Abstract
The serpinopathies result from the ordered polymerization of mutants of members of the serine proteinase inhibitor (serpin) superfamily. These polymers are retained within the cell of synthesis where they cause a toxic gain of function. The serpinopathies are exemplified by inclusions that form with the common severe Z mutant of α1-antitrypsin that are associated with liver cirrhosis. There is considerable controversy as to the pathway of serpin polymerization and the structure of pathogenic polymers that cause disease. We have used synthetic peptides, limited proteolysis, monoclonal antibodies, and ion mobility-mass spectrometry to characterize the polymerogenic intermediate and pathological polymers formed by Z α1-antitrypsin. Our data are best explained by a model in which polymers form through a single intermediate and with a reactive center loop-β-sheet A linkage. Our data are not compatible with the recent model in which polymers are linked by a β-hairpin of the reactive center loop and strand 5A. Understanding the structure of the serpin polymer is essential for rational drug design strategies that aim to block polymerization and so treat α1-antitrypsin deficiency and the serpinopathies.
Keywords: alpha-1-antitrypsin deficiency, protein folding, serpins, polymers, cirrhosis
An increasing number of disorders are recognized to result from the self-association and tissue deposition of misfolded proteins. These conformational diseases include Alzheimer, Huntington, and Parkinson disease as well as the amyloidoses and serpinopathies. The serpinopathies are characterized by the aggregation of members of the serine proteinase inhibitor or serpin superfamily of proteins (1). These include mutations in neuroserpin that cause the dementia FENIB and mutations in antithrombin, C1-inhibitor, and α1-antichymotrypsin associated with thrombosis, angio-oedema, and emphysema respectively. The best characterized of the serpinopathies results from the severe Z deficiency allele of α1-antitrypsin that is found in 4% of the Northern European Caucasian population. In homozygotes this mutation results in the retention of 85%–90% of synthesized α1-antitrypsin within the endoplasmic reticulum of hepatocytes, where it is either targeted for degradation or sequestered as ordered polymers. The retained protein causes hepatocellular damage and liver disease (2) while the lack of circulating α1-antitrypsin predisposes the Z homozygote to early onset emphysema (1).
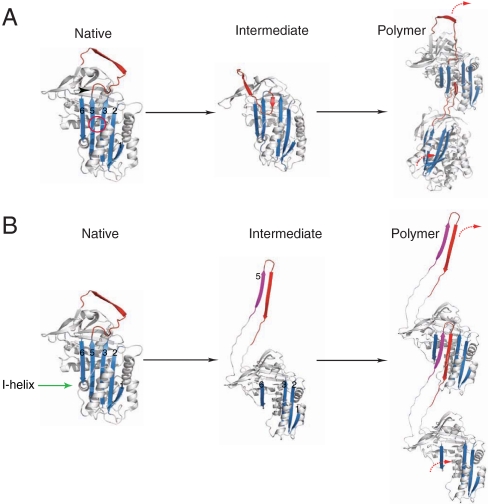
The structure of α1-antitrypsin is based on a central β-sheet A and a mobile reactive center loop (Fig. 1). The classical pathway of serpin polymerization suggests that the Z mutation of α1-antitrypsin distorts the relationship between the reactive center loop and β-sheet A. This perturbation in structure allows the formation of an unstable intermediate (M∗) and a sequential β-strand linkage between the reactive center loop of one molecule and β-sheet A of another (Fig. 1A) (3–8). However this model has recently been challenged by the crystal structure of a self-terminating dimer of another serpin, antithrombin (9). The closed dimer revealed a β-hairpin domain swap containing not only the reactive center loop but also strand 5 of β-sheet A (strand 5A; Fig. 1B).
Fig. 1.
The two proposed pathways of serpin polymerization. (A) The classical pathway of polymerization. The Z mutation (Glu342Lys-arrow head) or shutter domain mutations (red circle) destabilize β-sheet A (blue) to form an activated intermediate species (M*). Intermolecular linkage occurs via donation of the reactive loop (red) from one molecule to the open lower portion of the central β-sheet A channel of a second molecule. (B) The alternative β-hairpin pathway. The formation of an intermediate requires unraveling of the helix I (green arrow) and the extrusion of strand 5A (purple) from β-sheet A to generate a donor interface with the reactive center loop. Linkage occurs by the insertion of this hairpin domain into β-sheet A of a similarly activated molecule. The strands of β-sheet A are labeled.
We have used a range of techniques to evaluate these models of polymerization.
Results
Inhibition of α1-antitrypsin Polymerization by Strand 5A Peptides.
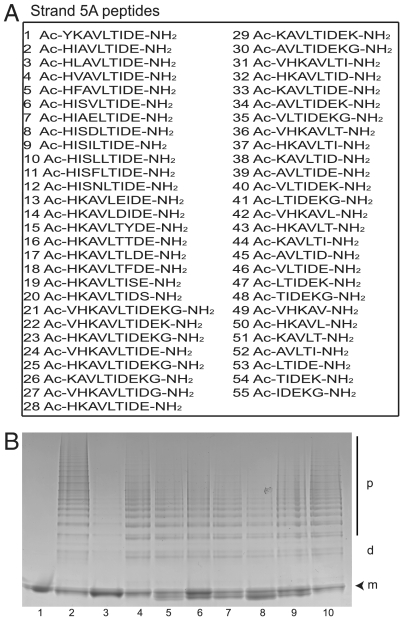
Synthetic peptides with homology to the reactive center loop block and reverse the polymerization of Z α1-antitrypsin (3, 10, 11). An extension of the new β-hairpin model of polymerization is that polymer formation should also be blocked by peptides that include strand 5A. Heat-induced polymers of Z α1-antitrypsin share an epitope with polymers formed in vivo (12). We assessed 55 peptides of 5–12 amino acid length based on the strand 5A sequence of α1-antitrypsin for their ability to block polymerization (Fig. 2A). Only the reactive loop tetrapeptide TTAI (11) blocked polymerization (Fig. 2B). Some s5A peptides formed a small amount of binary complex that migrated anodal to native α1-antitrypsin, however none was able to block the polymerization of Z α1-antitrypsin (Fig. 2B) or the wildtype (M) α1-antitrypsin.
Fig. 2.
Assessing the ability of strand 5A peptide analogues to inhibit the heat-induced polymerization of Z α1-antitrypsin. (A) The sequences of the 55 strand 5A analogue peptides assessed for their ability to block polymerization. The strand 5A sequence is KAVHKAVLTIDE. (B) 7.5% (w/v) acrylamide nondenaturing PAGE to evaluate the ability of strand 5A peptides to block the polymerization of Z α1-antitrypsin. The peptides were incubated at a molar ratio of 100∶1 with monomeric Z α1-antitrypsin (0.2 mg/mL) in PBS at 37 °C for 2 d. The temperature of the mixture was then raised to 41 °C for 5 d to form polymers. Lane 1: monomeric Z α1-antitrypsin; lane 2: control Z α1-antitrypsin polymers; lane 3: Z α1-antitrypsin incubated with the reactive loop blocking peptide TTAI; lanes 4–10: Z α1-antitrypsin incubated with strand 5A peptides (peptides 1–7 from A). The monomer (m), dimer (d) and polymer (p) are indicated. Each lane contains 4 μg of protein.
Limited Proteolysis of α1-antitrypsin Polymers.
The role of strand 5A in forming the intermolecular linkage of the polymer chain was further assessed by limited proteolysis of Z α1-antitrypsin polymers. The β-hairpin model proposes that helix I must unfold in order to release strand 5A and allow the formation of polymers. Indeed the exposure of cryptic sites in the helix I in polymers of wildtype M α1-antitrypsin was used to provide support for the β-hairpin model of polymerization (9).
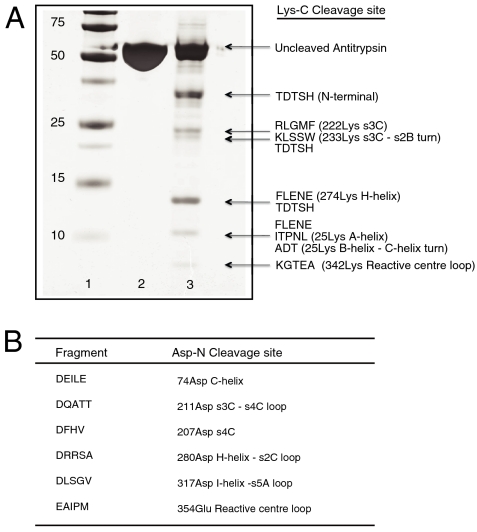
The endopeptidases Lys-C and Asp-N were used for the enzymatic digestion of heat-induced Z α1-antitrypsin polymers. These enzymes were selected as they have cleavage sites within the folded helix I that will only be exposed if the helix is unwound. The cleavage products were separated by SDS-PAGE (Fig. 3A) before blotting onto PVDF membranes for N-terminal sequencing. The Lys-C cleavage data were confirmed by separation of the products by reverse phase HPLC prior to N-terminal sequencing. All the cleavage sites in Lys-C digested polymeric Z α1-antitrypsin were in exposed regions in the crystal structure of monomeric α1-antitrypsin (13), except for cleavage at 222Lys in strand 3C, 274Lys in helix H, and 310Lys in the single-turn adjacent to helix I. There was no evidence of cleavage at 300Lys within helix I. The fragment pattern obtained on Lys-C digestion of the monomer was virtually identical to that obtained by digesting polymeric α1-antitrypsin, except for the lack of cleavage at 274Lys.
Fig. 3.
Endopeptidase cleavage of polymeric Z α1-antitrypsin. (A) 12% (w/v) SDS-PAGE to show the cleavage products of Lys-C digestion of polymeric Z α1-antitrypsin and their secondary structural location in the monomer. Lane 1: molecular mass markers (kDa); lane 2: undigested Z α1-antitrypsin; lane 3: Z α1-antitrypsin polymers incubated overnight with Lys-C. The gel was blotted onto PVDF and subjected to N-terminal sequencing. Longer incubation times did not change the cleavage profile. (B) Secondary structural location of cleavage fragments following Asp-N digestion of Z α1-antitrypsin polymers.
The endopeptidase cleavage profile was then repeated using polymers isolated from hepatocytes of an individual with Z α1-antitrypsin associated liver disease (14). There was no cleavage at 300Lys, which should be available if the helix I is unfolded. Limited proteolysis experiments were also undertaken using Asp-N to digest polymeric Z α1-antitrypsin. The cleavage sites are shown in Fig. 3B. There was no cleavage at 298Asp at the junction between strand 6A and helix I.
Conformational Analysis of Polymers Using Monoclonal Antibodies.
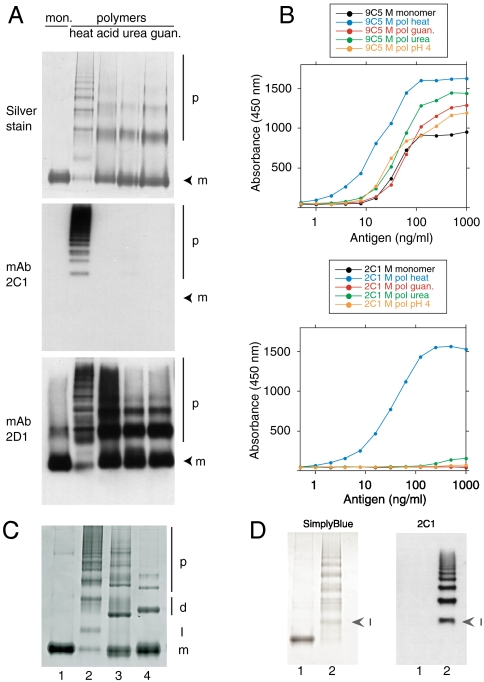
Polymers of α1-antitrypsin have been prepared under a variety of conditions but it is unclear which method best reproduces the polymers that form in vivo. The unique 2C1 mAb that recognizes polymers of α1-antitrypsin that cause disease (12) was used to assess α1-antitrypsin polymers formed under a range of conditions in vitro. M α1-antitrypsin polymers were prepared by heating, by incubation at low pH and in 4M urea, or 3M guanidine. They were separated by nondenaturing PAGE and probed by Western blot analysis (Fig. 4A). The mAb 2D1 detected all α1-antitrypsin conformers seen by silver staining while mAb 2C1 only recognized polymers formed by heating. The same results were obtained by sandwich ELISA using mAb 9C5 to detect all α1-antitrypsin conformers and mAb 2C1 to detect polymers (Fig. 4B). These results show that only heating of the monomeric protein produces polymers that share an epitope with those found in human disease.
Fig. 4.
Monoclonal antibody 2C1 recognizes polymers of α1-antitrypsin prepared by heating but not by other denaturing conditions. (A) 7.5% (w/v) nondenaturing PAGE to show polymer analyzed by silver stain (upper) or Western blot, first with mAb 2C1 (center) and then with mAb 2D1 (bottom) on the same membrane. mAb 2C1 recognized heat-induced polymers but gave no signal for polymers prepared by treating α1-antitrypsin at low pH, or with 1–3 M guanidine or 1–4 M urea. The monomer (m) and polymer (p) are indicated. (B) The same polymers assayed in sandwich ELISA, using either mAb 9C5 (upper graph) or 2C1 (lower graph) as the detecting antibodies. mAb 9C5 detected all species with high affinity but mAb 2C1 detected only heat-induced polymers. (C) 7.5% (w/v) nondenaturing PAGE. Lane 1: M α1-antitrypsin monomer; lane 2: heat-induced α1-antitrypsin polymer; lane 3: antithrombin hexapeptide-induced polymer of α1-antitrypsin; lane 4: P9-P10 reactive center loop-cleaved polymer of α1-antitrypsin. The monomer (m), dimer (d), polymer (p) and intermediate state (I) are indicated. The intermediate in lane 2 migrates between the monomeric and dimeric states of the non heat-induced polymers. (D) 7.5% (w/v) nondenaturing PAGE of Z α1-antitrypsin monomer (lane 1) and polymer (lane 2) visualized by SimplyBlue™ (left) or following Western blot analysis with mAb 2C1 (right). Arrowheads indicate the intermediate (I) in the polymer lanes. The 2C1 shows no signal for the monomer but recognizes the intermediate and higher order polymers.
There is no crystal structure of the pathological α1-antitrypsin polymer, however there are structures of polymers linked by a cleaved reactive center loop (15, 16). Polymers are also formed by incubating α1-antitrypsin with the antithrombin hexapeptide SEAAAS, which anneals to the upper portion of β-sheet A and allows the incorporation of an exogenous reactive loop into the lower part of β-sheet A (10). These polymers were used to provide structurally defined comparisons with polymers formed by heating. Analysis by nondenaturing PAGE demonstrated an additional band in the heat-induced polymers when compared to the hexapeptide-induced and the reactive loop-cleaved α1-antitrypsin polymers (Fig. 4C). Gel filtration chromatography indicated that this species was monomeric and its intensity waned with that of residual monomer at the concluding stages of polymerization. The mAb 2C1 recognized this monomeric intermediate as readily as the polymerized protein in Western blot analysis (Fig. 4D). It did not recognize monomeric Z α1-antitrypsin suggesting the intermediate shares a conformational epitope with the monomeric subunits in the polymer chain that are absent in the native protein.
Structural Analysis of the Polymer by Mass Spectrometry.
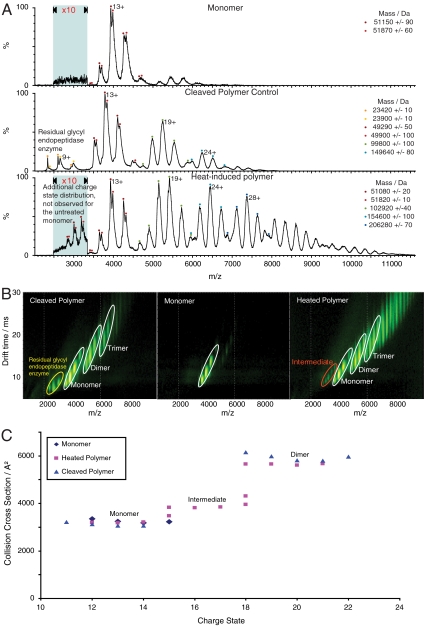
Mass spectrometry was used to further characterize the intermediate and the polymers of α1-antitrypsin. The mass spectra of monomeric, cleaved polymer, and heat-induced polymer of α1-antitrypsin are shown in Fig. 5A. A distribution of peaks from 12+ to 15+ was observed for monomeric M α1-antitrypsin. The splitting of the peaks observed for each charge state is due to the microheterogeneity at the N terminus of α1-antitrypsin (17). The reactive loop-cleaved spectrum contains four charge state distributions that are in excellent agreement with the species observed on nondenaturing PAGE (Fig. 4C, lane 4). The series of peaks at 2,250–3,000 m/z centering on the 9+ ion correspond to the enzyme glycyl endopeptidase (23 kDa) that was used to cleave the reactive center loop. Residual unpolymerized cleaved monomer has a similar charge distribution to the untreated monomer centered on the 13+ ion. The distribution of peaks around 5,250 m/z corresponds to the cleaved polymer dimer (100 kDa) and that around 6,250 m/z to the trimer (150 kDa). The heat-induced polymer spectrum has five distinct charge state distributions corresponding to five different species. The charge state distributions were comparable to those found for the cleaved polymer ladder except for the unique series of peaks at 2,775–3,250 m/z. These low intensity charge states (15+ to 18+) correspond to the mass of the monomer but have increased charge relative to the monomer distribution that is observed for all three samples. These peaks are consistent with the intermediate species observed on nondenaturing PAGE. Similar data were obtained by real time mass spectral analysis of M and Z α1-antitrypsin polymerization within temperature regulated electrospray capillaries. The polymerogenic intermediate of Z α1-antitrypsin predictably formed at an earlier time point than that of M α1-antitrypsin and its appearance was coincident with the formation of the higher order polymers.
Fig. 5.
α1-antitrypsin polymers studied by IM-MS. (A) Mass spectra of M α1-antitrypsin monomer, the heat-induced polymer, and P9-P10 reactive loop-cleaved polymer. The mass spectrum of the monomer has a charge state distribution centered on the 13+ ion (upper spectrum). The mass spectrum of the reactive loop-cleaved polymer has four charge state distributions (middle spectrum). The peaks at low m/z are attributed to residual enzyme and the series of peaks at 4,000 and 5,250 and 6,250 m/z represent the cleaved monomer, dimer and trimer respectively. Similar spectra were observed for the heat-induced sample (lower spectrum). However, this spectrum exhibits a second monomer population with higher charge states (15+ to 18+), which are consistent with the polymerogenic intermediate. (B) Ion mobility drift times observed for the M α1-antitrypsin monomer, the heat-induced polymer, and reactive loop-cleaved polymer (corresponding mass spectra shown in A). The monomer and polymer species are labeled and highlighted with white ellipses, the intermediate with a red ellipse, and the cleavage enzyme with a yellow ellipse. (C) Drift times from (B) converted into a collision cross sectional plot. The monomer collision cross sections for all the samples are almost identical and similarly the collision cross sections for the cleaved polymer and heat-induced polymer dimers also overlap. The intermediate in the heat-induced polymer sample shows an expanded collision cross section relative to the monomer and maintains the same electrophoretic relationship with the dimer as that observed on nondenaturing PAGE (Fig. 4C).
Structural Analysis of the Polymer by Ion Mobility Spectrometry.
IM-MS provides multidimensional separation of gas phase protein ions and has only recently been applied to large noncovalent protein complexes (18–20). The drift times of protein ions in a gas filled drift tube under the influence of a weak electric field can be converted into collision cross sections that depend directly on their size and geometry (21–23). More compact ions transverse the drift tube faster than elongated ions for a given mass and charge state, thus allowing a measure of the overall topology and conformation of a polymer ion in the gas phase and from these values the intermolecular distances for the components of the polymer chain can be estimated.
Fig. 5B shows the ion mobility spectra of the monomer, and both reactive loop-cleaved and heat-induced polymers. A monomeric intermediate, whose collision cross section is significantly larger than those observed for the other monomers, was again observed for the heat-induced polymers. The collision cross section plot (Fig. 5C) showed a striking resemblance to the nondenaturing PAGE of heat-induced α1-antitrypsin polymers, with the intermediate species eluting between the monomer and the dimer.
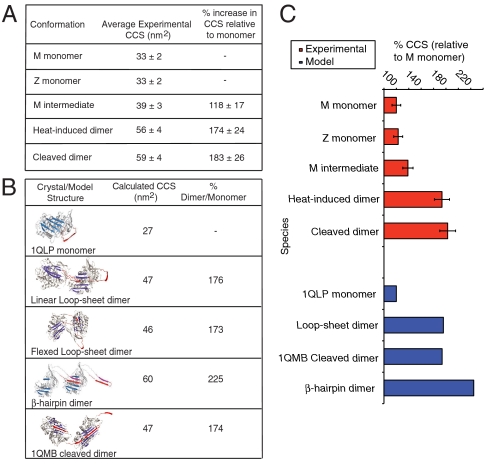
The theoretical collision cross sections for the α1-antitrypsin monomer 1QLP (13) and the cleaved dimer 1QMB (15) were calculated for comparison with the experimental data. The model for the β-hairpin dimer of α1-antitrypsin was generated using PyMOL and the domain swapped antithrombin dimer 2ZNH (9) as a template. The disparity between the experimental collision cross sections (Fig. 6A) and the calculated collision cross sections (Fig. 6B) is due to the three N-linked oligosaccharides that are not seen in the X-ray structures of α1-antitrypsin. However, a measure of the relative intermolecular arrangement and distance was obtained by the calculation of the percentage increase in collision cross section in the dimer relative to the monomer. The experimental data show that both the heat-induced and glycyl endopeptidase cleaved dimers display a similar increase in collision cross section when compared with the monomer (Fig. 6C). These data are indicative of comparable intermolecular separation. The percentage increase in the theoretical collision cross section for the cleaved dimer crystal structure is in good agreement with the experimental percentage increase in collision cross section for the glycyl endopeptidase cleaved dimer. Significantly the loop-sheet dimer model rather than the β-hairpin dimer model gave values consistent with the experimental increase in collision cross section.
Fig. 6.
Comparison of experimental and calculated collision cross sections (CCS) for models of α1-antitrypsin polymers. (A) Experimental collision cross sections for α1-antitrypsin monomers, the heat-induced and reactive loop-cleaved M α1-antitrypsin dimers. The dimer collision cross sections are indistinguishable. (B) Table of calculated collision cross section measurements for the X-ray crystallographic structures of monomeric α1-antitrypsin (1QLP), the cleaved dimer (1QMB), and the three-dimensional dimer models for the loop-sheet and β-hairpin pathways. The flexed loop-sheet, linear loop-sheet dimers and 1QMB cleaved dimers collision cross sections are very similar, whereas the β-hairpin dimer has a significantly larger collision cross section. (C) Plot comparing the percentage increases in collision cross section upon dimer formation for experimental (red) and calculated (blue) data. The relationship between the experimental heat-induced and reactive loop-cleaved dimer collision cross sections are consistent with a loop-sheet linkage and the crystallographic structure of cleaved dimers (1QMB). The calculated collision cross section of the β-hairpin dimer model is significantly larger than that observed experimentally for dimers formed by heating α1-antitrypsin.
Discussion
Point mutations can cause serpins to be retained as ordered polymers and lead to disease by a toxic gain of function in the cell of synthesis and a loss of function due to a lack of secreted protein (1). The structure of the pathological polymers was believed to result from the sequential linkage between the reactive center loop of one molecule and β-sheet A of another (3). However, this pathway was recently challenged by a model in which polymers are formed by a β-hairpin linkage containing the reactive center loop and strand 5A (9).
The new model proposes that polymers form while the protein is folding rather than from an intermediate with a near native structure. This model is difficult to reconcile with the spontaneous polymerization of folded mutant serpins under physiological conditions (3), the presence of polymers in tissues such as the skin from which they are not secreted (24), and in vivo polymer formation from wildtype protein (25). The exposed hydrophobic β-hairpin of the intermediate (Fig. 1B) would predictably activate the unfolded protein response. However the unfolded protein response is characteristically absent during the intracellular accumulation of serpin polymers (26–28). It is clearly critical to define the intermolecular linkage in order to develop strategies to block serpin polymerization and so treat the serpinopathies.
Previous in vitro and crystallographic studies have shown that exogenous peptides with homology to the reactive center loop can anneal to β-sheet A and so block polymerization (3, 10, 11, 29). The β-hairpin model would suggest that a similar blocking mechanism should be achieved by peptides based on strand 5A. Five-twelve mer peptides based on strand 5A of α1-antitrypsin interacted weakly, or not at all, with α1-antitrypsin and none was able to block the polymerization of either wildtype or Z α1-antitrypsin. This finding is inconsistent with a model of polymerization in which intermolecular insertion of strand 5A is critical.
The linkage in the serpin polymer was then assessed by limited proteolysis aimed at assessing the availability of cleavage sites in helix I. Helix I must unravel if linear and flexible polymers are formed by a reactive center loop/strand 5A-β-sheet A linkage, while helix I plays no role in polymers formed by a reactive center loop-β-sheet A interaction. Endopeptidase cleavage data showed that helix I remained protected from digestion in monomeric Z α1-antitrypsin, in polymers of Z α1-antitrypsin formed under physiological conditions in vitro and in polymers isolated from the endoplasmic reticulum of hepatocytes of a Z α1-antitrypsin homozygote. These data are consistent with hydrogen/deuterium exchange studies that showed no change in the helix I of α1-antitrypsin upon formation of the polymerogenic intermediate or chains of polymers (30).
Polymers of α1-antitrypsin can be prepared by a range of chaotropic conditions. The 2C1 polymer specific mAb demonstrated that only heating, and not low pH or denaturants, results in polymers that share a neoepitope that defines polymers formed in disease. Studies of heat-induced polymerization in vitro are therefore the most applicable to pathological polymerization. This finding questions the validity of the biochemical data that was used to support the β-hairpin model of polymerization as this was based on α1-antitrypsin conformers formed in the presence of guanidine (9). The heat-induced polymer shares the epitope recognized by mAb 2C1 with the polymerogenic intermediate that was not found in monomeric α1-antitrypsin.
IM-MS data were used to unambiguously assign the intermediate state to a monomer with an enlarged surface area relative to the native monomer. The percentage increase in collision cross section from the native monomer to this intermediate is of the same magnitude as that observed by Stokes radii using hydrogen-deuterium exchange (31), although these unit measures are not interchangeable. The experimental collision cross section data for the heat-induced and reactive center loop-cleaved dimers suggested comparable topographies that were in excellent agreement with calculated collision cross sections for the loop-sheet modeled dimer and the structure of the cleaved dimer 1QMB (15). However, the theoretical collision cross section for the β-hairpin α1-antitrypsin dimer model based on the 2ZNH template (9) is significantly larger than the collision cross sectional area of the heat-induced α1-antitrypsin dimer. These findings imply that the α1-antitrypsin polymer shares the short single strand of the cleaved dimer rather than the more extended and unfolded linker domain of the β-hairpin polymer. Moreover, the identical charge states of the reactive center loop-cleaved and heat-induced polymer species suggest that they share similar surface topologies. These data argue against the extensive structural unravelling proposed for the β-hairpin of the reactive center loop/strand 5A linkage, as this would reveal more residues for protonation.
In summary the single strand reactive loop-β-sheet A interaction best explains the data from exogenous reactive loop peptides, limited proteolysis, monoclonal antibodies, and mass spectrometry with α1-antitrypsin. These findings are in keeping with recent biophysical studies assessing the polymerization of another serpin, neuroserpin (32). It is likely that the fine details of the polymer linkage will vary with different mutations and in different serpins. Nevertheless the single strand linkage of serpin polymers should be used as the model to develop strategies for blocking polymerization in order to treat the serpinopathies.
Experimental Methods
Preparation of α1-antitrypsin Conformers.
Polymers were prepared by heating α1-antitrypsin (17) at 5 μM and 60 °C in 40 mM Tris, pH 8.0. Polymers for IM-MS were formed in 20 mM ammonium acetate buffer. P9-P10 reactive loop-cleaved α1-antitrypsin polymers were prepared by incubating the protein with glycyl endopeptidase (B.S.C Biochemical) at 37 °C for 4 h in 20 mM ammonium acetate buffer. The antithrombin hexapeptide-induced polymers were formed by incubating α1-antitrypsin with the SEAAAS hexapeptide (molar ratio α1-antitrypsin:peptide of 1∶10) in 40 mM Tris, pH 8.0 overnight at 37 °C. Acid pH polymerization of M α1-antitrypsin was performed by incubating M α1-antitrypsin in 0.1 M sodium acetate pH 4.5 at 25 °C for 24 h. Urea polymers were prepared by incubating M α1-antitrypsin in 1–4 M urea (buffered by 40 mM Tris-HCl pH 8) at 25 °C for 24 h. Guanidine polymers were prepared by incubating the protein in 1–3 M guanidine hydrochloride (buffered by 40 mM Tris-HCl pH 8) at 25 °C for 24 h. Pathogenic Z α1-antitrypsin polymers were isolated as described previously (14). All polymers were confirmed by 7.5% (w/v) acrylamide nondenaturing PAGE.
Endopeptidase Cleavage.
Residual monomer was removed from polymers formed at 41 °C by gel filtration (Superdex™ 200 GE healthcare) before overnight incubation with Lys-C or Asp-N in 40 mM Tris, pH 8.0 at molar ratios of 200∶1–10∶1 at 20 °C. Digestion fragments were either separated by 10 % (w/v) acrylamide SDS-PAGE before transfer onto a PVDF membrane for sequencing, or separated by reverse phase chromatography. After lyophilization, all peptide peaks were analyzed by N-terminal sequencing at the Department of Biochemistry, University of Cambridge.
Peptide Blockade of α1-antitrypsin Polymerization.
Strand 5A peptides and the TTAI peptide were kind gifts from Amicus Therapeutics Inc. and GlaxoSmithKline (Stevenage) respectively. Peptide blockade of α1-antitrypsin polymerization was assessed by incubation at a molar ratio of 100∶1 with monomeric Z α1-antitrypsin (0.2 mg/mL) in PBS at 37 °C for 2 d. The temperature of the mixture was then raised to 41 °C for 5 d to form polymers. Peptides were dissolved in DMSO (final concentration 0.38% v/v) and polymer formation assessed by 7.5% (w/v) nondenaturing PAGE.
Ion Mobility-Mass Spectrometry.
IM-MS experiments were performed on a Synapt HDMS quadrupole-ion trap-IM-MS instrument (Waters). Nanoflow electrospray capillaries were prepared with a nanoESI source as previously described (33). The collision cross sections acquired from fixed wave height data were calibrated externally (18). The error in the collision cross section measurements is 7%. Spectra were analyzed using MassLynx V4.1 and DriftScope V2.1 (Waters).
Acknowledgments.
We thank Dr. Aiwu Zhou (Department of Haematology, University of Cambridge), Dr. Helena Hernández, and Dr. Alan Sandercock (Department of Chemistry, University of Cambridge) for helpful advice. This work was supported by the Medical Research Council (United Kingdom), the Engineering and Physical Sciences Research Council, Papworth National Health Service Trust and the Royal Society. U.I.E. is an MRC Clinical Research Training Fellow. J.F. is an Engineering and Physical Sciences Research Council/Royal Society of Chemistry student and B.G. is a Wellcome Trust Intermediate Clinical Fellow. J.P. is supported by the Ministerio de Educación y Ciencia (Grant PR2007-0018 and BFU 2006 11754) and the Junta de Andalucía (Grant P07 CVI 03079). M.F.B is a Waters Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gooptu B, Lomas DA. Conformational pathology of the serpins: themes, variations and therapeutic strategies. Annu Rev Biochem. 2009;78:147–176. doi: 10.1146/annurev.biochem.78.082107.133320. [DOI] [PubMed] [Google Scholar]
- 2.Sveger T. The natural history of liver disease in α1-antitrypsin deficient children. Acta Paediatr Scand. 1988;77:847–851. doi: 10.1111/j.1651-2227.1988.tb10767.x. [DOI] [PubMed] [Google Scholar]
- 3.Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z α1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 4.James EL, Bottomley SP. The mechanism of α1-antitrypsin polymerization probed by fluorescence spectroscopy. Arch Biochem Biophys. 1998;356:296–300. doi: 10.1006/abbi.1998.0751. [DOI] [PubMed] [Google Scholar]
- 5.Dafforn TR, Mahadeva R, Elliott PR, Sivasothy P, Lomas DA. A kinetic mechanism for the polymerization of α1-antitrypsin. J Biol Chem. 1999;274:9548–9555. doi: 10.1074/jbc.274.14.9548. [DOI] [PubMed] [Google Scholar]
- 6.Gooptu B, et al. Inactive conformation of the serpin α1-antichymotrypsin indicates two stage insertion of the reactive loop; implications for inhibitory function and conformational disease. Proc Natl Acad Sci USA. 2000;97:67–72. doi: 10.1073/pnas.97.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahadeva R, Dafforn TR, Carrell RW, Lomas DA. 6-mer peptide selectively anneals to a pathogenic serpin conformation and blocks polymerization: implications for the prevention of Z α1-antitrypsin related cirrhosis. J Biol Chem. 2002;277:6771–6774. doi: 10.1074/jbc.C100722200. [DOI] [PubMed] [Google Scholar]
- 8.Purkayastha P, et al. α1-antitrypsin polymerization: a fluorescence correlation spectroscopic study. Biochemistry. 2005;44:2642–2649. doi: 10.1021/bi048662e. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 10.Zhou A, et al. How small peptides block and reverse serpin polymerization. J Mol Biol. 2004;342:931–941. doi: 10.1016/j.jmb.2004.07.078. [DOI] [PubMed] [Google Scholar]
- 11.Chang YP, Mahadeva R, Chang WS, Lin SC, Chu YH. Small molecule peptides inhibit Z α1-antitrypsin polymerization. J Cell Mol Med. 2008;13:2304–2316. doi: 10.1111/j.1582-4934.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda E, et al. Conformational specificity of polymers of α1-antitrypsin that are associated with liver disease. Hepatology. 2010;52:1078–1088. doi: 10.1002/hep.23760. [DOI] [PubMed] [Google Scholar]
- 13.Elliott PR, Pei XY, Dafforn TR, Lomas DA. Topography of a 2.0 Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000;9:1274–1281. doi: 10.1110/ps.9.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An JK, Blomenkamp K, Lindblad D, Teckman JH. Quantitative isolation of α1-antitrypsin mutant Z protein polymers from human and mouse livers and the effect of heat. Hepatology. 2005;41:160–167. doi: 10.1002/hep.20508. [DOI] [PubMed] [Google Scholar]
- 15.Huntington JA, et al. A 2.6 Å structure of a serpin polymer and implication for conformational disease. J Mol Biol. 1999;293:449–455. doi: 10.1006/jmbi.1999.3184. [DOI] [PubMed] [Google Scholar]
- 16.Dunstone MA, et al. Cleaved antitrypsin polymers at atomic resolution. Protein Sci. 2000;9:417–420. doi: 10.1110/ps.9.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lomas DA, Evans DL, Stone SR, Chang WS, Carrell RW. Effect of the Z mutation on the physical and inhibitory properties of α1-antitrypsin. Biochemistry. 1993;32:500–508. doi: 10.1021/bi00053a014. [DOI] [PubMed] [Google Scholar]
- 18.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 19.Pukala TL, et al. Subunit architecture of multiprotein assemblies determined using restraints from gas-phase measurements. Structure. 2009;17:1235–1243. doi: 10.1016/j.str.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Scarff CA, Patel VJ, Thalassinos K, Scrivens JH. Probing hemoglobin structure by means of travelling-wave ion mobility mass spectrometry. J Am Soc Mass Spectr. 2009;20:625–631. doi: 10.1016/j.jasms.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 21.von Helden G, Wyttenbach T, Bowers MT. Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science. 1995;267:1483–1485. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 22.Jarrold MF. Peptides and proteins in the vapor phase. Annu Rev Phys Chem. 2000;51:179–207. doi: 10.1146/annurev.physchem.51.1.179. [DOI] [PubMed] [Google Scholar]
- 23.Smith DP, et al. Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur J Mass Spectrom. 2009;15:113–130. doi: 10.1255/ejms.947. [DOI] [PubMed] [Google Scholar]
- 24.Gross B, et al. New finding in PiZZ α1-antitrypsin deficiency-related panniculitis. Demonstration of skin polymers and high dosing requirements of intravenous augmentation therapy. Dermatology. 2009;218:370–375. doi: 10.1159/000202982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagi R, et al. Novel serpinopathy in rat kidney and pancreas induced by overexpression of medsin. J Am Soc Nephrol. 2005;16:1339–1349. doi: 10.1681/ASN.2004070600. [DOI] [PubMed] [Google Scholar]
- 26.Graham KS, Le A, Sifers RN. Accumulation of the insoluble PiZ variant of human α1-antitrypsin within the hepatic endoplasmic reticulum does not elevate the steady-state level of grp78/BiP. J Biol Chem. 1990;265:20463–20468. [PubMed] [Google Scholar]
- 27.Hidvegi T, Schmidt BZ, Hale P, Perlmutter DH. Accumulation of mutant α1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB and BAP31 but not the unfolded protein response. J Biol Chem. 2005;280:39002–39015. doi: 10.1074/jbc.M508652200. [DOI] [PubMed] [Google Scholar]
- 28.Davies MJ, et al. Neuroserpin polymers activate NF-kappaB by a calcium signalling pathway that is independent of the unfolded protein response. J Biol Chem. 2009;284:18202–18209. doi: 10.1074/jbc.M109.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze AJ, et al. Structural transition of α1-antitrypsin by a peptide sequentially similar to β-strand s4A. Eur J Biochem. 1990;194:51–56. doi: 10.1111/j.1432-1033.1990.tb19425.x. [DOI] [PubMed] [Google Scholar]
- 30.Tsutsui Y, Kuri B, Sengupta T, Wintrode PL. The structural basis of serpin polymerization studied by hydrogen/deuterium exchange and mass spectrometry. J Biol Chem. 2008;283:30804–30811. doi: 10.1074/jbc.M804048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsutsui Y, Wintrode PL. Cooperative unfolding of a metastable serpin to a molten globule suggests a link between functional and folding energy landscapes. J Mol Biol. 2007;371:245–255. doi: 10.1016/j.jmb.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 32.Chiou A, et al. Probing neuroserpin polymerization and interaction with amyloid-β peptides using single molecule fluorescence. Biophys J. 2009;97:2306–2315. doi: 10.1016/j.bpj.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]