Abstract
Invertases are highly regulated enzymes with essential functions in carbohydrate partitioning, sugar signaling, and plant development. Here we present the 2.6 Å crystal structure of Arabidopsis cell-wall invertase 1 (INV1) in complex with a protein inhibitor (CIF, or cell-wall inhibitor of β-fructosidase) from tobacco. The structure identifies a small amino acid motif in CIF that directly targets the invertase active site. The activity of INV1 and its interaction with CIF are strictly pH-dependent with a maximum at about pH 4.5. At this pH, isothermal titration calorimetry reveals that CIF tightly binds its target with nanomolar affinity. CIF competes with sucrose (Suc) for the same binding site, suggesting that both the extracellular Suc concentration and the pH changes regulate association of the complex. A conserved glutamate residue in the complex interface was previously identified as an important quantitative trait locus affecting fruit quality, which implicates the invertase–inhibitor complex as a main regulator of carbon partitioning in plants. Comparison of the CIF/INV1 structure with the complex between the structurally CIF-related pectin methylesterase inhibitor (PMEI) and pectin methylesterase indicates a common targeting mechanism in PMEI and CIF. However, CIF and PMEI use distinct surface areas to selectively inhibit very different enzymatic scaffolds.
Keywords: active site, crystallography, posttranslational enzyme regulation, protein complex, sugar metabolism
The plant cell wall is a complex structural network playing a crucial role in all aspects of plant life. Environmental changes lead to apoplastic metabolic and signaling processes that drive differential gene expression and intracellular metabolism (1). The model genome of Arabidopsis harbors a large family of extracellular protein inhibitors characterized by a predicted four-helix bundle fold and four strictly conserved cysteine residues. Family members have been found to inhibit plant pectin methylesterases (PME) or invertases (2), extracellular enzymes of different sequence, architecture, and function.
Arabidopsis also contains at least three catalytically active acidic cell-wall invertases (CWIs), secreted apoplastic enzymes belonging to the glycoside hydrolase family 32 (http://www.cazy.org) (3, 4). Invertases catalyze the hydrolytic cleavage of the disaccharide sucrose (Suc; the main transport sugar in plants) into glucose (Glc) and fructose (Frc) (5). CWI activity strongly influences the apoplastic Suc:Glc ratio and the related Glc- and Suc-specific signaling pathways (6). Importantly, CWIs maintain Suc concentration gradients between photosynthetic source and sink tissues, a driving force for carbohydrate transport, partitioning, and storage in plants (7, 8). The activity of CWIs, including the well-characterized cell-wall invertase 1 (INV1) from Arabidopsis thaliana (9) (also abbreviated as AtcwINV1), is tightly regulated at both the transcriptional and the posttranscriptional level (10). At the posttranslational level, cell-wall and vacuolar invertases can be targeted by the above-mentioned compartment-specific inhibitor proteins (2). These invertase inhibitors efficiently regulate invertase activity and the downstream Glc-signaling cascades in vivo (11, 12), but it is not understood how they recognize and inhibit their enzyme targets. Interaction of tobacco CWI with the apoplasmic invertase inhibitor CIF (for cell wall inhibitor of β-fructosidase) is strictly pH-dependent (13), as is the activity of the enzyme itself (14). The inhibitors adopt a four-helix bundle core structure that is extended by an N-terminal α-hairpin module (15). Both motifs are stabilized by disulfide bonds and are required for structural stability (13, 15). Protein-engineering experiments suggest that the four-helix bundle core is critical for the interaction with invertase (16). Here we report the crystal structure of the complex between INV1 and the CWI inhibitor CIF from tobacco. The structure reveals that CIF directly inserts a small amino acid motif into the invertase substrate-binding cleft. Biochemical experiments support a model in which local pH variations and the substrate:inhibitor ratio modulate this highly specific protein–protein interaction in vivo. Comparison with a pectin methylesterase inhibitor (PMEI)–PME complex structure reveals that CIFs and PMEIs use a common mechanism to inactivate their target enzymes.
Results and Discussion
Architecture of the INV1–CIF Complex.
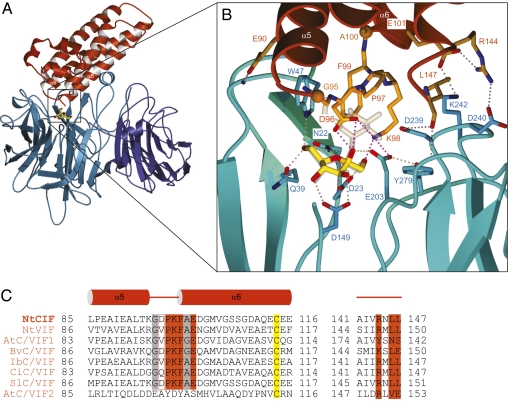
The invertase-inhibitor complex was formed by mixing purified INV1 and CIF, was crystallized, and the 2.6 Å structure solved by molecular replacement using the isolated INV1 and CIF structures as search models (9, 15) (Materials and Methods; Table S1). Our structure reveals that the four-helix bundle of CIF binds primarily to the five-bladed β-propeller module of INV1 (9) and shows only a few interactions with the β-sandwich module of the enzyme (Fig. 1A). The N-terminal α-helical hairpin in CIF does not participate in the interaction, consistent with earlier studies (16). The complex interface buries an area of ∼1,000 Å2 corresponding to ∼15% of the inhibitor and only ∼5% of the enzyme total solvent accessible area, respectively. We found a Frc molecule bound in the active site of INV1 (Fig. 1B; Fig. S1) that had not been observed in the isolated INV1 structure (9). Frc in our crystals possibly was produced during complex formation from traces of Suc used in the preparation of CIF (17) and stayed bound in a complex ensemble that was selected by the crystallization process. Such postcatalysis product states have been observed in crystal structures of other GH32 family members (18, 19). Although Frc is apparently stably bound in our crystals and undergoes favorable interactions also with the inhibitor component (Fig. 1B), we cannot be certain whether Frc is indeed required for CIF-mediated INV1 inhibition. In fact, the Asp residue in CIF that makes hydrogen bond contacts with Frc (see below) is not strictly conserved in other validated invertase inhibitors (Fig. 1 B and C). We think that our complex structure represents a reasonably good model for invertase inhibitor interaction on the basis of the high sequence similarity among the respective protein components. Comparative sequence analyses reveal that all INV1 residues that are in contact with tobacco CIF (Fig. 1; Fig. S2) are identical or similar in tobacco CWI, which shares 72% overall sequence identity with INV1 (Fig. S2). Moreover, homology modeling of Arabidopsis CWI inhibitor 1 (AtC/VIF1) (20), which is 41% sequence identical to tobacco CIF, suggests that most interactions are also conserved in the Arabidopsis system (Fig. S3 A and C). Although there may be sequence variations in different enzyme–inhibitor pairs (see below), homology modeling suggests that our complex structure can provide mechanistic insights into both the tobacco and the Arabidopsis system.
Fig. 1.
CIF targets the substrate-binding cleft of INV1. (A) Overview of the complex. Ribbon diagram of CIF (red) binding to the β-propeller module (cyan) of INV1 with only a few direct interactions to the noncatalytic β-sandwich domain (dark blue). (B) Close-up view of the active-site region depicting the bound Frc molecule and important interactions between enzyme and inhibitor. A Suc molecule derived from a INV1–substrate complex structure (PDB-ID 2qqw) (21) is shown in light yellow to indicate steric clashes of CIF with the Glc moiety of Suc. Polar interactions (distances <3.1 Å) are indicated as dotted lines. The sharply bent loop containing the PKF motif is shown in orange with small residues at the loop boundaries highlighted as spheres. Interactions between the C terminus of CIF and INV1 are included. (C) Sequence comparison of experimentally validated CIFs. The PKF motif is shaded by red, along with the conserved C terminus of CIF involved in complex stabilization. The small residues flanking the PKF motif are shaded by gray, an invariant cysteine by yellow.
CIF Targets the Invertase Substrate-Binding Cleft.
Complex interface residues contributed by CIF originate from the very C terminus of the inhibitor, from helices α5 and α6, and from a short loop connecting these helices (Fig. 1 A and B). From this loop in CIF, three amino acids (Pro97, Lys98, and Phe99) directly contact the INV1 substrate-binding cleft (21). In INV1, the Asp239/Lys242 pair is essential to keep the Glc moiety of Suc in a catalytically competent position, while Trp47 further stabilizes binding of the substrate (Fig. 1B) (22). Insertion of CIF into the active site of INV1 drastically alters these constellations. In the complex structure, Lys98 of CIF interacts with the acid–base catalyst Glu203 of INV1. Interestingly, similar interactions mediate complex formation between fungal xylanases and the wheat xylanase-inhibiting protein I (23). Whereas Asp239 occupies a similar position in our complex and in the free INV1 structure, Lys242 now interacts with Glu101 in CIF. Furthermore, Phe99 in CIF establishes an almost perfect stacking interaction with Trp47 (in INV1), thereby excluding Suc from its binding site. Small residues flanking the small inserting loop in CIF (Gly95 and Ala100 in Fig. 1C) favor a particularly sharply bent loop structure and allow the insertion of consecutive loop residues into the INV1 active-site cleft (Fig. 1B). We propose that this PKF motif defines a sequence fingerprint critical for invertase–inhibitor interaction because it is conserved in most biochemically validated invertase inhibitors (Fig. 1C). In addition to this motif in CIF, an ionic bond network contributes to the complex interface (Fig. 1B), with Glu101 (contacting Lys242 in INV1) and Arg144 (contacting Asp240) being highly conserved in the known invertase inhibitors (Fig. 1 B and C). CIF residue Asp96 appears to stabilize binding of the Frc molecule in the active site of the complex, thereby trapping the enzyme in a product-bound conformation (Fig. 1B). Our structure is consistent with earlier observations that suggested that CWI can be substrate-protected from interaction with CIF (24), further suggesting that, in excess of apoplastic Suc, the inhibitor cannot bind its target enzyme. Vice versa, in excess of secreted CIF, the inhibitor should efficiently compete with Suc for binding, thereby inhibiting INV1. Thus, the apoplastic sucrose:inhibitor ratio may be an important determinant regulating CWI activity in vivo.
pH Dependence of the Interaction.
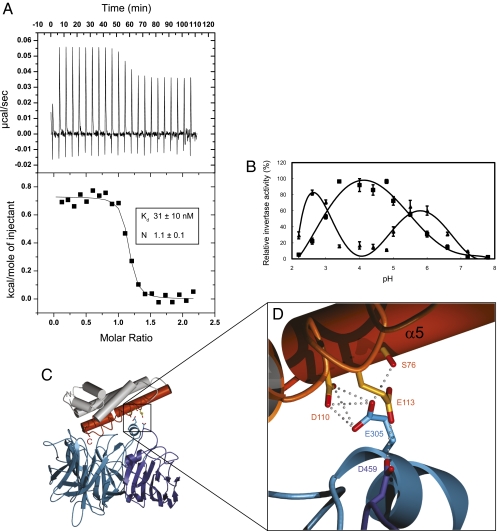
Earlier studies revealed that the activity of CWI is strictly pH dependent (with a maximum at about pH 4.5), as is the interaction with CIF (25). We previously ruled out major pH-induced structural changes in CIF to account for this feature (26). When we quantified complex formation using isothermal titration calorimetry (ITC; Materials and Methods), we found that CIF tightly binds INV1 with nanomolar affinity at pH 4.5 (Fig. 2A). The binding is endothermic (ΔH ∼700 cal·mol−1) with a significant entropic contribution (ΔS ∼35 cal·mol−1·deg−1), consistent with the small and rather hydrophobic complex interface described above. Consistently, we found that CIF inactivates INV1 best under acidic buffer conditions (pH range 3.5–5.5) in enzymatic assays (Fig. 2B). These experiments and earlier studies (13) together suggest that the interaction between CIF and its CWI target is tailored to the pH environment typically associated with the plant cell wall and to the pH optimum of the enzyme itself (Fig. 2B).
Fig. 2.
A pH-sensitive INV1–CIF complex interface. (A) Isothermal titration calorimetry profile for CIF binding to INV1 at pH 4.5. Shown are experimental values ± fitting errors. (B) Effect of pH on the relative activity (in percentage of maximal enzyme activity at pH 4.8) of INV1 in the absence (squares) and presence (triangles) of 250 nM CIF (Materials and Methods). (C) Ribbon diagram of CIF (red) establishing ionic contacts with both the β-propeller (light blue) and the β-sandwich (dark blue) modules of the enzyme. (D) Close-up view of the acidic network in the complex interface with distances <3.1 Å included as dotted lines.
More specifically, our complex structure reveals a surface area containing a small cluster of acidic residues that titrate in the relevant pH range (Fig. 2 C and D). These include Asp110 (side chain pKa ∼3.9) and Glu113 (side chain pKa ∼4.1) from CIF along with Glu305 from the β-propeller module and Asp459 from the β-sandwich domain (9) of INV1. Importantly, the resulting interactions in the crystal structure show interatomic distances that would require the functional carboxylate groups to be protonated for interaction (Fig. 2D). Consequently, a pH shift toward more basic conditions renders these contacts unfavorable, thus destabilizing the complex. Similar interactions are present in a tobacco CWI–CIF complex, where this phenomenon was first described (13), whereas in AtC/VIF1 Glu113 is replaced by a Val residue (Fig. S3 B and C). This suggests that moderate extracellular pH changes may modulate the interaction between CWIs and at least some of their cognate inhibitors. Taken together, our findings support a scenario in which the concentration of free Suc, the Suc:inhibitor ratio, and the apoplastic pH together determine the catalytic state of CWIs in vivo.
Complex Structure Rationalizes Genetic Studies.
A quantitative trait locus (QTL) was previously identified in the invertase LIN5 that increases sugar yield in tomato fruits (27). Associated with this QTL is the substitution of Glu348 in LIN5 (Glu305 in INV1) for aspartic acid. Mapping this alteration onto the structure of a bacterial invertase (28), the authors speculated that it would interfere with the catalytic activity of LIN5. Later, inspection of the more closely related Arabidopsis enzyme structure (9) revealed that the corresponding Glu305 in INV1 is more than 25 Å away from the active site, making a direct role in catalysis unlikely. In our INV1–CIF complex, Glu305 is a critical part of the invertase–inhibitor complex interface (see above; Fig. 2 C and D), and thus the QTL may not affect LIN5 activity directly, but rather the interaction with its invertase inhibitor. In this respect, it is noteworthy that a highly conserved sequence ortholog of tobacco CIF exists in tomato (sharing 88% sequence identity), which has been successfully used to silence CWI activity in transgenic tomato plants, leading to delayed senescence, superior Hex:Suc ratios, and an increase in seed weight (12). Consistently, RNA interference of LIN5 in tomato appears to have opposite effects, resulting in abnormal plants with lower fruit and seed weights and lower hormone levels (29). The identification of a QTL within the invertase–inhibitor complex interface indicates that it is not CWI itself but the invertase–inhibitor complex that should be regarded as a main regulator of carbon partitioning in plants.
Evolution of Different Inhibitor Specificities.
The cell-wall homogalacturonan content and the methylesterification status of this polymer control plant growth, development, and biomass production (30). De-methylesterification of homogalacturonan is regulated through the action of the ubiquitous PMEs, which in turn are controlled by PMEIs.
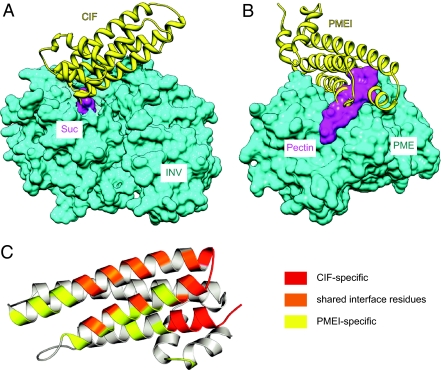
CIFs and PMEIs are sequence-related proteins with very similar core structures that target two distinct classes of plant enzymes, invertases and PMEs (15, 16). Comparing the interface residues in a PMEI–PME complex (31) with those of the INV1–CIF complex reveals that both inhibitors directly bind to the substrate-binding cleft of their respective target enzyme. Comparison of these complex structures (this study and ref. 31) with the respective substrate-bound enzymes (21, 32) further suggests that both CIF and PMEI act by preventing substrate binding to the enzyme component (Fig. 3 A and B). They thus share a common mode of action, also observed with the α-amylase/BASI system (33) and in a variety of other enzyme–inhibitor complexes (34, 35). However, PMEI and CIF have evolved partially overlapping and partially distinct surface areas, allowing them to recognize rather different enzymatic scaffolds. Complex interface residues in CIF originate mainly from helices α5 and α6, from the short loop connecting these helices (containing the PKF motif) and from the very C terminus of the inhibitor (Fig. 3B). Importantly, neither the corresponding loop regions nor the C terminus of PMEI are involved in the interaction with PME (31). Instead, PMEI provides additional contacts originating from two core helices and from the N-terminal helix hairpin module (36) (Fig. 3B). Because the inhibitor-enzyme interfaces are comparatively small, PMEI and CIF could evolve unique surface properties while maintaining significant sequence and structural homology. Although PMEI and CIF have a common evolutionary origin, they inhibit two completely different enzymes. Similarly, the wheat xylanase inhibitor (XIP-1) can also inhibit xylanase enzymes with different folds (23).
Fig. 3.
Invertase inhibitors and PMEIs share a common targeting mechanism. (A) CIF (yellow) is bound to the substrate-binding cleft of INV1 (surface representation in cyan). CIF binding interferes with coordination of the substrate Suc (surface representation in magenta), the position of which is inferred from the structure of an INV1–Suc complex structure (PDB-ID 2qqw) (21). (B) Structure of the PMEI-PME complex (PDB-ID 1xg2) (31). PMEI (yellow) inserts its four-helix bundle core into the pectin-binding cleft of PME (surface representation in cyan). Apparently PMEI interferes with the binding of the methylated pectin substrate, whose position has been inferred from a bacterial PME–pectin complex structure (PDB-ID 2nsp) (32). (C) Invertase inhibitors and PMEIs use partially overlapping surface areas to bind to very different target enzymes. Ribbon diagram of a PMEI–CIF hybrid with regions involved in complex stabilization indicated in yellow (specific to a PME–PMEI complex) (31) or red (specific to the INV1–CIF complex). Areas contributing to the respective complex interface in both complex structures are depicted in orange.
Concluding Remarks
Many plant genomes harbor large, expanded gene families. These families probably originated from gene duplication events and often have evolved to fulfill diverse functions. In this study, we have analyzed a family of protein inhibitors that has achieved functional diversity by evolving distinct surface properties on an ancestral scaffold. In the case of the invertase inhibitor CIF, we define a small set of conserved residues required to interact with CWI in a pH-dependent manner. The fact that a subset of these residues was found in quantitative trait analyses screening for tomato fruit quality (27) strongly suggests that formation of the CIF–invertase complex is an important mechanism in the regulation of invertase activity in vivo. Our study provides the mechanistic information that allows for rationalizing such genetic studies.
Although sequence comparisons with the known invertase inhibitors suggest that the complex interface is largely conserved, variations of the PKF motif may well be tolerated in different enzyme–inhibitor combinations (Fig. 1C). Thus, our structure represents one possible view of a system that has probably undergone extensive coevolution of the enzyme and inhibitor components. It can be expected that the related vacuolar invertase inhibitors use similar surface areas and essentially the same targeting mechanism to inhibit vacuolar isoenzymes. Nevertheless, very different residue combinations may define the respective complex interfaces. Other enzyme–inhibitor pairs thus remain to be functionally characterized to fully appreciate the biochemical, cellular, and developmental roles of the ∼180 invertase inhibitor-like proteins in the Arabidopsis genome.
Materials and Methods
Protein Expression, Purification, Crystallization, and X-Ray Data Collection.
Arabidopsis INV1 and Nicotiana CIF were expressed and purified as described (9, 17). The complex was formed by mixing enzyme and inhibitor in 1:1.5 molar ratio. Samples were dialyzed against 50 mM Na acetate (pH 5.0) and 100 mM NaCl and concentrated to 10 mg/mL using a Vivapore 10/20 mL concentrator (7.5 kDa molecular weight cut-off; Vivascience). Hexagonal crystals were grown at room temperature by vapor diffusion in hanging drops composed of equal volumes (1 + 1 μL) of protein solution and crystallization buffer [15% (vol/vol) PEG 5,000 monomethylether, 0.2 M (NH4)2SO4, 0.1 M Mes (pH 6.5)] suspended over 1 mL of the latter as reservoir solution. A small needle of ∼80 × 15 × 15 μm was transferred into the reservoir solution containing 20% (vol/vol) ethylene glycol and directly frozen in the cryo-stream. From this microcrystal, a dataset at 2.6 Å was recorded at beam-line ID-29 at the European Synchrotron Radiation Facility (Grenoble, France). Data processing and scaling was performed with XDS (37) (Version June 2008).
Structure Determination and Refinement.
The structure of the INV1–CIF complex was determined by molecular replacement with PHASER (38) using structures of INV1 (PDB ID 2AC1) (9) and CIF (PDB ID 1RJ1) (15) as search models. There are six complexes per asymmetric units, and the root mean square deviation between 683 corresponding Cα atoms is <0.2 Å. Twinning analysis in PHENIX.XTRIAGE (http://www.phenix-online.org) using data between 10.0 and 3.5 Å indicated the presence of a twin fraction of about 0.170 along -k, -h, and -l. The structure was completed in alternating cycles of model building in COOT (39) and restrained TLS refinement against merohedrally twinned data as implemented in PHENIX.REFINE and REFMAC5 (40) (version 5.5). Incorporation of the twin law in the refinement protocol resulted in a 5% and 6.5% drop in Rcryst and Rfree, respectively. The refined model includes residues 5–541 in INV1 and residues 2–147 in CIF, respectively. Structural validation of the refined model with MOLPROBITY (41) revealed 96.2% of the residues in the favored regions of the Ramachandran plot and no outliers (see Table S1 for a summary of the crystallographic analysis). Structural visualization was done with POVSCRIPT (42) and POVRAY (http://www.povray.org).
Isothermal Titration Calorimetry.
ITC was performed using a VP-ITC calorimeter (Microcal). Both proteins were dialyzed against ITC buffer (100 mM Na acetate, pH 4.5, 100 mM NaCl) before all titrations. The experiments were performed at 25 °C. A typical titration consisted of injecting 12-μL aliquots of CIF (100 μM) into a 10-μM INV1 solution at time intervals of 5 min to ensure that the titration peak returned to the baseline. ITC data were corrected for the heat of dilution by subtracting the mixing enthalpies for the titrant solution injections into protein-free buffer. ITC data were analyzed using program Origin (Version 5.0) as provided by the manufacturer. We used a single set of identical binding sites model. Due to the small endothermic signal, experiments were repeated three times and validated with independent protein preparations.
pH Dependence of CWI Inhibition by CIF.
Invertase activity was followed by quantifying Frc production using high performance anion exchange chromatography with pulsed amperometric detection (43). Reaction mixtures contained 20 mM Suc and 3 nM recombinant, purified INV1 at varying pH (pH 2.2–7.8) in the presence and absence of 250 nM recombinant, purified CIF. Buffering (pH 2.2–7.8) was accomplished with the McIlvaine buffer system (80 mM Na2HPO3, with 40 mM citric acid varying according to the required pH value) supplemented with 0.02% (wt/vol) NaN3. A preincubation of 15 min was performed as described (24). Samples were incubated for 60 min at 30 °C.
Homology Modeling of AtC/VIF 1.
(Uniprot http://www.uniprot.org; accession Q9C7Y8) and tobacco CWI (Uniprot; accession Q43799) was performed with the program MODELLER (44) using the CWI–CIF complex structure as template. Structure-based sequence alignments were done using TCOFFEE (45).
Supplementary Material
Acknowledgments
We thank Christoph Mueller-Dieckmann for help with data collection and J. Noel, M. Betts, R. Russell, and T. Rausch for discussion. M.H. is grateful to J. Chory for her generous support. W.V.d.E and W.L. thank R. Vergauwen for technical assistance. This work was supported by the Peter and Traudl Engelhorn Foundation (Penzberg, Germany), by a Human Frontier Science Program postdoctoral fellowship (to M.H.), and by grants from Fonds Wetenschappelijk Onderzoek-Vlaanderen (to W.V.d.E. and W.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2XQR).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004481107/-/DCSupplemental.
References
- 1.Seifert GJ, Blaukopf C. Irritable walls: The plant extracellular matrix and signaling. Plant Physiol. 2010;153:467–478. doi: 10.1104/pp.110.153940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rausch T, Greiner S. Plant protein inhibitors of invertases. Biochim Biophys Acta. 2004;1696:253–261. doi: 10.1016/j.bbapap.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Van den Ende W, Lammens W, Van Laere A, Schroeven L, Le Roy K. Donor and acceptor substrate selectivity among plant glycoside hydrolase family 32 enzymes. FEBS J. 2009;276:5788–5798. doi: 10.1111/j.1742-4658.2009.07316.x. [DOI] [PubMed] [Google Scholar]
- 4.De Coninck B, et al. Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant Cell Environ. 2005;28:432–443. [Google Scholar]
- 5.Roitsch T, González MC. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004;9:606–613. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Hanson J, Smeekens S. Sugar perception and signaling—An update. Curr Opin Plant Biol. 2009;12:562–567. doi: 10.1016/j.pbi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 7.Sturm A. Invertases: Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–8. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chourey PS, Jain M, Li QB, Carlson SJ. Genetic control of cell wall invertases in developing endosperm of maize. Planta. 2006;223:159–167. doi: 10.1007/s00425-005-0039-5. [DOI] [PubMed] [Google Scholar]
- 9.Verhaest M, et al. X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr. 2006;62:1555–1563. doi: 10.1107/S0907444906044489. [DOI] [PubMed] [Google Scholar]
- 10.Huang LF, Bocock PN, Davis JM, Koch KE. Regulation of invertase: a ‘suite’ of transcriptional and post-transcriptional mechanisms. Funct Plant Biol. 2007;34:499–507. doi: 10.1071/FP06227. [DOI] [PubMed] [Google Scholar]
- 11.Balibrea Lara ME, et al. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell. 2004;16:1276–1287. doi: 10.1105/tpc.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y, Ni DA, Ruan YL. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weil M, Krausgrill S, Schuster A, Rausch T. A 17-kDa Nicotiana tabacum cell-wall peptide acts as an in-vitro inhibitor of the cell-wall isoform of acid invertase. Planta. 1994;193:438–445. doi: 10.1007/BF00201824. [DOI] [PubMed] [Google Scholar]
- 14.Goetz M, et al. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hothorn M, D'Angelo I, Marquez JA, Greiner S, Scheffzek K. The invertase inhibitor Nt-CIF from tobacco: A highly thermostable four-helix bundle with an unusual N-terminal extension. J Mol Biol. 2004;335:987–995. doi: 10.1016/j.jmb.2003.10.066. [DOI] [PubMed] [Google Scholar]
- 16.Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K. Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell. 2004;16:3437–3447. doi: 10.1105/tpc.104.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hothorn M, Bonneau F, Stier G, Greiner S, Scheffzek K. Bacterial expression, purification and preliminary X-ray crystallographic characterization of the invertase inhibitor Nt-CIF from tobacco. Acta Crystallogr D Biol Crystallogr. 2003;59:2279–2282. doi: 10.1107/s0907444903021036. [DOI] [PubMed] [Google Scholar]
- 18.Alvaro-Benito M, Polo A, González B, Fernández-Lobato M, Sanz-Aparicio J. Structural and kinetic analysis of Schwanniomyces occidentalis invertase reveals a new oligomerization pattern and the role of its supplementary domain in substrate binding. J Biol Chem. 2010;285:13930–13941. doi: 10.1074/jbc.M109.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuankhayan P, et al. Crystal structures of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete subsites in the active site for catalysis. J Biol Chem. 2010;285:23251–23264. doi: 10.1074/jbc.M110.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link M, Rausch T, Greiner S. In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS Lett. 2004;573:105–109. doi: 10.1016/j.febslet.2004.07.062. [DOI] [PubMed] [Google Scholar]
- 21.Lammens W, Le Roy K, Van Laere A, Rabijns A, Van den Ende W. Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose. J Mol Biol. 2008;377:378–385. doi: 10.1016/j.jmb.2007.12.074. [DOI] [PubMed] [Google Scholar]
- 22.Le Roy K, et al. Unraveling the difference between invertases and fructan exohydrolases: A single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1- exohydrolase. Plant Physiol. 2007;145:616–625. doi: 10.1104/pp.107.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payan F, et al. Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson) Biochem J. 2003;372:399–405. doi: 10.1042/BJ20021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander A, Krausgrill S, Greiner S, Weil M, Rausch T. Sucrose protects cell wall invertase but not vacuolar invertase against proteinaceous inhibitors. FEBS Lett. 1996;385:171–175. doi: 10.1016/0014-5793(96)00378-x. [DOI] [PubMed] [Google Scholar]
- 25.Weil M, Rausch T. Acid invertase in Nicotiana tabacum grown-gall cells: molecular properties of the cell wall isoform. Planta. 1994;193:430–437. doi: 10.1007/BF00201824. [DOI] [PubMed] [Google Scholar]
- 26.Hothorn M, Scheffzek K. Multiple crystal forms of the cell-wall invertase inhibitor from tobacco support high conformational rigidity over a broad pH range. Acta Crystallogr D Biol Crystallogr. 2006;62:665–670. doi: 10.1107/S0907444906011693. [DOI] [PubMed] [Google Scholar]
- 27.Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science. 2004;305:1786–1789. doi: 10.1126/science.1101666. [DOI] [PubMed] [Google Scholar]
- 28.Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M. The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem. 2004;279:18903–18910. doi: 10.1074/jbc.M313911200. [DOI] [PubMed] [Google Scholar]
- 29.Zanor MI, et al. RNA interference of LIN5 in tomato confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 2009;150:1204–1218. doi: 10.1104/pp.109.136598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf S, Grsic-Rausch S, Rausch T, Greiner S. Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett. 2003;555:551–555. doi: 10.1016/s0014-5793(03)01344-9. [DOI] [PubMed] [Google Scholar]
- 31.Di Matteo A, et al. Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell. 2005;17:849–858. doi: 10.1105/tpc.104.028886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fries M, Ihrig J, Brocklehurst K, Shevchik VE, Pickersgill RW. Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J. 2007;26:3879–3887. doi: 10.1038/sj.emboj.7601816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallée F, et al. Barley alpha-amylase bound to its endogenous protein inhibitor BASI: Crystal structure of the complex at 1.9 A resolution. Structure. 1998;6:649–659. doi: 10.1016/s0969-2126(98)00066-5. [DOI] [PubMed] [Google Scholar]
- 34.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 35.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 36.Ciardiello MA, et al. The peculiar structural features of kiwi fruit pectin methylesterase: amino acid sequence, oligosaccharides structure, and modeling of the interaction with its natural proteinaceous inhibitor. Proteins. 2008;71:195–206. doi: 10.1002/prot.21681. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 38.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 41.Lovell SC, et al. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 42.Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
- 43.Vergauwen R, Van Laere A, Van den Ende W. Properties of fructan:fructan 1-fructosyltransferases from chicory and globe thistle, two Asteracean plants storing greatly different types of inulin. Plant Physiol. 2003;133:391–401. doi: 10.1104/pp.103.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 45.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.