Abstract
Aquaporins are transmembrane channels that facilitate the permeation of water and small, uncharged amphipathic molecules across cellular membranes. One distinct aquaporin subfamily contains pure water channels, whereas a second subfamily contains channels that conduct small alditols such as glycerol, in addition to water. Distinction between these substrates is central to aquaporin function, though the contributions of protein structural motifs required for selectivity are not yet fully characterized. To address this question, we sequentially engineered three signature amino acids of the glycerol-conducting subfamily into the Escherichia coli water channel aquaporin Z (AqpZ). Functional analysis of these mutant channels showed a decrease in water permeability but not the expected increase in glycerol conduction. Using X-ray crystallography, we determined the atomic resolution structures of the mutant channels. The structures revealed a channel surprisingly similar in size to the wild-type AqpZ pore. Comparison with measured rates of transport showed that, as the size of the selectivity filter region of the channel approaches that of water, channel hydrophilicity dominated water conduction energetics. In contrast, the major determinant of selectivity for larger amphipathic molecules such as glycerol was channel cross-section size. Finally, we find that, although the selectivity filter region is indeed central to substrate transport, other structural elements that do not directly interact with the substrates, such as the loop connecting helices M6 and M7, and the C loop between helices C4 and C5, play an essential role in facilitating selectivity.
Keywords: mutagenesis, transmembrane protein, X-ray structure
Aquaporins (AQPs) are integral membrane channels found throughout all kingdoms of life that selectively facilitate the permeation of water and small amphipathic solutes across cellular membranes (1). They display rates of conduction near the diffusion limit, but are remarkably selective and exclude all ions, including hydroxide and hydronium. AQPs are functionally divided into two subfamilies, the orthodox AQPs, which are selective for water and the more promiscuous aquaglyceroporins (AQGPs), which conduct both water and amphipathic solutes such as glycerol. This selectivity is central to biological function, and AQPs are involved in such disparate functions as water reabsorption in the kidney, glycerol efflux from adipocytes, and glycerol uptake by the causative agent of malaria, Plasmodium falciparum (1, 2). The Escherichia coli genome encodes one member of each subfamily, aquaporin Z (AqpZ) (3), an orthodox AQP, and the glycerol facilitator (GlpF) (4), an AQGP.
Atomic resolution X-ray structures of both AQPs and AQGPs have been determined and a comparison of these structures reveals the conserved structural architecture characteristic of the family and particular motifs that modulate function (5–7). All members are tetramers of four functional monomeric channels. The structure of the GlpF monomer (Fig. 1) is a right-handed bundle of six transmembrane and two half-spanning helices (M1–M8) that form a central channel. Helices M1–M4 are related by pseudotwofold symmetry to M5–M8 (7). The channel is roughly 20-Å long and has a diameter of less than 3 Å. At both the cytoplasmic and extracellular/periplasmic entrance, there are conically shaped vestibules, which give the protein an hourglass shape. The channel is amphipathic: Its lining of hydrophobic side chains is split longitudinally by two symmetry related sets of four main-chain carbonyls evenly spaced throughout the length of the channel. These eight carbonyls act as hydrogen bond acceptors to successive waters that are, in turn, capable of hydrogen bonding with each other. This arrangement creates a single-file chain of water-binding sites running the length of the pore. In addition, the half-spanning helices M3 and M7 meet at the center of the channel and are capped by a signature Asn-Pro-Ala (NPA) motif common to all AQPs. The asparagines each provide a hydrogen bond donor that polarizes the orientation of the central water(s) (8).
Fig. 1.
Superposition of AqpZ and GlpF. AqpZ (orange) is superposed on GlpF (green) and both structures are displayed in cartoon representation. Helices are labeled M1–M8. The dashed boxes denote symmetry-related sets of water-coordinating carbonyls, and the solid box denotes the selectivity filter near the periplasmic entrance to the channel.
The most significant and conserved difference between the two subfamilies occurs at a region we defined as the selectivity filter (SF), the narrowest constriction point of the channel (7, 8). In orthodox AQPs, the SF radius (∼1.1 Å) is slightly smaller than the radius of water (1.4 Å), whereas in AQGPs, it is slightly larger (∼1.7 Å) than water. The SF is located roughly halfway between the NPA motifs and the extracellular vestibule and is composed of a strictly conserved arginine and three other amino acids that are conserved only within each subfamily and which distinguish AQPs from AQGPs. In water-selective AQPs, this region is more polar and contains a conserved histidine. In AQGPs, it is more hydrophobic, with two conserved aromatic residues, usually tryptophan and tyrosine, whose side chains provide a hydrophobic corner that interacts favorably with the carbon backbone of linear alditols (Fig. S1). Because of the correlation between SF sequence and function—larger, hydrophobic channels conduct solutes and smaller, hydrophilic channels conduct water—we proposed that this region forms the basis for substrate selectivity in AQPs. Removal of one of the conserved aromatic residues in GlpF leads to an increase in water permeability (8) and widening of the AQP1 SF by alanine mutations results in a significant increase in glycerol permeability (9). Molecular dynamic simulations have also implicated a steric barrier at the SF region as important to selectivity (10). In order to further investigate the structural determinants of substrate selectivity in AQPs, we sequentially engineered the most conserved element of the GlpF SF into AqpZ.
AqpZ has been functionally characterized as an orthodox AQP (11) and its X-ray crystal structure has been determined to 2.5 Å in our laboratory (12). AqpZ and GlpF therefore together constitute an excellent model system to probe structure–function relationships (13). We used site-directed mutagenesis to investigate the role of pore diameter and polarity on conduction of both water and glycerol in reconstituted proteoliposomes. Replacing three SF residues in AqpZ by their counterparts in GlpF resulted in a decrease in water permeability, but no increase in glycerol conduction. To understand this result, we determined the X-ray crystal structures of all the mutant channels. Unexpectedly, the pore sizes of all channels were similar to the pore size of the parent WT AqpZ. This complete size conservation allowed us to dissect the effects of SF polarity and size on the energetics of water and glycerol conduction, respectively, in AQPs. Finally, we find that the differences in pore size among AQPs are not determined by the SF alone but are also promoted by significant changes in structural architecture from one AQP to another, as earlier proposed based on molecular dynamics simulations (10).
Results and Discussion
The structures of GlpF and AqpZ show a high degree of similarity (α-carbon root mean square deviation of 1.7 Å), but are quite different at the SF (Fig. 1). To test the role of the SF in glycerol conduction, we sequentially mutated the three AQP-specific SF residues of AqpZ into their AQGP-specific counterparts in GlpF (Fig. S1). Mutants F43W, H174G/T183F, and F43W/H174G/T183F were expressed, purified in the detergent n-octyl-β-D-glucopyranoside (OG), and functionally and structurally characterized. WT protein expressed at 10 mg/L of culture and the mutants expressed to roughly 5 mg/L of culture. GlpF was expressed and purified as described previously (7).
Water Permeability of AqpZ and GlpF.
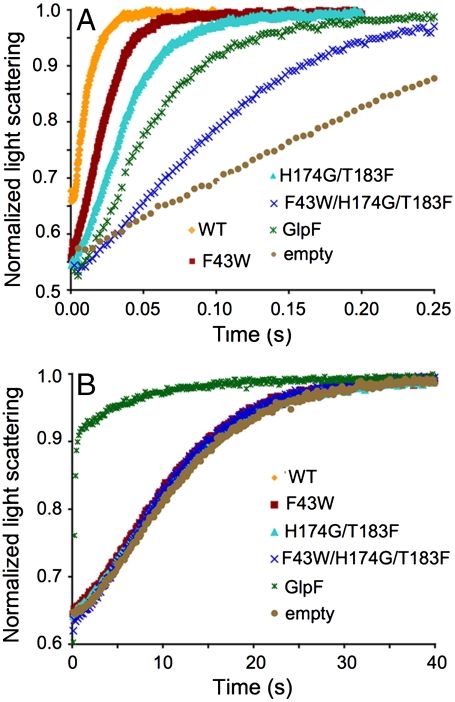
Flux through the AQP channel was measured using osmotically driven proteoliposome permeability assays (13). AqpZ, AqpZ mutants, and GlpF were purified and reconstituted into liposomes. Permeabilities were measured in a stopped-flow device by challenging the proteoliposomes with a hyperosmolar reconstitution buffer to drive water efflux. The resulting proteoliposome shrinkage rate was measured by light scattering and the curves fit to an exponential equation with a single rate constant (kwat). Raw light scattering curves can be seen in Fig. 2A and the values for kwat are listed in Table 1. Values are 102.9 ± 1 s-1, 56.8 ± 3 s-1, 34.8 ± 0.2 s-1, 12.3 ± 0.1 s-1, 20.2 ± 0.4 s-1, and 6.3 ± 0.3 s-1 for WT, F43W, H174G/T183F, F43W/H174G/T183F, GlpF, and control liposomes, respectively. Stepwise mutation of the AqpZ SF introduced a more hydrophobic SF, removed the polar imidazole moiety, and resulted in a significant decrease in water permeability. Strikingly, the triple mutant F43W/H174G/T183F, which should have diminished water permeability similar to that of GlpF, had a water permeability that was much less than GlpF and nearly equivalent to that of empty liposomes.
Fig. 2.
Proteoliposome conduction assays. (A) Curves of the raw data obtained in an osmotic challenge assay measuring water permeability. The color code is WT (orange), F43W (maroon), H174G/T183F (cyan), F43W/ H174G/T183F (blue), GlpF (green), and empty liposomes (tan). These colors are used throughout the paper to denote their respective protein. (B) Raw data for glycerol conductivity.
Table 1.
Proteoliposome permeability for water and glycerol
| kwat, s-1 | kgly, s-1 | |
| WT AqpZ | 102.9 ± 1 | 0.102 ± 0.001 |
| F43W | 56.8 ± 3 | 0.105 ± 0.001 |
| H174G/T183F | 34.8 ± 0.2 | 0.100 ± 0.001 |
| F43W/H174G/T183F | 12.3 ± 0.1 | 0.107 ± 0.004 |
| GlpF | 20.2 ± 0.4 | 4.82 ± .06 |
| Empty liposomes | 6.3 ± 0.3 | 0.094 ± 0.002 |
Glycerol Permeability of AqpZ and GlpF.
Glycerol permeability was measured in a similar manner to water permeability. Proteins were reconstituted into liposomes and flux was measured in a stopped-flow device by challenging glycerol-loaded proteoliposomes with an isoosmolar buffer containing impermeant sucrose. The glycerol efflux rate was measured by light scattering and the curves fit to an exponential equation with a single rate constant (kgly). Fig. 2B shows the raw light scattering data and the values for kgly are listed in Table 1. Values are 0.102 ± 0.001 s-1, 0.105 ± 0.001 s-1, 0.100 ± 0.001 s-1, 0.107 ± 0.004 s-1, 4.82 ± .06 s-1, and 0.094 ± 0.002 s-1 for WT, F43W, H174G/T183F, F43W/H174G/T183F, GlpF, and control liposomes, respectively. Thus, although GlpF had significant glycerol permeability, AqpZ and all of the mutants converting AqpZ into a more GlpF-like channel had glycerol permeabilities similar to control protein-free liposomes. Although the mutants diminished water permeability, they did not restore glycerol permeability at all.
Structures of the Engineered Selectivity Filter Mutants.
To understand the decrease in water flux coupled with a lack of increase in glycerol permeability, we determined the X-ray crystal structures of the three AqpZ mutants. Following purification, the mutants were crystallized and atomic resolution diffraction data were collected (Table 2). All mutants crystallized in the space group P4 with two monomers (chain A and chain B) in the asymmetric unit. WT AqpZ crystallized in the same space group with similar unit cell dimensions. The structures were solved by molecular replacement using the 1.9-Å L170C AqpZ structure (PDB ID 209G) as the search model (14). The final resolution cutoffs were 2.40, 2.50, and 3.10 Å, and the Rfree factors for the refined structures were 21.8%, 22.0%, and 22.1% for F43W, H174G/T183F, and F43W/H174G/T183F, respectively (Table 2).
Table 2.
Crystallographic data and refinement statistics
| F43W | H174G/T183F | F43W/H174G/T183F | |
| Data Collection | |||
| Space group | P4 | P4 | P4 |
| Unit cell | |||
| a, Å | 92.4 | 92.8 | 92.9 |
| c, Å | 78.7 | 78.9 | 79.5 |
| Resolution range,* Å | 20–2.40 (2.46–2.40) | 20–2.50 (2.56–2.50) | 20–3.10 (3.18–3.10) |
| Unique reflections | 24,848 | 21,743 | 10,963 |
| Completeness* | 95.4 (86.6) | 93.8 (82.4) | 88.0 (90.4) |
| Rsym, *, † | 0.074 (0.700) | 0.063 (0.645) | 0.115 (0.679) |
| I/σ* | 15.4 (1.3) | 17.4 (1.6) | 10.6 (1.5) |
| Refinement statistics | |||
| Rwork/Rfree, % | 18.1/22.0 | 18.6/22.0 | 19.8/22.2 |
| rmsd‡ bonds, Å | 0.020 | 0.021 | 0.016 |
| rmsd‡ angles, Å | 1.71 | 1.73 | 1.54 |
| Number of protein atoms | 3,356 | 3,357 | 3,359 |
| Number of OG molecules | 4 | 4 | 3 |
| Number of solvent atoms | 136 | 84 | 13 |
| Average B factor, Å2 | 21.7 | 22.7 | 29.1 |
| PDB ID code | 3NK5 | 3NKA | 3NKC |
*Values in parenthesis refer to the highest-resolution shell.
†Σ|I—〈I〉|/ΣI, where I equals observed intensity and 〈I〉 equals average intensity for symmetry-related reflections.
‡Rmsd of bond lengths and angles from ideal values.
Superposition of the structures indicated little main-chain variation between WT and the mutants (Fig. S2A). Electron density for the mutant SF residues was well defined in 2Fo - Fc difference maps. Representative density is shown for H174G/T183F in Fig. S2B. In this mutant, two glycerol molecules (denoted Cry1 and Cry2) were located just outside the selectivity filter in a similar location to glycerol G1 in the original GlpF structure. The F43W and H174G/T183F structures included 136 and 84 solvent atoms, respectively, and there was a continuous single-file chain of water present in the A monomers for both proteins. As in the WT structure, three OG molecules clustered around the conserved aromatics of helix M8 near the periplasmic leaflet (12). These detergents molecules were in nearly the same conformation for all structures, indicative of a high-affinity binding site.
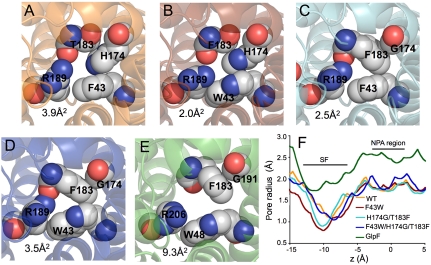
The van der Waals surfaces of the pore cross-sections at the SFs were, surprisingly, similar in size to WT AqpZ (Fig. 3 A–D). Channel radii were calculated as a function of distance along the channel axis (z axis) and revealed that F43W and H174G/T183F mutants are extremely narrow at the SF (z = -10 Å), with a radius under 1.0 Å (15) (Fig. 3F). In contrast, the GlpF radius for this region is approximately 1.7 Å (7). Cross-sectional areas for the SF are 3.9, 2.0, 2.5, 3.5, and 9.3 Å2 for WT, F43W, H174G/T183F, F43W/H174G/T183F, and GlpF, respectively. Thus, the series of mutations expected to provide a more hydrophobic, GlpF-like pore introduced the mutated side chains into the SF region, but instead of dilating the pore they actually decreased its size.
Fig. 3.
The SF of AqpZ, AqpZ mutants, and GlpF. (A) Structure of the WT AqpZ SF. Residues of the SF are drawn as spheres with van der Waals radii. The area of the selectivity filter measured with the program HOLE2 (23) is shown in the lower left-hand corner. (B) F43W SF. (C) H174G/T183F SF. (D) F43W/H174G/T183F SF. (E) GlpF SF. (F) Channel radii determined by HOLE2 for all proteins plotted as a function of channel axis position. The NPA region is the origin.
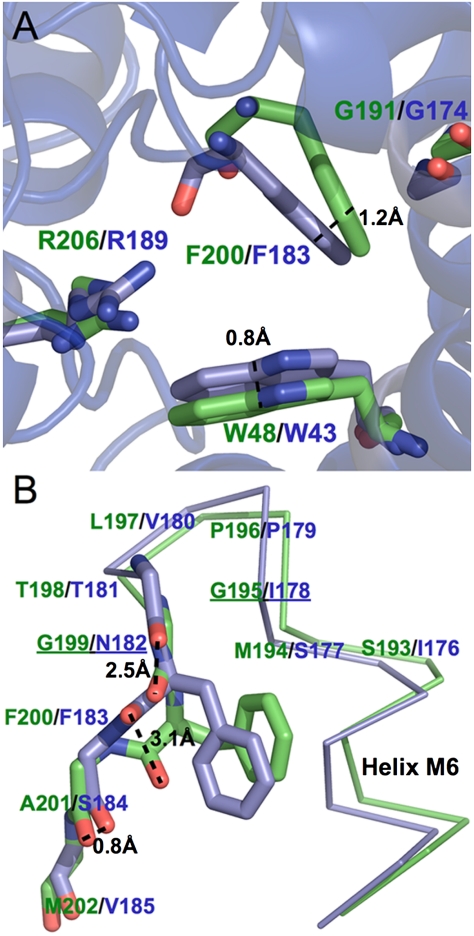
The smaller pore sizes of AqpZ mutants compared to that of GlpF could be partly attributed to differences in helix–helix packing, which are the result of differences in the nonconserved loops of the proteins predominantly outside the pore region. Superposition of AqpZ and GlpF showed a difference in helix M2 orientation, resulting in a ∼1-Å difference in α-carbon position for residue 43. This difference in M2 orientation persisted after the F43W mutation so that the indole rings of Trp-43 in F43W and in F43W/H174G/T183F were laterally shifted 0.8 Å into the pore, relative to the corresponding Trp in GlpF (Fig. 4A). The larger size of the indole ring of Trp compared to the phenyl ring of Phe resulted in a F43W SF nearly 50% smaller by area than the WT AqpZ SF.
Fig. 4.
The F43W/H174G/T183F SF is structurally different than GlpF. (A) Superposition of the mutant F43W/H174G/T183F (blue) on GlpF (green) reveals significant differences in pore size despite the identical residues (shown in stick format). (B) The M6–M7 loops are conformationally distinct. Spatial differences between carbonyls are indicated with a dashed line and positions containing glycines in GlpF alone are underlined.
The differences in pore sizes of AqpZ mutants and GlpF could also be accounted for by different conformations of the M6–M7 linkers in AqpZ and GlpF (Fig. 4B). The M6–M7 linker in GlpF is an α-turn whose ends are two glycines (Gly-195 and Gly-199), followed by SF residue Phe-200, which is the first residue of an unstructured loop leading to helix M7. In contrast with GlpF, the α-turn glycines are replaced by bulky residues in AqpZ (Ile-178 and Asn-182) leading to highly strained conformation of the corresponding SF residue Thr-183 (ϕ = 66°, ψ = 116°). In addition, the Cα of AqpZ Thr-183 is shifted ∼2 Å away from the pore relative to Phe-200 in GlpF.
The small, hydrophilic side chain of Thr-183 accommodates the strictly conserved (in AQPs), water-selective His-174 of helix M6 at the SF pore, and a small hydrophilic residue paired with His at the SF is a signature of orthodox AQPs. In GlpF, the corresponding covariant pair of residues is Phe-200 and Gly-191, allowing the bulky Phe-200 phenyl group to pack tightly against helix M6 (Fig. 4A). Thus His-174 and Thr-183 must be comutated to Gly and Phe, respectively, in the engineering of the AqpZ selectivity filter.
However, comutation of these SF positions is unable to compensate for the strain induced on Thr-183 via the M6–M7 α-turn. In the AqpZ double mutant H174G/T183F and in the triple mutant F43W/H174G/T183F, the Phe-183 backbone retains the same strained geometry as Thr-183 in wild-type AqpZ. The Phe-183 side chain packs against Gly-174 of M6, analogously to the Phe-200 side chain in GlpF, but adopts a different side chain rotamer (Chi-1 = 177° compared to Chi-1 = 60° in GlpF). As a result, the phenyl side chain is positioned 1.2 Å further into the channel in the AqpZ mutants than in GlpF (Fig. 4 A and B). Thus the M6–M7 linker in AqpZ appears to have a rather inflexible backbone conformation that is insensitive to single-site mutations and is responsible for SF side-chain orientation.
The different backbone geometries of the M6–M7 linkers in AqpZ and GlpF also lead to significant differences in the orientations of the main-chain carbonyls comprising the C-terminal water-binding motifs. In GlpF, these carbonyls (i.e., the M6–M7 linker post α-turn) are positioned away from the pore and are as far as 3.1 Å away from their respective positions in WT AqpZ and in the AqpZ mutants (Fig. 4B). This structural difference could play a role in the faster water permeation kinetics observed for AqpZ over GlpF as reported here and previously (13). In contrast, the quasi-twofold related N-terminal water-binding motifs have virtually identical conformations in GlpF, AqpZ, and AqpZ mutants.
As a further consequence of the different architectures of GlpF and AqpZ, the conformations of Arg-189, an invariant SF residue, are also significantly different in all AqpZ mutants than in GlpF. In our wild-type AqpZ structure and in the AqpZ mutants, Arg-189 is in a conformation referred to as “down” (Cβ-Cγ-Cδ-Nε dihedral angle = -101.0°) (Fig. 5A), which orients the guanidinium group toward the pore (10). This conformation is stabilized by a network of hydrogen bonds with the carbonyls of the C-terminal water-binding motif (distance of 2.7 Å) described above and by a hydrogen bond to the carbonyl of residue Ala-117 in the C loop (distance of 2.6 Å), a loop between helices M4 and M5 that dips into the extracellular vestibule near the SF (Fig. 5B). Notably, in AQGPs the C loop tends to be longer and extend further into the vestibule (Fig. S1). In GlpF, the invariant Arg is in a conformation referred to as “up” (Cβ-Cγ-Cδ-Nε dihedral angle = -179.8°), in which the positively charged guanidinium moiety is positioned out of the pore (Fig. 5A). This structure is stabilized specifically by a hydrogen bond with the carbonyl of C-loop residue Phe-135 (distance of 2.9 Å) (Fig. 5B). The inward orientations of the guanidinium groups of AqpZ and AqpZ mutants contribute significantly to their narrower pores relative to GlpF.
Fig. 5.
Structural differences in the SF arginine. (A) Superposition of the WT (orange) and GlpF (green) SF arginine conformations. In transparency are the other structures also superposed, showing a consistent conformation between WT and all mutants. (B) Role of the C loop and carbonyls in positioning the arginine. Potential hydrogen bonding partners are shown as dashed lines.
In simulations, Arg-189 in AqpZ was highly dynamic, transitioning among the up and down conformations, and a conformation midway between the two, on a nanosecond timescale. The down conformation impinged on and effectively closed the channel, suggesting Arg-189 is a gate that modulates conductance (10, 16). Consistent with this, in a 3.2-Å structure of AqpZ, Arg-189 was found in the up conformation in one of the four molecules in the asymmetric unit and in the down conformation in the other three (17). In contrast, conformational changes were not observed for the SF Arg in GlpF, which remained in the up position during simulations (8). Thus, compared to GlpF, AqpZ would incur an entropic cost in ordering Arg-189 in the open conformation required for glycerol transport. The down conformation of Arg-189 has only been seen in the crystal structures of AqpZ and AqpZ mutants. Perhaps crystallization conditions failed to capture the down conformation in other orthodox AQPs. Alternatively, the proposed gating function of this residue may be unique to AqpZ.
It has also been proposed that hydrogen bonds between the Arg-189 guanidinium group and the protein decrease the desolvation penalty for the Arg side chain, facilitating water conductance. Mutation of Glu-125 (Ser-136 in GlpF) to Ser in the Plasmodium AQGP, abolishes water conduction with no significant change in glycerol permeability (18). The recent structure of the Plasmodium AQGP suggests that Glu-125 in the C loop plays an essential role in orienting the carbonyl group of Trp-124 to accept hydrogen bonds from NH1 and NH2 of the SF Arg, thereby enhancing water conduction (19). Thus, the SF arginine plays a major role in selectivity and, as described above, is oriented by specific structural interactions with the C loop, which is not conserved in length or structure between AqpZ and GlpF.
Model for Selectivity.
The AQP family of channels is divided into orthodox water-selective AQPs and AQGPs, selective for both water and other small amphipathic solutes. Selectivity is central to biological function and an understanding of the motifs necessary and sufficient for a particular selectivity is fundamental to elucidating the role of AQPs in biological processes. The functional dichotomy is thought to be encoded by the SF, the narrowest region of the channel, which is typically smaller and more polar in orthodox AQPs. This divergence is embodied in AqpZ and GlpF, and so to investigate the determinants of selectivity, we sequentially mutated the SF residues of AqpZ to the corresponding residues in GlpF, assayed the mutants for both water and glycerol conduction, and solved their X-ray crystal structures.
Mutants F43W, H174G/T183F, and F43W/H174G/T183F displayed a serial decrease in water permeability, an expected result considering the increased hydrophobicity of their SFs. We hypothesized that sequential mutation would lead to an increase in size of the SF versus WT AqpZ, but the structures revealed that the mutant SFs are similar in size to WT AqpZ. Though unexpected, this result is fortuitous because it allows functional comparison between channels in which SF polarity is modified while size is kept constant. We postulate the following general model for AQP selectivity, based on three regimes of decreasing channel size.
It has previously been shown that mutation of the SF residues to alanine in AQP1, which drastically increases SF size (modeled at 8 Å in the AQP1 FA/HA mutant), increases glycerol permeability and has little affect on WT AQP1 water permeability (9). In this regime, the SF environment resembles bulk water and, as it cannot influence channel sterics or participate significantly in water/solute solvation, does little to influence selectivity. Indeed, such mutations have been shown to allow for ion conduction (9). These amino acid substitutions do not appear in naturally occurring SF sequences and are, most likely, not physiologically relevant.
The second regime is that of the AQGPs, such as GlpF, which display an SF slightly larger (∼2 Å) than the radius of water (1.4 Å). In this situation, solute permeability is sterically allowed and is facilitated by the conserved aromatic residues of the SF interacting favorably with the alkyl backbone of the substrate. Water conduction is also allowed, as water molecules moving through the pore can favorably interact with polar SF moieties and each other, resulting in a minor bulk-water desolvation penalty. Although it has not been demonstrated experimentally, computer simulations indicate that the AQGP SF construction conducts water at higher rates than the orthodox AQP SF, suggesting this penalty may be nonexistent (16). Finally, because the SF environment does not resemble bulk water, ions are excluded by a high desolvation penalty.
The last regime is that of the orthodox AQPs, in which SF size is comparable to that of water and the chemical environment is therefore strongly dependent on SF residues. In this case, such as in WT AqpZ, solutes are sterically excluded and conduction can only occur upon severe rearrangement of the SF (10). Water conduction is facilitated by the SF’s hydrophilic moieties, which lower the free-energy penalty for desolvating water from the bulk solvent. Any decrease in polarity, such as in the mutant F43W/H174G/T183F, diminishes transport.
Materials and Methods
Expression and Purification.
Mutants of AqpZ were generated by site-directed-mutagenesis of the pET28b-AqpZ construct used for the structure determination of WT AqpZ. The E. coli strain C43(DE3) was transformed, grown to 0.6–1 OD at 600 nm at 37 °C in 2 × LB media, 0.5% glycerol (vol/vol), 1 × M9 salts, and 25 mg/L kanamycin, and induced with 1 mM isopropyl-D-thiogalactoside (Anatrace).
All purifications were carried out at 4 °C or on ice. Cells from 6 L of culture were harvested and lysed by a microfluidizer in 20 mM Tris pH 7.4, 100 mM NaCl, 0.5 mM phenylmethylsulforyl fluoride, and 5 mM β-mercaptoethanol (BME). Membranes were recovered from supernatant by 100,000 × g centrifugation for 2 h. Protein was solubilized from membranes by agitation in 20 mM Tris pH 7.4, 100 mM NaCl, 5 mM BME, 10% glycerol, and 270 mM OG (Anatrace) for 12–16 h at 4 °C. Solubilized protein was bound in batch to Ni-NTA resin (Qiagen) for 1 h, washed with 25 resin volumes of 20 mM Tris pH 7.4, 100 mM NaCl, 5 mM BME, 10% glycerol, 40 mM OG, and 20 mM imidazole, and eluted with 20 mM Tris pH 7.4, 100 mM NaCl, 5 mM BME, 10% glycerol, 40 mM OG, and 250 mM imidazole. Imidazole was removed using a Biorad Econo-Pac 10DG desalting column and the histidine tag was removed by digestion with 5 μg of trypsin (Worthington Biochemical) for 12 h at 4 °C. Trypsin was removed by passing over a benzamidine-sepharose matrix (GE Healthcare), and the protein sample was injected onto a Pharmacia Superose 12 gel filtration column running a mobile phase of 20 mM Tris pH 7.4, 100 mM NaCl, 2 mM dithiothreitol, 10% glycerol, and 40 mM OG. Except as noted, all materials were purchased from Sigma or Fisher. The sample was judged pure and homogenous by Coomassie-stained denaturing gels and gel filtration chromatography, respectively.
Crystallization and Data Collection.
Following gel filtration chromatography, the protein was concentrated to 25 mg/mL using a 30 kDa cutoff Amicon Ultra-15 Centrifugal Filter. Crystals were grown by hanging drop vapor diffusion at room temperature by 1∶1 addition of protein and 25–30% polyethylene glycol monomethyl ether 2000 (Fluka), 100 mM sodium cacodylate pH 6.5, and 50–100 mM MgCl2. Crystals grew to roughly 300 × 300 × 150 μm over the course of several days and were flash frozen in liquid nitrogen following a brief washing in the mother liquor plus 15% glycerol for cryoprotection. Diffraction intensities were collected on Advanced Light Source Beamline 8.3.1 using an ADSC Quantum-Q210 CCD detector.
Structure Determination, Refinement, and Analysis.
Data were processed with Elves (20) and CCP4 (21) (using MOSFLM ref. 22) and the structures were solved by molecular replacement with Phaser (23) using the 1.9 Å L170C AqpZ structure (PDB ID code 209F) as the search model. The structures were refined with iterative cycles of manual building with Coot (24) and positional refinement with Refmac5 including, alternately, translation, libration, screw-motion refinement and individual B-factor refinement, and using tight noncrystallographic symmetry restraints (25). Mutations were verified and side chains positioned using omit maps calculated in Refmac5.
Pore cross-sectional areas and pore radii were measured from the crystal structures using HOLE2, which determines the largest sphere that fits in the channel space without overlapping the van der Waals surfaces of surrounding residues (15). Superposition of structures was done in Coot.
Proteoliposome Reconstitution.
Before removing the 6xHis tag with trypsin, aliquots of protein were set aside for proteoliposome reconstitution. E. coli polar lipids were sonicated to clarity and the reconstitution cocktail was prepared by sequentially adding 100 mM MOPS pH 7.5, 1.5% (wt/vol) OG, 50 μg/μL of purified protein, and 10 mg/mL E. coli polar lipids (Avanti) (13). To reduce oxidation, lipid stocks were stored in 2 mM BME and all buffers were under argon atmosphere. Following cocktail incubation for 1 h at room temperature (RT), proteoliposomes were formed by diluting the mixture 50-fold into a running buffer of 20 mM Hepes pH 7.5 and harvested by centrifugation at 100,000 × g for 2 h. Pelleted liposomes were resuspended into 1 mL of running buffer (RB) (20 mM Hepes pH 7.5) and stored at 4 °C. Liposomes monodispersity and size were verified by dynamic light scattering, with liposomes having a mean diameter of 90 nm.
To analyze the kinetics of water conduction through the channel, we diluted the proteoliposomes 7.5-fold in RB and subjected them to an osmotic gradient by mixing 1∶1 proteoliposomes (final AqpZ monomer concentration of 0.27 uM) and RB with osmolyte (RBS) (20 mM Hepes pH 7.5, 570 mM sucrose; a 285 milliosmolarity final gradient) and measured water efflux (liposome shrinkage) by light scattering in a stopped-flow device (Applied Photophysics) at 440 nm. All assays were conducted at 12 °C. Resulting curves were fit to a single-exponential rate constant (kwat) as a measure of permeability to use in comparison between proteins. All errors represent the standard deviation calculated from independent proteoliposome preparations.
To analyze glycerol kinetics, pelleted proteoliposomes were resuspended in 1 mL of RB with 570 mM glycerol (RBG). They were then diluted 7.5-fold in RBG and mixed 1∶1 with RBS in a similar manner as above. The impermeant sucrose then drives glycerol efflux and light scattering detects liposome shrinkage. Resulting curves were fit to a single-exponential rate constant (kgly) as a measure of glycerol permeability.
Supplementary Material
Acknowledgments.
We thank James Holton for his assistance at the Advanced Light Source Beamline 8.3.1 supported by National Institutes of Health (NIH) Grant GM074929 (to R.M.S.). D.F.S. was supported by a Burroughs Wellcome Graduate Trainee Fellowship. Research was supported by NIH Grant GM24485 (to R.M.S.) and by the NIH Roadmap Center Grant P50 GM073210. The computer cluster and computation was supported from a scientific computing grant from Microsoft Corporation (R.M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3NK5, 3NKA, and 3NKC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009864107/-/DCSupplemental.
References
- 1.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem. 1999;68:425–458. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Hansen M, Kun JF, Schultz JE, Beitz E. A single, bi-functional aquaglyceroporin in blood-stage Plasmodium falciparin malaria parasites. J Biol Chem. 2002;277:4874–4882. doi: 10.1074/jbc.M110683200. [DOI] [PubMed] [Google Scholar]
- 3.Calamita G, Bishai WR, Preston GM, Guggino WB, Agre P. Molecular cloning and characterization of AqpZ, a water channel from Escherichia coli. J Biol Chem. 1995;270:29063–29066. doi: 10.1074/jbc.270.49.29063. [DOI] [PubMed] [Google Scholar]
- 4.Heller KB, Lin EC, Wilson TH. Substrate specificity and transport properties of the glycerol facilitator of I. J Bacteriol. 1980;144:274–278. doi: 10.1128/jb.144.1.274-278.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 6.Walz T, et al. The three-dimensional structure of aquaporin-1. Nature. 1997;387:624–627. doi: 10.1038/42512. [DOI] [PubMed] [Google Scholar]
- 7.Fu D, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 8.Tajkhorshid E, et al. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science. 2002;296:525–530. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- 9.Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc Natl Acad Sci USA. 2006;103:269–274. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Schulten K, Tajkhorshid E. What makes an aquaporin a glycerol channel? A comparative study of AqpZ and GlpF. Structure. 2005;13:1107–1118. doi: 10.1016/j.str.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Borgnia MJ, Kozono D, Calamita G, Maloney PC, Agre P. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J Mol Biol. 1999;291:1169–1179. doi: 10.1006/jmbi.1999.3032. [DOI] [PubMed] [Google Scholar]
- 12.Savage DF, Egea PF, Robles-Colmenares Y, O’Connell JD, Stroud RM. Architecture and selectivity in aquaporins: 2.5 Å X-ray structure of aquaporin Z. PLoS Biol. 2003;1:334–340. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borgnia MJ, Agre P. Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli. Proc Natl Acad Sci USA. 2001;98:2888–2893. doi: 10.1073/pnas.051628098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savage DF, Stroud RM. Structural basis of aquaporin inhibition by mercury. J Mol Biol. 2007;368:607–617. doi: 10.1016/j.jmb.2007.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen MØ, Mouritsen OG. Single-channel water permeabilities of Escherichia coli AqpZ and GlpF. Biophys J. 2006;90:2270–2284. doi: 10.1529/biophysj.105.073965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Daniels BV, Fu D. Crystal structure of AqpZ tetramer reveals two distinct Arg-189 conformations associated with water permeation through the narrowest constriction of the water-conducting channel. J Biol Chem. 2006;281:454–460. doi: 10.1074/jbc.M508926200. [DOI] [PubMed] [Google Scholar]
- 18.Beitz E, Pavlovic-Djuranovic S, Yasui M, Agre P, Schultz JE. Molecular dissection of water and glycerol permeability of the aquaglyceroporin from Plasmodium falciparum by mutational analysis. Proc Natl Acad Sci USA. 2004;101:1153–1158. doi: 10.1073/pnas.0307295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newby ZE, et al. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat Struct Mol Biol. 2008;15:619–625. doi: 10.1038/nsmb.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci USA. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 23.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr, Sect D: Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.