Abstract
Covalent modification of proteins by small ubiquitin-like modifier (SUMO) regulates various cellular activities in yeast and mammalian cells. In Arabidopsis, inactivation of genes encoding SUMO or SUMO-conjugation enzymes is lethal, emphasizing the importance of SUMOylation in plant development. Despite this, little is known about SUMO targets in plants. Here we identified 238 Arabidopsis proteins as potential SUMO substrates because they interacted with SUMO-conjugating enzyme and/or SUMO protease (ESD4) in the yeast two-hybrid system. Compared with the whole Arabidopsis proteome, the identified proteins were strongly enriched for those containing high-probability consensus SUMO attachment sites, further supporting that they are true SUMO substrates. A high-throughput assay was developed in Escherichia coli and used to test the SUMOylation of 56% of these proteins. More than 92% of the proteins tested were SUMOylated in this assay by at least one SUMO isoform. Furthermore, ADA2b, an ESD4 interactor that was SUMOylated in the E. coli system, also was shown to be SUMOylated in Arabidopsis. The identified SUMO substrates are involved in a wide range of plant processes, many of which were not previously known to involve SUMOylation. These proteins provide a basis for exploring the function of SUMOylation in the regulation of diverse processes in Arabidopsis.
Keywords: diverse functions, post-translational modification, small protein modifiers
In eukaryotes, posttranslational modification by the attachment of small ubiquitin-like modifier (SUMO) alters the activity of many substrate proteins. In yeast and mammalian cells, this regulatory mechanism is involved in diverse cellular processes, including nuclear–cytoplasmic shuttling, DNA and chromatin activities, transcriptional regulation, RNA transport, protein–protein interaction, and various other biological processes (1–5). Many of the SUMO substrate proteins involved in these processes were identified by systematic screening (6–13). Despite genetic demonstration of the importance of SUMOylation in plants, few SUMO substrates have been identified, and the extent to which these are conserved with other eukyarotes is unclear. Here we describe a proteome-wide identification of SUMO substrates in Arabidopsis.
The enzymatic activities required for protein SUMOylation are well characterized. Three enzymes mediate covalent attachment of SUMO to substrate proteins: SUMO-activating enzyme (SAE or E1), SUMO-conjugating enzyme (SCE or E2), and SUMO ligase (E3) (14, 15). SAE, a heterodimer (SAE1 and SAE2), forms a thioester bond between a reactive cysteine residue in its large subunit (SAE2) and the C-terminal end of SUMO. SCE binds both SUMO and the potential substrate and mediates the transfer and conjugation of SUMO from SAE to the substrate. Specific residues in SCE interact with a sequence motif present in the substrate called the SUMO attachment site (SAS) (16). A SAS consensus sequence (ΨKXE/D) consists of a lysine residue to which SUMO is attached (position 2), flanked by a hydrophobic amino acid (position 1), any amino acid (position 3), and an acidic amino acid (position 4). SCE catalyzes the formation of an isopeptide bond between the ε-amino group of the lysine residue of the substrate and the C-terminal glycine residue of SUMO (14, 16). In Arabidopsis, SCE and SAE2 are encoded by single genes, whereas SAE1 is encoded by two genes (SAE1a and SAE1b) (17). In plants and mammals, SUMO, SUMO ligases, and SUMO proteases are encoded by multigene families (15, 17). Different SUMO isoforms are conjugated to specific substrates, and this is regulated by specific ligases and proteases. Whereas SUMO ligases aid in the conjugation reaction, SUMO proteases cleave SUMO from substrates (deconjugation) and cleave a C-terminal extension in precursor SUMO proteins to expose a glycine residue (processing) that can be conjugated to the substrate protein (14, 15).
SUMOylation also has been evaluated by mutation studies. This approach demonstrated that in several model systems, SUMO, SUMO-conjugation genes, and SUMOylation are essential for normal growth and development (1, 14, 15, 18). In Arabidopsis, mutants of SAE2, SCE, or both SUMO1 and SUMO2 cause embryonic lethality (18), and mutations that impair SUMO ligase or protease functions cause developmental and physiological defects (19–25). Arabidopsis mutants of the SUMO ligase SIZ1, for example, exhibit several phenotypes, including symptoms of phosphate starvation (19), reduced tolerance to freezing and drought (20, 21) and early flowering (22). Similarly, mutations that impair the SUMO-specific protease ESD4 lead to accumulation of SUMO protein conjugates and a pleiotropic phenotype that includes extreme early flowering (23, 24). This suggests that maintaining a regulated pool of SUMOylated proteins is essential for normal growth and development, and that identifying and characterizing SUMO substrates will be key to understanding how SUMO manifests its effects.
SUMO substrates have been identified in several model systems, including yeast, Caenorhabditis elegans, Drosophila, Arabidopsis, and mammals (5–13, 26, 27). Systematic proteome-scale attempts have been successful only in yeast and cell cultures and are lacking for multicellular higher eukaryotes, however. In Arabidopsis, candidate gene approaches have identified two transcription factors involved in cold tolerance and abscisic acid signaling (20, 28), whereas enrichment of epitope-tagged SUMO conjugates yielded 14 potential substrates (27). Although these and other reports established the importance of identifying Arabidopsis SUMO substrates as a prerequisite to studying the mechanisms through which SUMO modification of proteins regulates cellular and developmental processes, no large-scale screen for these substrates has yet been described. Immunologic approaches to identifying SUMO substrates in Arabidopsis have been hampered by low levels of SUMOylated proteins and by the SUMO protease activity associated with cell lysis, which is likely to reduce the levels of conjugated proteins further.
Because SCE and ESD4 interact with substrates to catalyze SUMO conjugation and deconjugation, we performed yeast two-hybrid screening using the two proteins as bait, and identified 238 potential SUMO substrates. We used a high-throughput assay to test the SUMOylation of 56% of these proteins, and found that >92% of them are likely bona fide SUMO substrates. One of these was confirmed in transgenic plants. The biological processes in which these substrate proteins act were documented. The identification of such a high number of SUMO substrates in Arabidopsis will help elucidate the roles of SUMOylation in the regulation of various cellular and biological activities in plants, and also will allow a comparison of the processes regulated by SUMOylation in different eukaryotic kingdoms.
Results and Discussion
Identification of Arabidopsis SUMO Substrates by Yeast Two-Hybrid Screening.
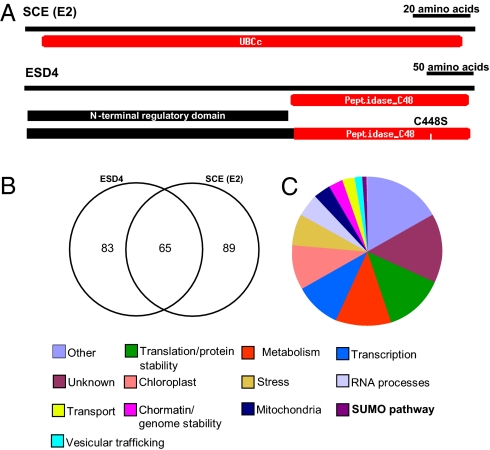
To identify putative SUMO substrates by yeast two-hybrid screening, we used SCE and ESD4 as bait in two different screens. Related approaches have been used previously to identify or characterize SUMO substrates in yeast and C. elegans (5, 12). These proteins were used because ESD4 is a major SUMO protease, so that a large number of SUMO conjugates accumulate in the esd4 mutant (24), and SCE is the only SUMO-conjugating enzyme in Arabidopsis (17). Attempts to use full-length ESD4 were impeded by our inability to maintain it in yeast cells; thus, we used catalytically inactive (C448S) and truncated (spanning the N-terminal regulatory domain) forms of ESD4 (Fig. 1A). The yeast two-hybrid screens identified a total of 238 unique interacting proteins. Using ESD4 (C448S) as bait recovered 24 proteins and the N-terminal domain identified 124 proteins, consistent with this regulatory domain being involved in determining substrate specificity (29) (Table S1). We also identified 154 potential substrates using SCE as bait (Table S1). None of the proteins interacting with either bait had been previously identified as putative SUMO substrates using different approaches (Table S1) (20, 27, 28), as observed in yeast, where distinct sets of substrates were identified by different methods (12). The large number of proteins identified using ESD4 and SCE is consistent with the central role of these proteins in SUMOylation in Arabidopsis. Interestingly, 65 proteins (28% of all proteins identified in these screens) interacted with both ESD4 and SCE (Fig. 1B), further supporting that they are indeed SUMO substrates.
Fig. 1.
Identification of Arabidopsis SUMO substrates by yeast two-hybrid screening. (A) The SCE (E2; Upper), and a catalytically inactive version (C448S) and the N-terminal region of ESD4 (Lower) were used as baits in different yeast two-hybrid screens. (B) A total of 238 potential SUMO substrates were identified, 65 of which (28%) interacted with both ESD4 and SCE. (C) SUMO substrates cover a wide range of functions and are found in the nucleus as well as the cytoplasm, chloroplast, and mitochondria.
We used a combination of BLAST, Gene Ontology, and literature searches to identify the molecular functions of SCE- and ESD-interacting proteins (Fig. 1C). We identified SUMO pathway components (SAE2, SCE, and SUMO1); metabolic enzymes; and proteins involved in translation, protein folding, and stability (translation initiation and elongation factors, 40S and 60S ribosomal protein subunits, calnexins, proteases, and subunits of the 26S proteasome); RNA splicing and processing (U2 and U5 SnRNP components, DEAD/DEAH helicases, ribonucleases, and RNA recognition motif proteins); chromatin and genome stability (DNA topoisomerase, chromatin-associated kinesin, chromomethylase 2, arginine N-methyltransferase); and stress response or protein folding (mostly annotated as heat-shock proteins), as well as many transcription factors (Fig. 1C and Table S1). SUMO substrates belonging to similar categories of proteins also have been identified in yeast and human cell cultures (7, 12, 13). Because we used yeast two-hybrid libraries of whole plant tissues and SCE (the only SUMO E2) and ESD4 (the major SUMO protease) as bait, many proteins involved in normal growth and development likely were identified, and were therefore not limited to those induced by specific conditions, such as environmental stress.
Although many of these proteins are localized to the nucleus, we also identified chloroplast proteins (photosystem I and II subunits; CAB-binding proteins 2, 3, and CP29; protein import receptors; GTP-binding proteins; ferredoxins; FtsH protease; ribosome recycling/releasing factor; ADP/ATP translocase; and metabolic enzymes) and mitochondrial proteins (porins, ADP/ATP translocase, AAA-type ATPase, ABC transporter protein and metabolic enzymes) (Fig. 1C and Table S1). The identification of metabolic enzymes, as well as other cytoplasmic and organellar proteins, supports an emerging body of evidence suggesting that SUMOylation is involved in functions in other cell compartments besides the nucleus (1, 30, 31).
Potential SUMO Substrates Are Enriched in High-Probability SUMO Attachment Sites.
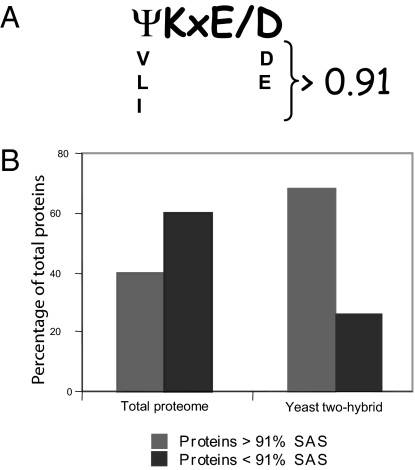
Unlike ubiquitin, SUMO attaches to target proteins at a sequence motif known as the SAS. Residues in SCE interact with the hydrophobic and acidic residues at the first and fourth positions of the SAS, respectively (16). Algorithms that assign values for the probability at which a specific SAS actually may be used for SUMO attachment use hydrophobicity and acidity values for the amino acids at these positions (32). We searched the Arabidopsis proteome for proteins containing an SAS with an arbitrary cutoff value of 91% (hpSAS) (Fig. 2A and Materials and Methods). This identified more than 10,000 proteins, constituting ≈40% of the Arabidopsis proteome (Table S2 and Fig. 2). We obtained a similar estimate (48%) when we searched the yeast proteome. SUMOylation at noncanonical sites has been reported previously (33, 34). The 3D context of SAS determines whether SUMO actually attaches to the lysine residue (16); however, a dataset of proteins with hpSAS offers the possibility of testing candidate proteins for SUMOylation. Of the proteins that interacted with SCE, 70% were found to contain hpSAS (Table S1 and Fig. 2), a significantly higher proportion than that obtained from an unbiased proteome-wide search (40%) (Fig. 2B), demonstrating that yeast two-hybrid screening with SCE enriched for proteins containing hpSAS. The identification of these proteins by two independent approaches increases the likelihood that they are indeed SUMO substrates.
Fig. 2.
SUMO substrates are enriched in hpSAS. (A) The SAS consists of a hydrophobic residue (Ψ), the lysine residue to which SUMO is attached (K), any amino acid (x), and an acidic amino acid (D or E). An hpSAS (≥91%) is shown (I/V/LKx D/E). (B) Proteins identified by yeast two-hybrid screening using the SCE as bait are enriched in hpSAS. Whereas 40% of the Arabidopsis proteome contains hpSAS, 70% of SCE interactors contain hpSAS.
High-Throughput SUMOylation Assay Confirms SUMO Attachment of Substrate Proteins.
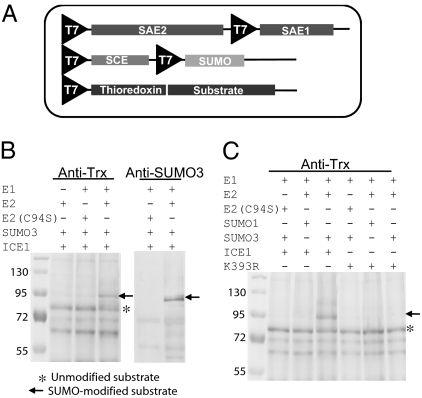
To test SUMOylation of proteins identified in the yeast two-hybrid screens, we developed a high-throughput SUMOylation assay. In vitro SUMOylation assays necessitate the purification of SAE1, SAE2, SCE, SUMO, and potential substrate proteins (35), which makes them difficult to use in a high-throughput manner. For this reason, we developed E. coli strains in which the Arabidopsis SUMO conjugation pathway was engineered (Fig. 3A). Similar strains expressing the mammalian and Arabidopsis pathways have been described previously (36, 37). Our system is optimized for the high-throughput analysis of substrates through the incorporation of a gateway-compatible bacterial expression vector to express putative substrate proteins and to express those harboring an N-terminal fusion with bacterial thioredoxin (Trx), which increases solubility and allows detection on immunoblot analysis. In this system, expression of proteins and SUMOylation of the substrate proteins occurs in the bacterial cells. As a control, we used this system to test the SUMOylation of a known Arabidopsis SUMO substrate (ICE1) (20). Immunoblot analysis using anti-Trx antisera showed that ICE1 is SUMOylated by SUMO3, as indicated by the presence of a higher–molecular weight form consistent with the attachment of one SUMO molecule (Fig. 3B, lane 4; Fig. 3C, lane 4). The presence of this form depended on the expression of both subunits of SAE (Fig. 3B, lane 2), the catalytic activity of SCE (Fig. 3B, lane 3), and the presence of an appropriate lysine residue, because SUMOylation of ICE1 was prevented by a mutation that replaced the lysine residue previously shown to be required for SUMOylation (K393) (20) with arginine (Fig. 3C, lane 7). Immunoblot analysis using anti-SUMO3 antisera confirmed that this higher–molecular weight form is indeed a SUMOylated form of ICE1 (Fig. 3B, lane 6). As negative controls, we searched the Arabidopsis proteome for proteins that contain lysine residues not located within a recognizable SAS, and used five of these in the E. coli assay (Fig. 4). None of these proteins was SUMOylated (Fig. 4 E and F). These experiments indicate that the E. coli system recreates the specificity of endogenous Arabidopsis SUMOylation.
Fig. 3.
Reconstitution of the SUMO conjugation pathway in E. coli. (A) The two subunits of SAE, the SCE, and either SUMO1 or SUMO3 were cloned into two vectors and introduced into E. coli expression strains. Genes encoding putative substrates are cloned into a gateway-compatible vector as N-terminal fusions with bacterial Trx and introduced into the same strain. (B) SUMOylation of a model substrate protein (ICE1) requires the activity of both SAE and SCE. Western blot analysis with anti-Trx antibody (Left) and anti-SUMO3 antibody (Right). (C) The E. coli SUMOylaytion system can be used to identify the SAS in ICE1. Western blot analysis using anti-Trx antibody shows that ICE1 is SUMOylated by SUMO3 (lane 4) and that this is abolished in a K393R mutant version of ICE1 (lane 7). Genotypes of the strains used are detailed on top of each blot, and the molecular weights (in kDa) are indicated on the left.
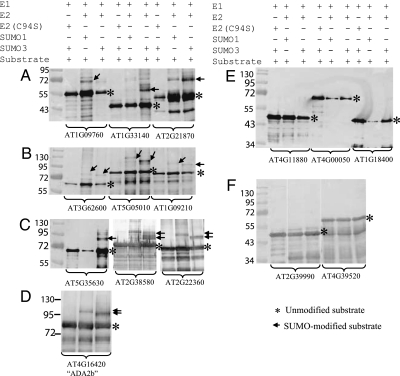
Fig. 4.
SUMOylation of potential SUMO substrates in E. coli. (A, B, C, and D) Ten potential substrates are SUMOyated by SUMO1, SUMO3, or both. (E and F) Five proteins (used as negative controls) containing lysine residues that are not located within SAS and are not SUMOylated in the E. coli assay. Genotypes of the strains used and the Arabidopsis Genome Initiative names of the proteins tested are detailed above and below the panels, respectively. Molecular weights (in kDa) are indicated on the left.
We then used this assay to test the SUMOylation of a large proportion of the putative SUMO substrates identified in the yeast two-hybrid screens (Fig. 4 and Table S1). We introduced into E. coli plasmids expressing full-length cDNAs encoding 54 proteins that interacted with SCE, 51 proteins that interacted with ESD4, and 42 proteins that interacted with both SCE and ESD4, which compose 62% of all yeast two-hybrid interactors. Of these 147 proteins, 134 were successfully expressed and purified, and 124 were SUMOylated by SUMO1, SUMO3, or both, whereas 10 proteins were not SUMOylated (Fig. 4 A–D and Table S1). Only one of the 10 proteins that were not SUMOylated contained an hpSAS, whereas the remaining nine proteins contained no obvious SAS or only a medium- or low-probability SAS. Similarly, of the 124 proteins that were SUMOylated in this experiment, 67 contained an hpSAS, whereas 57 contained a medium- or low-probability SAS. This suggests that although the presence of an hpSAS increases the likelihood that a protein will be SUMOylated, this is not an absolute requirement for SUMOylation. This conclusion is consistent with previous reports noting the importance of the 3D context of the SUMO attachment lysine and also with evidence of SUMOylation at noncanonical sites (16, 33, 34). The E. coli experiments indicated that ≈92.5% (124/134) of the yeast two-hybrid interactors are probable SUMO substrates.
The E. coli assay also differentiated between the two SUMO isoforms used in this study (SUMO1 and SUMO3), as demonstrated for the SUMOylation of ICE1 (Fig. 3) and many of the potential substrates tested here (Fig. 4 and Table S1). Arabidopsis encodes eight SUMO isoforms, of which only SUMO1, 2, 3, and 5 are highly expressed, and SUMO3 and 5 are distinct from the other isoforms (17, 27). In addition to general amino acid sequence differences, SUMO3 contains a methionine residue instead of the otherwise conserved glutamine at position 90 (17). Differences between SUMO isoforms at this residue do not influence conjugation efficiency but might influence isoform properties (27) in ways that can regulate substrate specificity. Our data suggest that SUMO substrates in Arabidopsis are often SUMOylated by specific SUMO isoforms. A similar pattern of differential SUMOylation of a model substrate (yeast PCNA) by Arabidopsis SUMO1 and SUMO3 has been reported previously (38).
ADA2B, a Putative SUMO Substrate, Is SUMOylated in Vivo.
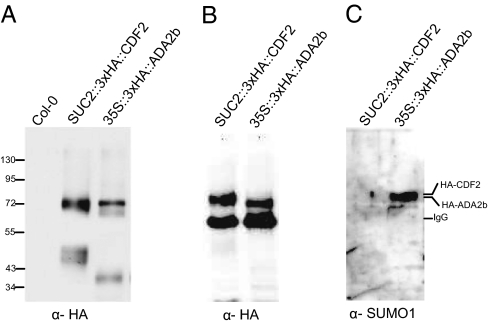
To test whether one of the potential SUMO substrates is SUMOylated in Arabidopsis, we generated transgenic lines overexpressing ADA2b. Also known as PRZ1, ADA2b is a component of a histone acetyltransferase complex required for histone acetylation and activation of gene expression (39, 40). ADA2b interacted with ESD4 in yeast two-hybrid screens (Table S1) and was SUMOylated by both SUMO1 and SUMO3 in the E. coli SUMOylation assay (Fig. 4D). We developed transgenic plants overexpressing an N-terminal triple HA-tagged version of ADA2b under the control of the cauliflower mosaic virus 35S promoter in an ada2b null background (39). Western blot analysis using anti-HA antibody and protein extracts from these plants revealed a major ADA2b form of ≈70 kDa (Fig. 5A), 14 kDa larger than the expected molecular weight of 56.1 kDa. For comparison, we also used extracts from transgenic plants overexpressing an epitope-tagged version of CYCLING DOF FACTOR 2 (3× HA-CDF2) (41), which migrates at a similar size (72 kDa) to ADA2b in Western blots (Fig. 5A) (41). We used the anti-HA antibody to precipitate HA-ADA2b and HA-CDF2 (70 and 72 kDa, respectively) (Fig. 5B). When the same immunoprecipitates were probed with anti-SUMO1 antisera, ADA2b was detected, but CDF2 was not (Fig. 5C), suggesting that ADA2b is SUMOylated in vivo by SUMO1. Interestingly, yeast ADA2b also was previously identified as a SUMO substrate (7), suggesting that SUMOylation may play an important conserved role in the function of this protein.
Fig. 5.
ADA2b is SUMOylated in vivo. (A) Western blot analysis using total extracts from transgenic lines overexpressing 3× HA-tagged ADA2b or CDF2 and probed with anti-HA antibody. (B and C) Total extracts used in (A) were used for immunoprecipitation using the anti-HA antibody followed by Western blot analysis using anti-HA antibody (B) or anti-SUMO1 antibody (C). CDF2- and ADA2b-reacting bands, as well as IgG (which reacts with the secondary antibody shown in B) are indicated on the right. Molecular weights (in kDa) are indicated on the left.
Many SUMO Substrates Are Involved in Stress Responses.
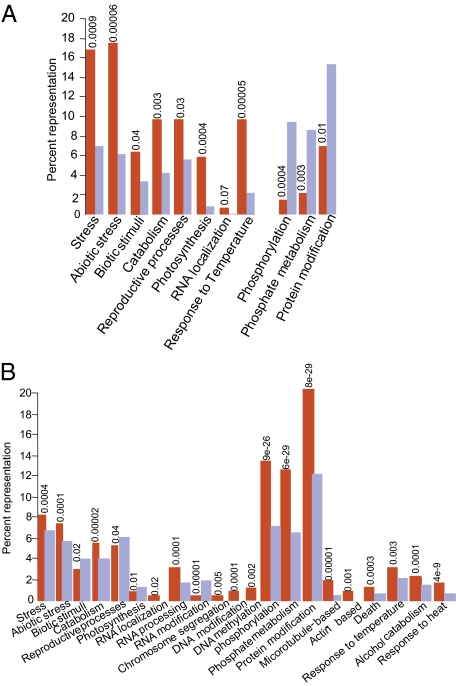
We used Gene Ontology searches to characterize the putative Arabidopsis SUMO substrates based on the biological processes in which they participate, and tested whether any specific category is differentially represented compared with the rest of the genome. P value indicate the probability with which the enrichment could occur at random (Fig. 6) (42). Proteins involved in stress responses were overrepresented in both the yeast two-hybrid interactors and the proteins identified computationally as containing hpSAS (P = 0.0009 and 0.0004, respectively) (Fig. 6 A and B). Previous reports suggest that SUMO conjugation of proteins is induced by various stress stimuli (13, 43, 44). In Arabidopsis, treatment with H2O2, canavanine, ethanol, or heat (37 °C) led to increased SUMO1 and SUMO2 conjugation of substrate proteins (43). Consistent with this, proteins identified computationally as containing hpSAS are enriched in proteins involved in response to temperature and heat (P = 0.003 and 4 × 10−9, respectively) and those involved in alcohol catabolism (P = 0.0001) (Fig. 6B), and substrates identified in yeast two-hybrid screens are enriched in proteins that respond to temperature (P = 0.00005) (Fig. 6A). Although proteins involved in abiotic stress are clearly overrepresented among putative substrates identified by both approaches (P = 0.00006 and 0.0001; Fig. 6 A and B), those involved in biotic stress are only marginally enriched (P = 0.04) or underrepresented (P = 0.02) among proteins identified by yeast two-hybrid screening or computationally. These findings are consistent with previous reports indicating that SUMO modification of proteins is important in responses to abiotic stress (13, 43).
Fig. 6.
Enrichment of SUMO substrates in proteins involved in specific biological processes. (A) Putative SUMO substrates identified by yeast two-hybrid analysis. (B) Putative SUMO substrates identified computationally as containing hpSAS. The y axis represents the percentage of all SUMO substrates (red columns) or of whole Arabidopsis proteome (blue columns); the x axis represents specific biological processes. P values are given above each category and indicate the probability with which this pattern would be expected to occur at random. P < 0.05 are highly significant.
Diverse Functions of Protein SUMOylation Revealed by Gene Ontology Searches.
Other functional categories in which putative SUMO substrates are clearly enriched include proteins involved in RNA localization and processing, chromosome segregation, and DNA modification (Fig. 6 A and B). Arabidopsis mutants of the nuclear pore complex (nua) that phenocopy the SUMO protease mutant esd4 and accumulate SUMO conjugates are defective in nuclear RNA export (2). Proteins involved in RNA processing and nuclear export also were identified in pull-down experiments performed to identify SUMO substrates in Arabidopsis (27), suggesting that SUMO modification of proteins indeed may be important for RNA processing and transport. Similarly, putative SUMO substrates are enriched in proteins involved in chromosome segregation and DNA modification (Fig. 6B). Roles for SUMO modification in sister chromatid cohesion, condensation, and topoisomerase functions have been suggested (45, 46).
In addition to these categories, we also detected significant enrichment in proteins involved in catabolism (observed with both datasets), photosynthesis (yeast two-hybrid; Fig. 6A), and actin- and microtubule-based movement (putative substrates identified computationally; Fig. 6B). Proteins involved in phosphorylation, phosphate metabolism, and posttranslational protein modification are highly enriched among proteins containing hpSAS (Fig. 6B), with the lowest P values (9 × 10−26 to 6 × 10−29). However, the same categories are underrepresented among proteins identified in yeast two-hybrid screens (Fig. 6A), perhaps due to constraints associated with expression of these proteins in yeast cells.
Conclusions
We have identified a large number of potential SUMO substrates in Arabidopsis and developed high-throughput assays to test their SUMOylation. The identified SUMO substrates are enriched in proteins involved in certain biological processes, which allowed us to further characterize the importance of SUMOylation in abiotic stress response and implicate it in such processes as photosynthesis and metabolism. Some of the substrates (eg, ADA2b) are homologs of yeast proteins previously shown to be SUMOylated, suggesting a conserved requirement for SUMOylation, whereas others are plant-specific proteins. The availability of this dataset and the E. coli SUMOylation strains that we have developed will be valuable in elucidating the role of protein SUMOylation in plant growth, development, and response to external stimuli.
Materials and Methods
A full description of the materials and methods used is provided in SI Materials and Methods. Yeast two-hybrid screens were performed using the SCE and ESD4 baits and Arabidopsis total cDNA libraries in the yeast strain PJ694a. E. coli SUMOylation strains were constructed in BL21(DE star) using the Novagen Duet cloning system. cDNAs encoding putative substrate proteins were cloned into the gateway-compatible bacterial expression vector pET32b-GW and introduced into these strains. Transgenic plants expressing the ADA2b under the control of CaMV 35S promoter were generated and used to assess the SUMOylation of ADA2B in vivo. Database and Gene Ontology searches were performed as described previously (42).
Supplementary Material
Acknowledgments
We thank Steven Triezenberg, Christian Luschnig, Andreas Bachmair (University of Vienna, Vienna), Fabio Fornara, and RIKEN BRC for materials, and Maria Elisa de Ansorena Pablos for excellent technical assistance. This work was funded by grants from the Max Planck Society and the Sonderforschungsbereich 635.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005452107/-/DCSupplemental.
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Xu XM, et al. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palancade B, Doye V. Sumoylating and desumoylating enzymes at nuclear pores: Underpinning their unexpected duties? Trends Cell Biol. 2008;18:174–183. doi: 10.1016/j.tcb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, et al. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat Genet. 2004;36:507–511. doi: 10.1038/ng1336. [DOI] [PubMed] [Google Scholar]
- 6.Li T, et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc Natl Acad Sci USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 8.Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 9.Vertegaal AC, et al. A proteomic study of SUMO-2 target proteins. J Biol Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA. Broad-spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J Biol Chem. 2004;279:20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 11.Wykoff DD, O'Shea EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 13.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 14.Dohmen RJ. SUMO protein modification. Biochim Biophys Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 16.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 17.Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A. SUMO conjugation in plants. Planta. 2004;220:1–8. doi: 10.1007/s00425-004-1370-y. [DOI] [PubMed] [Google Scholar]
- 18.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura K, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catala R, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin JB, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid–mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves PH, Murtas G, Dash S, Coupland G. Early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development. 2002;129:5349–5361. doi: 10.1242/dev.00113. [DOI] [PubMed] [Google Scholar]
- 24.Murtas G, et al. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti L, et al. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talamillo A, Sánchez J, Barrio R. Functional analysis of the SUMOylation pathway in Drosophila. Biochem Soc Trans. 2008;36:868–873. doi: 10.1042/BST0360868. [DOI] [PubMed] [Google Scholar]
- 27.Budhiraja R, et al. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 2009;149:1529–1540. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miura K, et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li SJ, Hochstrasser M. The Ulp1 SUMO isopeptidase: Distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol. 2003;160:1069–1081. doi: 10.1083/jcb.200212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: A Web server for sumoylation site prediction. Nucleic Acids Res. 2006;34(Web server issue):W254–W257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 34.Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell. 2009;35:669–682. doi: 10.1016/j.molcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner A, Moutty MC, Möller U, Melchior F. Performing in vitro sumoylation reactions using recombinant enzymes. Methods Mol Biol. 2009;497:187–199. doi: 10.1007/978-1-59745-566-4_12. [DOI] [PubMed] [Google Scholar]
- 36.Uchimura Y, Nakao M, Saitoh H. Generation of SUMO-1 modified proteins in E. coli: Towards understanding the biochemistry/structural biology of the SUMO-1 pathway. FEBS Lett. 2004;564:85–90. doi: 10.1016/S0014-5793(04)00321-7. [DOI] [PubMed] [Google Scholar]
- 37.Okada S, et al. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: Functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 2009;50:1049–1061. doi: 10.1093/pcp/pcp056. [DOI] [PubMed] [Google Scholar]
- 38.Colby T, Matthäi A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachonasios KE, Thomashow MF, Triezenberg SJ. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell. 2003;15:626–638. doi: 10.1105/tpc.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieberer T, Hauser MT, Seifert GJ, Luschnig C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr Biol. 2003;13:837–842. doi: 10.1016/s0960-9822(03)00327-0. [DOI] [PubMed] [Google Scholar]
- 41.Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 42.Al-Shahrour F, et al. FatiGO+: A functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35(Web server issue):W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurepa J, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 44.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 45.Watts FZ. The role of SUMO in chromosome segregation. Chromosoma. 2007;116:15–20. doi: 10.1007/s00412-006-0079-z. [DOI] [PubMed] [Google Scholar]
- 46.Lee MT, Bachant J. SUMO modification of DNA topoisomerase II: Trying to get a CENse of it all. DNA Repair (Amst) 2009;8:557–568. doi: 10.1016/j.dnarep.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.