibodies. Western blot analysis with anti-IKKα and anti-IKKβ antibodies show the endogenous IKKα and IKKβ. (D) Detection of InlC by immunofluorescence in HeLa cells infected for 3 h with the WT strain. Actin was labeled with Alexa Fluor 488–conjugated phalloidin. InlC was detected with anti-InlC antibodies, and bacteria were detected with anti-Listeria antibodies.
inlC is transcribed as a monocistronic mRNA from a single promoter displaying a typical consensus PrfA-binding site at position −40 from the transcription start site. The expression of inlC has been shown to be highly induced intracellularly at rather late stages of infection (12–14). A recent analysis of the entire Listeria transcriptome in various in vitro, ex vivo, and in vivo conditions of growth confirmed stronger inlC expression in the intestine and blood than in rich broth media (15).
An inlC deletion mutant is significantly attenuated when tested in the mouse model of infection by the i.v. route (10, 16). As reported recently, although the inlC deletion does not affect bacterial internalization and intracellular proliferation, it does impair cell-to-cell spread in polarized epithelial cells (17). InlC has been shown to bind the mammalian adaptor protein Tuba, thereby preventing its interaction with N-WASP. Impairment of Tuba–N-WASP interaction by InlC would relieve cortical tension at cell–cell junctions and promote protrusion formation and bacterial spreading.
Analyses of the transcriptional host responses in cultured human intestinal epithelial cells, murine macrophages, and intestinal tissues infected with L. monocytogenes have shown that MAP kinases and NF-κB/Rel pathways are the predominant host responses to a Listeria infection (13, 18, 19). More specifically, the virulence factor LLO has been shown to induce the NF-κB–mediated transcription of the proinflammatory cytokine IL-8 in endothelial cells (20), whereas InlB induces TNF-α and IL-6 in macrophages (21).
The eukaryotic transcription factor NF-κB consists of a dimeric complex of two subunits, including p65/RelA, c-Rel, RelB, p100/52, or p105/50. In resting cells, NF-κB dimers are sequestered in the cytoplasm and kept inactive through their binding to IκB (22–24). IκB is expressed as different isoforms, of which the alpha isoform (IκBα) is the most abundant and most ubiquitously expressed (24). Certain bacterial infections or other stimuli, such as TNF-α, can activate the IκB kinase complex (IKK), which is composed of two catalytic subunits (IKKα and IKKβ) and a regulatory subunit (IKKγ/NEMO). Activation of the IKK complex leads to phosphorylation of IκBs, followed by their polyubiquitination and degradation by the 26S proteasome. These events allow translocation of the NF-κB dimers to the nucleus and activation of NF-κB–regulated genes, which are involved mainly in innate immune responses. In the alternative nonclassical pathway, which is involved primarily in the development and maintenance of secondary lymphoid organs, there is no sequestration of NF-κB by IκB proteins. Instead, one of the precursors of NF-κB—the p100 or the p105 subunit before maturation into the p52 or the p50 subunit, respectively—acts as an inhibitor of the complex. In this pathway, the kinase complex comprises only two subunits of IKKα.
To investigate the function of InlC in the infectious process, we searched for its potential host partners and performed a large-scale two-hybrid screen in yeast. Among the potential cellular targets, we identified IKKα. In this study, we found that InlC binds to IKKα, impairs the phosphorylation of IκBα, delays the degradation of P-IκBα, prevents NF-κB nuclear translocation, and dampens the NF-κB–associated proinflammatory response both in vitro and in vivo.
Results
InlC Is a Bona Fide Virulence Factor Overexpressed Intracellularly.
Because the inlC mutant analyzed previously was still expressing a residual polypeptide (10, 16), we generated a complete inlC-deletion mutant (ΔinlC) in the L. monocytogenes strain EGD and reexamined the contribution of the inlC gene in the infectious process in vivo. As the original inlC mutant, our mutant was growing as well as the WT both in broth medium and intracellularly. It infected cells as efficiently as the WT. Intravenous injections of C57BL/6 female mice with 1 × 105 of the WT bacteria and the ΔinlC strain resulted in 100% mortality with the WT bacteria, but only 20% mortality with the inlC mutant (Fig. S1). Moreover, the LD50 of the ΔinlC mutant after i.v. injection in C57BL/6 female mice was 5.6 × 105 CFU, compared with 1.5 × 104 CFU for the WT strain. Together, these results confirm the role of InlC in Listeria virulence.
To examine the production of InlC, we generated antibodies against a recombinant InlC protein and examined InlC expression during bacterial infection of HeLa cells. As shown in Fig. 1B, in contrast to LLO, which is already expressed at the onset of infection, InlC expression gradually increased inside the cells over time, in perfect agreement with the previously reported transcriptional analysis of the inlC gene (12, 13). Moreover, as shown by immunofluorescence analysis, the InlC protein was detected in both the cytosol and the nucleus of infected cells (Fig. 1D).
InlC Binds IKKα.
To identify partners interacting with InlC in the eukaryotic cell, we used InlC as bait in a large-scale yeast two-hybrid screen. This screen identified IKKα, which had a very high interaction score. To confirm the interaction of InlC with IKKα, InlC tagged with a C-terminal Myc epitope and IKKα tagged with an HA epitope were expressed independently or simultaneously in HEK-293 T-REx cells. Cell extracts were immunoprecipitated with anti-InlC antibodies, and precipitates were analyzed by immunoblotting with anti-Myc, anti-HA, anti-IKKα, and anti-IKKβ antibodies. As shown in Fig. 1C, Myc-tagged InlC coimmunoprecipitated with HA-tagged IKKα as revealed by the anti-Myc and anti-HA antibodies. Using an anti-IKKα antibody recognizing both the transfected and the endogenous forms of IKKα, we showed that both forms coimmunoprecipitated with InlC (Fig. 1C, lanes 3 and 4). Moreover, by binding IKKα, InlC pulled out IKKβ, in agreement with the known IKKβ–IKKα interaction. Together, these results demonstrate that InlC interacts with the kinase subunit IKKα in mammalian cells.
InlC Impairs the Phosphorylation of IκBα and Delays Its Degradation in Infected and Transfected Cells.
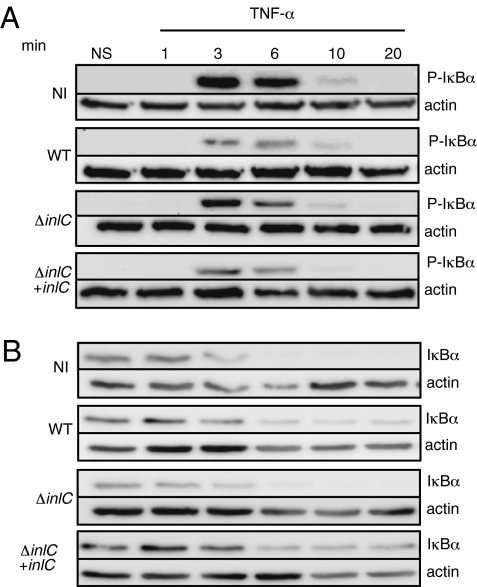
To investigate whether InlC, by binding the kinase IKKα, affects IκBα phosphorylation and degradation, we infected HeLa cells with the WT strain, the ΔinlC mutant, and the complemented strain. We then stimulated the cells with TNF-α to induce IκBα phosphorylation via the classical pathway, and compared the kinetics of IκBα phosphorylation and degradation in infected and noninfected cells. As shown in Fig. 2A, the phosphorylation of IκBα was detected as early as 3 min after stimulation with TNF-α, with smaller amounts found in WT-infected cells than in noninfected cells. The ΔinlC-infected cells showed higher amounts of phospho-IκBα compared with cells infected with WT or the complemented strains. The degradation of the IκBα form (Fig. 2B) began concomitantly with the appearance of phospho-IκBα in noninfected cells and in ΔinlC mutant–infected cells, but was significantly delayed (by up to 20 min when InlC was present), strongly suggesting that the interaction of InlC and IKKα blocks the phosphorylation of IκBα induced by TNF-α and, consequently, its degradation.
Fig. 2.
InlC delays the degradation of IκBα in infected cells. HeLa cells noninfected (NI) or infected by L. monocytogenes WT or by the ΔinlC mutant (ΔinlC) and the complemented-ΔinlC mutant (ΔinlC + inlC), and stimulated by TNF-α (10 ng/mL) were lysed at the indicated times points after stimulation. Extracts were submitted to immunoblot analysis with antibodies recognizing phosphorylated-IκBα (A) and IκBα (B). Membranes were reprobed using antibodies to actin for loading control. NS, nonstimulated.
All cells are not infected during infection of tissue cultured cells, precluding a precise evaluation of events occurring in each cell—in our case, the effect of InlC on the phosphorylation of IκBα. Therefore, to definitively demonstrate the effect of InlC on the phosphorylation of IκBα, we analyzed the fate of phospho-IκBα in human HEK-293 T-Rex cells cotransfected with plasmids expressing InlC and mouse IκBα, taking advantage of the fact that mouse IκBα and human IκBα migrate differently in acrylamide SDS/PAGE gels (25, 26). In this scenario, all cells expressing InlC also express the mouse IκBα. After stimulation by TNF-α, (Fig. S2A), lower amounts of mouse phospho-IκBα were detected in cells cotransfected with the plasmids expressing inlC or IκBα compared with cells cotransfected with IκBα and the control plasmid. Moreover, the phosphorylation of mouse IκBα was delayed, appearing only after 6 min. Similarly, IκBα’s degradation also was delayed, and it was still detectable 20 min after stimulation in cells expressing mouse IκBα cotransfected with the plasmid expressing InlC compared with the control plasmid (Fig. S2B). These findings, in full agreement with the results obtained with the Listeria-infected cells, reinforce the data showing that InlC prevents the phosphorylation and degradation of IκBα.
Taken together, these results establish that InlC, by binding IKKα, prevents the phosphorylation of IκBα induced by TNF-α and delays the degradation of phospho-IκBα.
InlC Prevents Translocation of NF-κB to the Nucleus in Infected Cells.
To assess whether InlC could block the translocation of NF-κB to the nucleus after stimulation by TNF-α, we performed immunofluorescence assays in noninfected cells (NI) and in cells infected with WT Listeria, the ΔinlC mutant, and the complemented strain (ΔinlC + inlC). Cells were fixed and stained with anti-p65 antibodies to label NF-κB, with anti-InlC antibodies to identify the cells where InlC was secreted, and with DAPI to visualize nuclei and bacteria. As shown in Fig. 3 A and B, at 30 min after stimulation by TNF-α, all of the noninfected cells were positive for p65 in the nucleus, confirming that TNF-α had activated the classical NF-κB pathway and that the p65 subunit of NF-κB had translocated to the nucleus. In cells infected with WT Listeria, the NF-κB complex was present only in the cytoplasm and was not detected in the nucleus. In ΔinlC-infected cells, activated NF-κB was detectable in the nucleus. In ΔinlC + inlC–infected cells, the NF-κB complex was not detectable in the nucleus. Together, these immunofluorescence findings unambiguously demonstrate that InlC expression prevents NF-κB translocation to the nucleus.
Fig. 3.
InlC prevents the translocation of NF-κB-p65 to the nucleus in infected cells. (A) HeLa cells noninfected (NI) or infected by Listeria monocytogenes WT, the ΔinlC mutant (ΔinlC), or the complemented-ΔinlC mutant (ΔinlC + inlC) were stimulated with TNF-α (50 ng/mL) for 30 min, fixed, and labeled by immunofluorescence with DAPI for the nucleus and bacteria, anti-InlC antibodies to detect the secretion of InlC in the cytoplasm of cells, and anti-p65 to recognize the NF-κB subunit. (B) To quantify the results shown in A, the percentage of nuclear translocation of NF-κB (P65) was determined by counting at least 100 noninfected or WT, ΔinlC, or ΔinlC+inlC infected cells using fluorescence microscopy. The experiment was performed twice, and typical results are shown. ***P < 0.0005, χ2 test.
InlC Inhibits the NF-κB–Regulated Promoter Response on TNF-α Activation.
To investigate whether InlC through its interaction with IKKα could modify the expression of NF-κB–regulated genes, we transfected cells with a plasmid encoding InlC and the reporter plasmid Igκ3-luciferase in which the luciferase gene is under the control of a NF-κB–regulated promoter. Luciferase activity was assayed at 6 h after TNF-α stimulation (Fig. 4) and found to be 35-fold greater in cells stimulated by TNF-α compared with unstimulated cells. Transfection of the InlC-encoding plasmid significantly decreased the total luciferase activity, with decreases ranging from 60% to 85% depending on the amount of plasmid used for transfection. We found no effect of InlC on luciferase activity when the cells were not stimulated by TNF-α. Moreover, neither the basal luciferase activity of nonstimulated cells nor that of TNF-α–stimulated cells was affected by the expression of LacZ used as a control. Together, these results indicate that InlC blocks TNF-α–induced activation of NF-κB and NF-κB–regulated downstream events.
Fig. 4.
InlC inhibits activation of an NF-κB–regulated promoter. HEK-293 T-REx cells were cotransfected with different plasmids, an NF-κB–dependent luciferase reporter, pRL-Tkluc, and pCDNA4-inlC or pCDNA4-lacZ at the indicated amount per well. The cells were then stimulated with TNF-α (10 ng/mL). Firefly luciferase activity was normalized against Renilla luciferase activity. Bars indicate fold induction compared with unstimulated cells. Values are mean ± SD of six wells. Each experiment was repeated three times. ***P < 0.0005, Welch's t test. NS, not significant.
InlC Decreases the Proinflammatory Cytokine Response to Infection.
Because NF-κB plays a role in regulating various genes involved in inflammation (24), we next addressed whether InlC could control the expression of NF-κB–dependent proinflammatory cytokines during infection. We used ELISA to quantify the expression of TNF-α and IL-6 in supernatants of RAW 264.7 macrophages infected by Listeria WT, ΔinlC, or complemented strains. As shown in Fig. 5, IL-6 and TNF-α production was increased in cells infected by the inlC mutant compared with those infected by the WT. Complementation of ΔinlC restored the capacity of bacteria to down-regulate IL-6 and TNF-α production. Together, these results indicate that inactivation of inlC leads to the induction of a stronger proinflammatory response on infection in vitro.
Fig. 5.
InlC impairs the proinflammatory response in vitro. RAW264.7 cells were infected with Listeria monocytogenes WT, the ΔinlC mutant, or the complemented ΔinlC mutant (ΔinlC + inlC) (MOI10), and levels of IL-6 and TNF-α in the supernatant were measured by ELISA at the indicated times points. Cytokines released from RAW264.7 exposed to LPS (50 ng/mL) were used as positive controls (data not shown). Values are mean ± SEM; n = 6. *P < 0.01; **P < 0.005; ***P < 0.0005, Student t test.
InlC Counteracts the Inflammatory Process Induced by Listeria and LPS in Vivo.
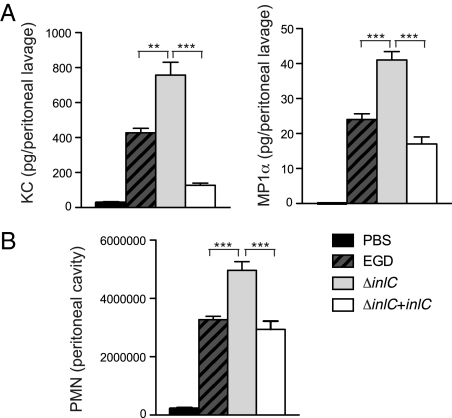
To further investigate the function of InlC in vivo, we used a mouse model of peritonitis in which WT Listeria, the ΔinlC mutant, and the complemented ΔinlC strains were injected i.p. in BALB/c mice. We first analyzed the cytokine response in the peritoneal cavity after 12 h of infection. Despite the reduced number of viable mutant bacteria (30% less than WT bacteria), the inlC mutant induced a stronger cytokine response than the WT, particularly for the chemokines KC (the murine IL8 homolog) and MIP1α (Fig. 6A). This enhanced cytokine response was abolished in mice infected with the complemented strain, in which CFU recovery was similar to that in mice infected with the WT bacteria. We next investigated the recruitment of leukocytes in the peritoneal cavity after infection. We found significantly increased recruitment of neutrophils in response to infection with the inlC mutant compared with infection with the WT or the complemented strain (Fig. 6B). These findings indicate that InlC down-regulates the inflammation induced in response to Listeria infection.
Fig. 6.
InlC modulates the inflammatory process in vivo. (A) Mice were inoculated i.p. with PBS or with L. monocytogenes WT (EGD), the ΔinlC mutant, or the complemented ΔinlC mutant (ΔinlC + inlC), and levels of KC and MIP1α in the peritoneal lavage fluid were measured by ELISA at 12 h postinfection. (B) Leukocytes present in the peritoneal lavage fluid were pelleted and processed by flow cytometry, and the number of neutrophils (PMN) was determined. Values are mean ± SEM; n = 6. *P < 0.01; **P < 0.005; ***P < 0.0005, Student t test.
Discussion
To overcome host defenses, bacterial pathogens or viruses often interfere with the NF-κB pathway, the major pathway involved in the regulation of innate host responses (27). In this pathway, stimulation with an agonist, such as TNF-α, activates the trimeric IKK complex (i.e., IKKα, IKKβ, and NEMO). Various other pathways, including TLR and NOD pathways, also lead to activation of the IKK complex. This complex then phosphorylates IκB inhibitor proteins, which normally sequester the NF-κB proteins in the cytosol. Phosphorylated IκBs then become targets for ubiquitination and degradation, allowing the translocation of free NF-κB to the nucleus and consequent activation of downtream genes. Strikingly, several viral and bacterial pathogens interfere with the NF-κB pathway; however, only a few interfere with the degradation of IκB to inhibit the activation of NF-κB (28). Among these, cowpox virus, racoon pox virus, and certain strains of vaccinia virus can prevent degradation of phosphorylated IκBα (29). It has been proposed that this inhibition may result from dephosphorylation of IκB or interference with its degradation after phosphorylation. Similarly, HIV uses various strategies to manipulate NF-κB activation. The HIV type I Vpu protein competitively inhibits the β-TrCP/ubiquitin ligase–dependent degradation of IκB, thereby keeping NF-κB in the cytosol and resulting in inhibition of NF-κB activity in T cells (30, 31). The Salmonella SseL effector protein deubiquitinates IκBα and prevents its degradation, thereby impairing NF-κB activation (32, 33); another Salmonella effector, AvrA, also has an IκBα deubiquitinase activity (34). Similarly, ChlaDub1 of Chlamydia deubiquitinates IκB (35). These three bacterial effector proteins are secreted by a type III secretion system (T3SS) and translocated through the Salmonella or Chlamydia vacuolar membrane into the cytosol. Their mode of action differs from that of YopP/YopJ protein of Yersinia, which acetylates IKKα and IKKβ, thereby preventing phosphorylation of IκB (36, 37). Enteropathogenic Escherichia coli (EPEC) also has been shown to prevent the phosphorylation of IκB through the use of two T3SS effectors, NleE and NleB, which interact with the TAK1-IKK pathway, albeit in an unknown fashion (38). Finally, Shigella has at least two effectors, OspG and IpaH9.8, that interfere with the NF-κB pathway (25, 39). IpaH9.8 is an ubiquitin ligase that interacts and interferes with NEMO, thereby impairing IκB phosphorylation. In contrast, OspG acts directly on IκBα. This T3SS is a kinase that binds various ubiquitin-conjugating enzymes, including UbcH5, which promotes ubiquitination of IκBα. Thus, OspG prevents the degradation of IκBα and negatively controls the infammatory response (25).
Here we have shown that InlC, a protein of the L. monocytogenes internalin family whose gene is regulated by the global virulence gene activator PrfA, is overexpressed and secreted in the cytosol by Listeria once the bacterium has escaped from the internalization vacuole. InlC interacts with IKKα. Interestingly, the InlC–IKKα interaction decreases phosphorylation of the inhibitory component IκB and delays degradation of its phosphorylated form, thereby impairing NF-κB translocation to the nucleus and subsequent activation of NF-κB–regulated genes, particularly genes encoding proinflammatory cytokines. Like all bacteria, Listeria expresses a number of components that stimulate nonspecific innate immune responses via the NF-κB pathway. As in many other Gram-positive bacteria, lipoteichoic acids in the Listeria cell wall stimulate the NF-κB pathway and the production of proinflammatory cytokines (40). The virulence factors LLO and InlB also have been reported to activate the NF-κB pathway (20, 21).
Nevertheless, similar to other bacterial pathogens [e.g., Shigella (41)], Listeria has evolved several mechanisms to counteract proinflammatory processes. We previously reported that deacetylation of the peptidoglycan by the deacetylase PgdA is a major anti-inflammatory mechanism that down-regulates the production of proinflammatory cytokines (42). We also have shown that the dephosphorylation of histone H3 leads to down-regulation of a subset of genes including genes involved in immune responses (43), and that LLO plays a critical role in this process. As reported herein, InlC also contributes to down-regulate the inflammatory response both in vitro and in vivo. Similar to several other bacterial factors, InlC dampens the inflammatory response by acting on one of the most appropriate pathway, the NF-κB pathway. InlC is highly induced inside eukaryotic cells and interacts with a key component of the NF-κB pathway. It is highly likely that IKKα interacts with InlC via the InlC LRR domain, given that InlC is a small protein composed almost entirely of LRRs. It will be interesting to examine whether other bacterial LRR-containing factors behave like InlC. It also will be interesting to investigate whether InlC, which was shown here to interfere with the classical NF-κB pathway, also modulates other pathways regulated by IKKα (e.g., histone phosphorylation), especially in vivo. Interestingly, InlC was recently shown to also interact with the cytoskeleton-associated protein Tuba. This interaction also occurs late in infection and affects the efficiency of the cell-to-cell spread of Listeria (17). The finding that InlC has several functions is not unexpected for a virulence factor. Indeed, it is well known that bacterial pathogen effectors can be multifunctional, be modular, or display cooperative activities. InlC is another example of such a bacterial effector that exerts several activities during infection. However, its effect on virulence is subtler than that of bona fide virulence factors, such as LLO and ActA (3, 44). Thus, InlC might be more appropriately considered a “virulence modulator” that acts at different stages of the infectious process. In conclusion, this study reinforces the importance of the internalin family in listerial pathogenesis, highlighting that this family is involved not only in the entry of Listeria into cells, but also in other critical events in the infectious process.
Materials and Methods
Bacterial Strains, Reagents, Antibodies, and Immunofluorescence Microscopy.
The bacterial strains and growth conditions, reagents, antibodies, and microscopy procedures used in this study are described in SI Materials and Methods and Table S1.
Cell Culture and Infections, Luciferase Reporter Assay, and ELISA.
The cell lines and protocols for this study are described in SI Materials and Methods.
Plasmids and Oligonucleotides.
The plasmids and oligonucleotides used in this study are listed in Tables S2 and S3. The construction of plasmids pcDNA4-inlC, pGEX-4T-inlC, and pAD-inlC is described in SI Materials and Methods.
Mutant Construction.
The ΔinlC isogenic deletion mutant was constructed as described in SI Materials and Methods.
Identification of InlC Interactor by Yeast Two-Hybrid Screening.
The Y2H screen was performed on a whole human genome cDNA placental library as described in SI Materials and Methods.
Immunoprecipitation and Immunoblotting.
HEK-293 T-REx or HeLa cells were transfected or infected, and cells lysates were prepared. Details of immunoprecipitation and Western blot analysis are provided in SI Materials and Methods.
Murine Infection Experiments and Cell Counting.
Infection procedures (i.v. and i.p.) and leukocyte counting in peritoneal lavage fluid are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. M. Cavaillon and C. Fitting for their constant support; R. Weil (Unité de Signalisation Moléculaire et Activation Cellulaire, Institut Pasteur, Paris, France) for a critical reading of the manuscript and for providing the Igκ3-conaluc, HA-IKKα, and pmIκBα-myc plasmids; C. Parsot for helpful discussions; R. Zenon for his help in obtaining polyclonal anti-InlC antibodies in rabbit; and the students of the Microbiology Course of the Pasteur Institute for construction of the pMAD-inlC plasmid. The work in the P.C. laboratory was supported by Institut Pasteur, Institut National de la Santé et de la Recherche Médicale, Institut National de la Recherche Agronomique, European Research Council (Advanced Grant 233348), and Fondation le Roch Les Mousquetaires. P.C. is an international research scholar of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007765107/-/DCSupplemental.
References
- 1.Cossart P, Toledo-Arana A. Listeria monocytogenes, a unique model in infection biology: An overview. Microbes Infect. 2008;10:1041–1050. doi: 10.1016/j.micinf.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 2.Dussurget O. New insights into determinants of Listeria monocytogenes virulence. Int Rev Cell Mol Biol. 2008;270:1–38. doi: 10.1016/S1937-6448(08)01401-9. [DOI] [PubMed] [Google Scholar]
- 3.Hamon M, Bierne H, Cossart P. Listeria monocytogenes: A multifaceted model. Nat Rev Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 4.Bierne H, Cossart P. Listeria monocytogenes surface proteins: From genome predictions to function. Microbiol Mol Biol Rev. 2007;71:377–397. doi: 10.1128/MMBR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierne H, Sabet C, Personnic N, Cossart P. Internalins: A complex family of leucine-rich repeat–containing proteins in Listeria monocytogenes. Microbes Infect. 2007;9:1156–1166. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Glaser P, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 7.Disson O, et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 8.Lecuit M, Cossart P. Genetically modified animal models for human infections: The Listeria paradigm. Trends Mol Med. 2002;8:537–542. doi: 10.1016/s1471-4914(02)02413-9. [DOI] [PubMed] [Google Scholar]
- 9.Lingnau A, Chakraborty T, Niebuhr K, Domann E, Wehland J. Identification and purification of novel internalin-related proteins in Listeria monocytogenes and Listeria ivanovii. Infect Immun. 1996;64:1002–1006. doi: 10.1128/iai.64.3.1002-1006.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelbrecht F, et al. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 11.Ooi A, Hussain S, Seyedarabi A, Pickersgill RW. Structure of internalin C from Listeria monocytogenes. Acta Crystallogr D Biol Crystallogr. 2006;62:1287–1293. doi: 10.1107/S0907444906026746. [DOI] [PubMed] [Google Scholar]
- 12.Bubert A, et al. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol Gen Genet. 1999;261:323–336. doi: 10.1007/pl00008633. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee SS, et al. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGann P, Ivanek R, Wiedmann M, Boor KJ. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae. Appl Environ Microbiol. 2007;73:2806–2814. doi: 10.1128/AEM.02923-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 16.Domann E, et al. Identification and characterization of a novel PrfA-regulated gene in Listeria monocytogenes whose product, IrpA, is highly homologous to internalin proteins, which contain leucine-rich repeats. Infect Immun. 1997;65:101–109. doi: 10.1128/iai.65.1.101-109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajabian T, et al. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol. 2009;11:1212–1218. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldwin DN, Vanchinathan V, Brown PO, Theriot JA. A gene-expression program reflecting the innate immune response of cultured intestinal epithelial cells to infection by Listeria monocytogenes. Genome Biol. 2003;4:R2. doi: 10.1186/gb-2002-4-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecuit M, Sonnenburg JL, Cossart P, Gordon JI. Functional genomic studies of the intestinal response to a foodborne enteropathogen in a humanized gnotobiotic mouse model. J Biol Chem. 2007;282:15065–15072. doi: 10.1074/jbc.M610926200. [DOI] [PubMed] [Google Scholar]
- 20.Kayal S, et al. Listeriolysin O secreted by Listeria monocytogenes induces NF-kappaB signalling by activating the IkappaB kinase complex. Mol Microbiol. 2002;44:1407–1419. doi: 10.1046/j.1365-2958.2002.02973.x. [DOI] [PubMed] [Google Scholar]
- 21.Mansell A, Braun L, Cossart P, O'Neill LA. A novel function of InIB from Listeria monocytogenes: Activation of NF-kappaB in J774 macrophages. Cell Microbiol. 2000;2:127–136. doi: 10.1046/j.1462-5822.2000.00038.x. [DOI] [PubMed] [Google Scholar]
- 22.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 25.Kim DW, et al. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weil R, Whiteside ST, Israël A. Control of NF-kappa B activity by the I kappa B beta inhibitor. Immunobiology. 1997;198:14–23. doi: 10.1016/s0171-2985(97)80023-x. [DOI] [PubMed] [Google Scholar]
- 27.Tato CM, Hunter CA. Host-pathogen interactions: Subversion and utilization of the NF-kappa B pathway during infection. Infect Immun. 2002;70:3311–3317. doi: 10.1128/IAI.70.7.3311-3317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shames SR, Auweter SD, Finlay BB. Co-evolution and exploitation of host cell signaling pathways by bacterial pathogens. Int J Biochem Cell Biol. 2009;41:380–389. doi: 10.1016/j.biocel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Oie KL, Pickup DJ. Cowpox virus and other members of the orthopoxvirus genus interfere with the regulation of NF-kappaB activation. Virology. 2001;288:175–187. doi: 10.1006/viro.2001.1090. [DOI] [PubMed] [Google Scholar]
- 30.Akari H, Bour S, Kao S, Adachi A, Strebel K. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor kappaB–dependent expression of antiapoptotic factors. J Exp Med. 2001;194:1299–1311. doi: 10.1084/jem.194.9.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bour S, Perrin C, Akari H, Strebel K. The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP–mediated degradation of Ikappa B. J Biol Chem. 2001;276:15920–15928. doi: 10.1074/jbc.M010533200. [DOI] [PubMed] [Google Scholar]
- 32.Le Negrate G, et al. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J Immunol. 2008;180:5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 33.Rytkönen A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171:882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Negrate G, et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 36.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 38.Nadler C, et al. The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog. 2010;6:e1000743. doi: 10.1371/journal.ppat.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashida H, et al. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB–mediated inflammatory response. Nat Cell Biol. 2009;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauf N, Goebel W, Fiedler F, Sokolovic Z, Kuhn M. Listeria monocytogenes infection of P388D1 macrophages results in a biphasic NF-kappaB (RelA/p50) activation induced by lipoteichoic acid and bacterial phospholipases and mediated by IkappaBalpha and IkappaBbeta degradation. Proc Natl Acad Sci USA. 1997;94:9394–9399. doi: 10.1073/pnas.94.17.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arbibe L, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 42.Boneca IG, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamon MA, et al. Histone modifications induced by a family of bacterial toxins. Proc Natl Acad Sci USA. 2007;104:13467–13472. doi: 10.1073/pnas.0702729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnupf P, Portnoy DA. Listeriolysin O: A phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.