Almost all antibiotics used in clinical medicine owe their origin to natural compounds. Discovery of these compounds has traditionally been driven by an outward-looking Saganesque conviction (somewhere, something incredible is waiting to be known) that broad screening of many hundreds of species of bacteria and fungi would unearth novel antimicrobials. Indisputably, this approach has yielded results. More than 200 species of free-living actinomycete bacteria have been shown to produce antimicrobial compounds. Many of these compounds have been of medical value, and their usefulness could be expanded by chemical tweaking to improve pharmacological properties. The discovery and development of antimicrobial drugs has, however, been an arduous and expensive process. For instance, the macrolide antibiotic erythromycin originates in the actinomycete Saccharopolyspora erythraea and has been derivatized by numerous pharmaceutical companies over the past 50 y to yield thousands of compounds (1); a few promising leads were then subjected to lengthy pharmacokinetics testing, toxicological studies, and clinical trials in preparation for the approval process. After finally entering the clinic, the lifespan of an antimicrobial drug might be short and risks being curtailed at any time by bacterial resistance. Such considerations have caused major drug companies to opt out of antimicrobial development (2), leaving medical science without an adequate flow of new antibiotics. The drug discovery process obviously needs streamlining but not at the expense of drug testing, safety, and approval. Two studies in PNAS (3, 4) indicate how the antimicrobial pipeline might be fed with likely drug candidates that target the bacterial ribosome.
Ribosomes are essential catalytic structures in all living cells and carry out the task of synthesizing new proteins. Key structural differences between bacterial and human ribosomes make it possible to direct therapy against the pathogen, and consequently, a large proportion of clinically approved antibiotics target the bacterial ribosome (5). Antibiotic use over recent years has exerted a strong selective pressure on bacteria, and resistance to all of the ribosome-targeting antibiotics has arisen in one bacterial pathogen or another (6); some pathogens now have such a comprehensive collection of resistance mechanisms that they have become virtually untreatable with antibiotics. The present challenge is to design novel compounds that inhibit new targets in bacteria or that are capable of side-stepping resistance mechanisms to reuse the old targets, and for this, atomic resolution structures of the drug targets are indispensable tools.
The first atomic models of the ribosome structure were elucidated by X-ray crystallography about a decade ago (7). Since then, continual refinements have revealed the folding of the RNA and protein components within the two subunits of the ribosome and how they interact with each other, as well as with auxiliary factors and antimicrobials. A noteworthy milestone for this work was the award of the 2009 Nobel Prize in Chemistry (8) to three of four major researchers in the field. The history of ribosome research is, in fact, much older, stretching back to the era of microscopists, biochemists, and geneticists who determined the rough ribosome architecture, how antibiotics work, where they bind, and how resistance occurs (9). The rRNA was shown to be essential and the probable catalytic moiety in ribosomes (10), and antibiotics block protein synthesis by binding to specific rRNA nucleotides (11). Many actinomycete bacteria, including the erythromycin producer, avoid committing suicide during antibiotic production by protecting their own ribosomes through rRNA methylation (12), and this same mechanism has been picked up by many pathogenic bacteria (13). Methylation, or nucleotide mutation, can confer resistance to chemically dissimilar compounds, indicating that there is overlap between antibiotic sites (10, 12). The subsequent contribution of the crystallographers was to provide accurate tertiary structures that revealed the orientations and molecular contacts of antibiotics within these ribosomal binding sites.
Medical considerations would dictate a preference for structures of ribosomes from bacteria linked with diseases such as meningitis, septicemia, or tuberculosis; however, technical practicality led to the use of ribosomes that produce the best diffracting crystals. As a result, ribosomes were recruited from Deinococcus radiodurans (14), a radiation-resistant bacterium; Thermus thermophilus (15), a thermophilic bacterium that requires temperatures around 70 °C to grow optimally; and Haloarcula marismortui (16), an archaeon that is evolutionarily distant from bacteria as it is from humans. Despite the varied and nonpathogenic origins of these ribosomes, most of the antibiotic binding data have proved remarkably relevant and robust.
On delving deeper into the atomic details, however, there was a lack of consensus. The Deinococcus and Haloarcula structures proposed different conformations for the lactone ring of erythromycin and related macrolides (14, 16); the orientation of part of clindamycin (a lincosamide) was flipped by 180°, as was the entire chloramphenicol molecule, compared with an analogous inhibitor anisomycin. Furthermore, the crystallographic models disagreed, not only with each other but also with genetic and biochemical data (17), over the position of a pair of heterocyclic (imidazolo-pyridyl) substituents that had been added to erythromycin to form its latest clinically approved derivative, telithromycin (18). The question on all ribosomologists’ lips was as follows: were these discrepancies caused by fundamental species differences in the ribosomes or problems with the resolution and interpretation of the data? The answer seems to be that it was a bit of the former (4) but mainly the latter (3, 4).
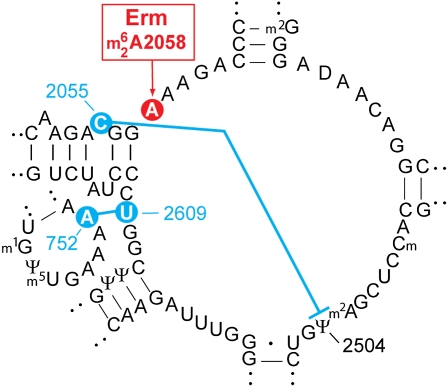
Several classes of antibiotics interact with key nucleotides at the bacterial peptidyl transferase center (the catalytic site of peptide bond formation) in the large subunit 23S rRNA. The most notable contact is at nucleotide 2058, which is conserved as an adenosine in all bacteria (Fig. 1). Methylation of A2058, or its substitution for a guanosine, sterically interferes with the binding of macrolide, lincosamide, and streptogramin B (MLSB) compounds, conferring collective resistance to these drugs. Eukaryotes, from yeast to humans, and archaea, to which Haloarcula belongs, all have a guanosine at 2058 and thus are naturally resistant to MLSB drugs. Remarkably, some of the larger macrolides, such as tylosin and carbomycin, at high concentration could be bound to Haloarcula ribosomes, giving clear changes in the electron density (16). Substituting an adenosine at 2058 in the Haloarcula rRNA greatly improved binding and enabled study of the smaller macrolide erythromycin (19). However, a single mutation does not a bacterial ribosome make; this was underlined by yeast ribosomes, which still do not bind erythromycin after changing their G2058 to an A (20). Therefore, despite the high quality of the Haloarcula structures, reservations remained about whether the results reflected the true state of affairs on the bacterial ribosome.
Fig. 1.
Secondary structure of the peptidyl transferase center in E. coli 23S rRNA. The site of erythromycin resistance methylation at A2058 is shown in red. Key nucleotide interactions (blue) that support antibiotic binding are the same in T. thermophilus but different in H. halobium rRNA. D. radiodurans rRNA has C752 (21) and thus lacks the 752–2609 bp onto which the telithromycin heterocycles would stack.
These antibiotic interactions have now been revisited on ribosomes from Thermus (3) and Escherichia coli (4), the latter organism being a bona fide pathogen. These two independent studies are in excellent agreement with each other and vindicate the earlier Haloarcula data concerning the conformation of the macrolide lactone ring and the orientations of chloramphenicol/anisomycin and clindamycin in their binding sites. This is not the same as saying that Haloarcula is an optimal model for antibiotic binding—the rRNA sequence variations (Fig. 1) make a difference in clindamycin's nucleotide contacts, displace chloramphenicol from its authentic site, and determine where the telithromycin heterocycles come to rest.
These are, of course, minor details. However, it is precisely the minor details that need to be understood when designing compounds that slot specifically and with high affinity into niches that are unique to the bacterial ribosome. These latest datasets now invite an inward-directed search for new compounds to block ribosomal sites and discover that, right there, something interposable is waiting to be conceived.
Acknowledgments
My research on rRNA, antimicrobial interaction, and resistance is funded by the Danish Research Agency (FNU-rammebevilling 272-07-0613) and the Danish Grundforskningsfond.
Footnotes
References
- 1.Schönfeld W, Kirst HA, editors. Macrolide Antibiotics. Basel: Birkhauser; 2002. [Google Scholar]
- 2.Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6:427–430. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Bulkley D, et al. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunkle JA, et al. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 6.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonath A. Antibiotics targeting ribosomes: Resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 8.Nierhaus KH. Nobel Prize for the elucidation of ribosome structure and insight into the translation mechanism. Angew Chem Int Ed Engl. 2009;48:9225–9228. doi: 10.1002/anie.200905795. [DOI] [PubMed] [Google Scholar]
- 9.Hill WE, et al., editors. The Ribosome: Structure, Function, and Evolution. Washington, DC: American Society for Microbiology; 1990. [Google Scholar]
- 10.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 11.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 12.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 13.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 15.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 16.Hansen JL, et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 17.Xiong L, Shah S, Mauvais P, Mankin AS. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol Microbiol. 1999;31:633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 18.Bryskier AJ, Denis A. Ketolides: Novel antibacterial agents designed to overcome resistance to erythromycin A within gram-positive cocci. In: Schönfeld W, Kirst HA, editors. Macrolide Antibiotics. Basel: Birkhauser; 2002. pp. 97–140. [Google Scholar]
- 19.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Bommakanti AS, Lindahl L, Zengel JM. Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Sacchromyces cerevisae. RNA. 2008;14:460–464. doi: 10.1261/rna.786408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannone JJ, et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]