Abstract
The structure of the protein-translocating channel SecYEβ from Pyrococcus furiosus at 3.1-Å resolution suggests a mechanism for chaperoning transmembrane regions of a protein substrate during its lateral delivery into the lipid bilayer. Cytoplasmic segments of SecY orient the C-terminal α-helical region of another molecule, suggesting a general binding mode and a promiscuous guiding surface capable of accommodating diverse nascent chains at the exit of the ribosomal tunnel. To accommodate this putative nascent chain mimic, the cytoplasmic vestibule widens, and a lateral exit portal is opened throughout its entire length for partition of transmembrane helical segments to the lipid bilayer. In this primed channel, the central plug still occludes the pore while the lateral gate is opened, enabling topological arbitration during early protein insertion. In vivo, a 15 amino acid truncation of the cytoplasmic C-terminal helix of SecY fails to rescue a secY-deficient strain, supporting the essential role of this helix as suggested from the structure.
Keywords: nascent protein chain, secretion, translocation, membrane protein folding
Living cells are compartmentalized by membranes that act as hydrophobic physical barriers at the outer membrane or in organelles. Proteins destined for transport across or insertion into biological membranes are usually targeted by cleavable signal sequences or transmembrane segments that act as surrogate signal sequences. Several membrane protein complexes are involved in the transport and membrane integration of polypeptides. Protein translocation is mediated by an evolutionarily conserved membrane protein complex, the translocon, called SecY in Bacteria and Archaea and Sec61 in Eukaryotes (1). The complex consists of a large multiple transmembrane-spanning subunit Sec61α/SecY and two small subunits Sec61γ/SecE and Sec61β/SecG. Although the minimal Sec61αγ/SecYE assembly constitutes the functional and essential core of the translocating channel (2, 3), Sec61β/SecG seems to be dispensable. The Sec61/Y channel associates with different partners that provide the driving force for translocation. During cotranslational protein targeting, the Sec61/SecY complex interaction with a translating ribosome is mediated by the signal recognition particle (SRP) and its receptor (4). In posttranslational translocation, the eukaryotic Sec61 complex associates with Sec62/63, another membrane-embedded complex, and the luminal ATPase binding protein (BiP) in the endoplasmic reticulum. In Bacteria, SecY associates with the ATPase SecA to translocate polypeptides (5); SecA then functions by pushing the polypeptide through the translocon. The electrochemical gradient across the membrane is essential in vivo and was shown to stimulate translocation in vitro (6). Archaea, however, lack the equivalent of the bacterial SecA or the eukaryotic Sec62/63, and it is unclear how they perform posttranslational protein translocation.
The first X-ray structure of the Methanococcus jannaschii (Mja) SecYEβ suggested that the functional channel was composed of a single heterotrimer (7); this was subsequently confirmed by the homologous structure of Thermus thermophilus (Tth) SecYE (8). In the case of bacterial posttranslational targeting mediated by the motor ATPase SecA (9), cross-linking suggested that protein translocation is mediated by oligomers of the SecY complex, with the active pore formed by just a single copy of SecY. However, the recent crystal structure of the Thermotoga maritima (Tma) SecYEG•SecA complex (10) combined with cysteine cross-linking experiments (11) supports a monomer of SecA acting to translocate proteins posttranslationally through a single SecYEG channel. The structure of a eukaryotic translocon seemingly caught in the act was recently revealed by single-particle cryo-EM in a complex formed between ribosomes harboring a nascent polypeptidic chain and purified Sec61 (12). This showed that a single Sec61 complex is the active state in cotranslational protein translocation. Three regions of the SecY subunit have been shown to be functionally important and are referred to as the central plug, the lateral gate, and the ring region. It is not clear whether the plug and the lateral gate are essential to the function of the channel. Here, we report a translocon X-ray structure that provides insights into fundamental aspects of cotranslational protein translocation. Our structure shows a portion of the C-terminal helix of an adjacent symmetry-related molecule that is inserted into the pore as if mimicking a polypeptide in transit.
Results and Discussion
Structure Determination of the Translocon from Pyrococcus furiosus.
The SecYEβ complex from the hyperthermophile archaeon Pyrococcus furiosus (Pfu) was expressed in Escherichia coli and purified as a heterotrimeric complex stabilized in n-octyl-β-D-glucopyranoside. An initial 3.5-Å resolution multiwavelength anomalous dispersion (MAD) X-ray diffraction dataset collected on one crystal of seleno-methionine substituted protein was used to determine the structure combining molecular replacement and MAD phasing methods. A 2.9-Å resolution native dataset corrected for anisotropic diffraction was used to complete the refinement (Materials and Methods, SI Materials and Methods, Tables S1–S3, and Movie S1).
Architecture of the Pfu-SecYE Complex.
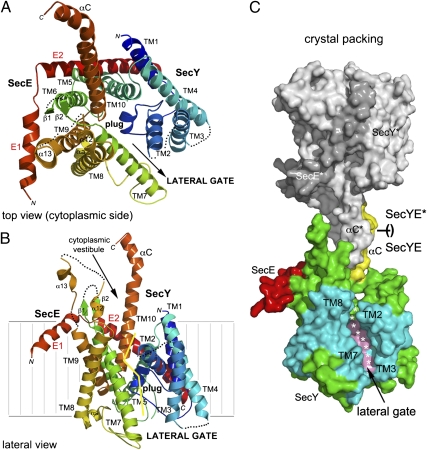
The two essential subunits, SecY and SecE, constituting the evolutionarily conserved core (2) of all Sec-translocases are visible in the structure (Fig. 1 and Fig. S1). Although the β subunit (13) is not present, the translocation channel is fully functional without this subunit (3, 8). The Pfu-SecY subunit retains the overall architecture described in homologs from Mja (7), Tth (8), and Tma (10) arranged such that the first five transmembrane helices (TMs 1–5) are related by a quasi-symmetrical 2-fold symmetry in the plane of the membrane to the second five helices (TMs 6–10). The SecE subunit is composed of a 33-residue-long highly tilted transmembrane helix (E2) linked to a 16-residue-long amphipathic helix (E1) that would probably lie along the membrane surface. A bend is located at a conserved Lys-Lys-Pro motif (Pro25/Pro29 in Pfu/Mja) such that SecE wraps around the SecY subunit to act as a clamp holding the two halves together (Fig. 1 A and B). Despite the dynamic nature of the SecY complex, the localization of all TMs was unambiguous in the initial electron density map (Fig. S2 and Movie S1).
Fig. 1.
Overview of the structure of Pfu-SecYE and the crystal packing. SecY is colored using a rainbow pattern. The plug (α3) and C-terminal (αC) helices are labeled. (A) Top view showing the cytoplasmic vestibule and the pore occluded by the plug. The two halves constituting SecY are delineated with an arrow indicating the lateral opening. (B) View showing the lateral gate opening between transmembrane helices TM2 and TM3 (in the N-terminal one-half) and TM7 and TM8 (in the C-terminal one-half). The yellow line delineates the lateral gate on the SecY subunit. (C) Surface representation of two complexes as they pack in the crystal. A SecYE complex interacts through the insertion of its C-terminal αC-helix in the cytoplasmic vestibule of a crystallographically related complex (SecYE* in gray). The plug and αC-helices are colored in pink and yellow, respectively. TMs delineating the opened lateral gate are colored in cyan.
Crystal Packing Suggests the Path Followed by a Nascent Chain and the Reason That the Lateral Gate of the Channel Is Opened.
Two adjacent SecYE complexes are brought together and face each other through both of their SecY cytoplasmic loops, whereas SecE is not involved in crystallographic packing. The most prominent feature is the interaction between the long cytoplasmic C-terminal helix (TM10-αC), the cytoplasmic loops formed between TMs 6/7 and TMs 8/9, and the αC-helix of an opposing symmetry-related molecule (Fig. 1C and Movie S2). Thus, this neighboring C-terminal αC-helix acts a nascent chain mimic.
This differs from the Mja-SecY structures, where the packing is completely different and does not involve the interaction shown in Fig. 1C. The same observation applies to the Tth-SecYE structure, where the use of an antibody for crystallization prevented such packing. In the Mja structure, the tracing of the SecY chain ends prematurely at position Leu426 (Met458 in Pfu), whereas in Pfu, the structure is perfectly defined up to the last residue, Ala468, folding at the solvent-exposed C-terminal αC-helix. In the Tma-SecYEG•SecA structure, this region is extended and featureless, buried at the interface between the SecA ATPase and the translocation channel. In our structure, a surface area of about 1,000 Å2 is buried on the face-to-face interaction between two crystallographically related SecY complexes. Entry of this pseudo-protein substrate is in concert with widening of the cytoplasmic vestibule and opening of the lateral gate.
Comparison with the Mja-SecYEβ, Tth-SecYE, and Tma-SecYEG•SecA Structures.
Translocon structures and henceforth, mechanisms are likely to be conserved; thus, we hypothesize that differences observed in structures from different species probably reflect the trapping of particular conformational states from a highly dynamic process. Although the fold and organization of subunits are similar among all known translocon structures, superposition reveals that our structure differs significantly from others in several aspects. First, the first helix of Pfu-SecE is tilted away from the SecY core by another 30° away from vertical, thus releasing the clamp that it exerts on SecY as observed in Mja-SecY (Fig. 2 A and B). Second, the C-terminal helix of the Pfu-SecY subunit is completely defined and extends much farther than seen in the Mja, Tth, or Tma structures. The selenium of seleno-Met458 in this element of secondary structure was readily placed in the initial maps, enabling unambiguous tracing of the helix (Fig. 2B). As a result, the cytoplasmic face of this translocon structure exposes all three cytoplasmic loops contributed by the two-stranded β-sheet between TMs 6/7, helices α13 and α14 between TMs 8/9, and the C-terminal helix αC (or α16). The corresponding surfaces create a paw-like structure overlooking the vestibule of the channel (Fig. 1 and Fig. S1).
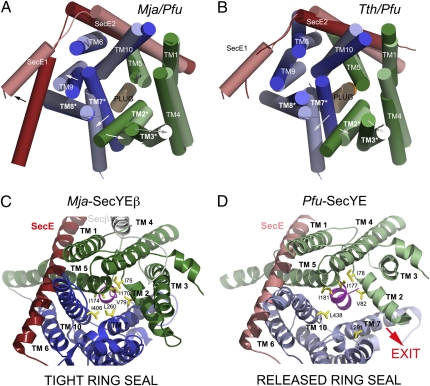
Fig. 2.
Conformational changes observed in the translocons from Pyrococcus, Thermus, and Methanococcus. Superposition of the Mja and Pfu channels (A) and the Tth and Pfu channels (B). The top view from the cytoplasmic side emphasizes the rearrangement of the N terminus of SecE and the rigid body motions of SecY TMs. The plug helix remains in place. TMs 2, 3, 7, and 8 delineate the lateral gate. The SecE subunit is colored in red. The arrows show movements from Mja or Tth to Pfu. The hydrophobic seal provided by the ring region is compromised on lateral gate opening. Cytoplasmic view along the channel pore showing the rearrangement of the hydrophobic ring that seals the cytoplasmic vestibule of the channel above the plug region. (C) Mja structure with a closed ring. (D) Pfu structure with an open ring. The N- and C-terminal halves of SecY are colored in green and blue, respectively. TMs and ring residues are labeled. The plug helix is colored in pink. Pfu is in light colors, and Mja or Tth is in dark colors.
The SecY subunit undergoes changes even more dramatic than those observed in SecE. These changes affect both its TM segments and its cytoplasmic loop regions. TMs 2 and 3 in the N-terminal one-half and TMs 7 and 8 in the C-terminal one-half translate as rigid bodies 4–6 Å away from the pore axis (Fig. 2 A and B). However, TMs 5 and 6 do not move but rather, act as a hinge delineating the N- and C-terminal halves. These shifts are the result of the hinge motion that opens the lateral gate defined by TMs 2/3 and 7/8.
The Hydrophobic Seal Provided by the Ring Region Is Compromised on Lateral Gate Opening.
In contrast to the Mja-SecY, the lateral rearrangements of the TMs compromise the seal that would normally be provided by residues Ile78, Val82, Ile177, Leu181, Leu291, and Leu438 (Fig. 2 C and D and Fig. S1). In the tightly sealed Mja pore, the narrowest constriction is 4.6 Å; the corresponding dimension in the Pfu pore is increased to 13.6 Å in the direction of the lateral exit site. The widest point in the Mja pore is 7.9 Å, with the corresponding point in Pfu increased to 10.3 Å. The most dramatic consequence of the partial opening of the lateral gate and the disruption of the seal is the creation of a cavity above the plug helix that still occludes the center of the channel. This cavity is large enough to accommodate the helix of a signal sequence packed against the lateral gate helix TM 7 in direct contact with the phospholipids, and it could potentially act as an Anfinsen cage, allowing proper topological arbitration of the incoming nascent chain in cooperation with the ribosome. Although the hydrophobic core of a signal sequence forms an α-helix of about two turns and is contacted on opposite sides by TMs 2 and 7, its N terminus contacts TM8 (14). Furthermore, during translocation, residues of the signal peptide are in contact with membrane lipids as shown by cross-linking. Based on the present structure, which may resemble an intermediate in protein insertion, we propose a model in which a bound-signal sequence contacts all three TMs, and membrane lipids gain access through the lateral opening of the channel.
The Plug Occludes the Pfu-SecY Channel While the Lateral Gate Opens.
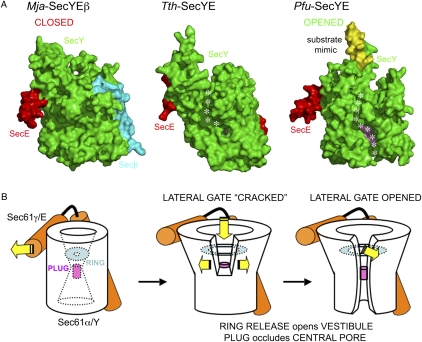
The region between TMs 1 and 2 corresponds to the plug region. In E. coli, the plug domain was shown to stabilize the closed state and to be involved in the maintenance of a membrane seal. Together, the helical plug and ring regions are required to maintain a membrane seal (15) and prevent ion and solute leakage (16). In our structure, as in the Mja and Tth structures, the plug remains in place, and thus, the channel is occluded; however, unlike the Mja and Tth structures, the lateral gate is entirely open in the Pfu structure (Fig. 3 and Fig. S3 A and B).
Fig. 3.
The lateral gate of the channel of the Pfu-SecYE channel is entirely opened while its plug remains in place, still occluding the central conduit. (A) Surface representations of the translocons from Mja, Tth, and Pfu showing the different states of the lateral gate. The SecY, SecE, and Secβ subunits are colored in green, red, and cyan, respectively. The αC-helix contributed by a symmetry-related SecYE complex is shown as a yellow surface to emphasize its insertion into the cytoplasmic vestibule. In Tth and Pfu, the so-called hydrophobic crack observed in Tth-SecYE (8) and the complete lateral gate opening in Pfu-SecYE are shown as a dotted white line. The plug helix is colored in pink and can only be clearly seen in the Pfu structure because of the extensive lateral gate. (B) Schematics of the three conformations shown in A.
The crystal structures of two plug deletion mutants in Mja-SecY show that the overall structure is maintained and new plugs are recruited (15), thus pointing to remarkable plasticity and structural adaptability of the channel. This is confirmed in Pfu-SecY, because the plug occupies the middle of the conduction path while the lateral gate is open. However, in the Mja deletions, the new plugs no longer stabilize the lateral gate. Plug deletion mutants are strong suppressors of defective or missing signal sequences.
In yeast, the plug seems to be important for efficient protein translocation but is not essential for cell viability (17). The Tth-SecYE structure revealed a partial opening of the lateral gate, referred as the hydrophobic crack (8), opening solely on the cytoplasmic side of the channel. In the Tma-SecYEG•SecA complex, the lateral gate is also partially opened on the cytoplasmic side but to a lesser extent. In Pfu-SecYE, the lateral gate of the channel is open, with the hydrophobic crevice now spanning the entire thickness of the membrane (Movie S2). This seems to be a direct effect of the insertion of the C-terminal helix from a neighboring SecY molecule into the cytoplasmic vestibule, thus acting as substrate mimic or nascent chain surrogate. The resulting crevice is about 11 Å wide and large enough to accommodate lipids or a 15 amino acid-long α-helical signal sequence.
In Vivo Rescue of SecY-Deficient E. coli by P. furiosus or E. coli SecY Requires the Cytoplasmic C-Terminal Helix of the SecY Subunit.
The structure suggests that the C-terminal helix may function in recruiting the signal sequence to the entry portal or guiding the nascent chain through the conduit. To investigate the possible functional importance of the C-terminal residues of SecY in Pfu, we performed in vivo complementation tests in E. coli using a thermosensitive secY mutant strain (secY24 DHB7302; SI Materials and Methods) (15). We generated C-terminal deletion mutants of SecY from P. furiosus (Pfu-YΔ) and E. coli (Ec-YΔ), in which the 15 (Pfu) or 20 (Ec) residues that terminate the cytoplasmic extension of the C-terminal helix of SecY were deleted (yielding species Met1-Glu453 in Pfu and Met1-Leu423 in Ec) (Fig. S1). Such deletion only removes the most C-terminal part of the cytoplasmic end of SecY and does not affect TM10 that remains intact (Fig. 4). Wild-type and truncated translocons expressed at similar levels and could be solubilized from membranes as complexes stable in detergent (Fig. S4). Although the secY24 mutant was rescued by the wild-type archaeal or bacterial SecY, the corresponding C-terminal truncated mutants failed to support growth at the nonpermissive temperature (Fig. 4A).
Fig. 4.
In vivo rescue of the secY-deficient bacterial strain by the archaeal SecY requires its C-terminal cytoplasmic helix. Complementation of the secY24 ts mutant E. coli strain with vectors expressing no gene (empty), the E. coli-SecY or Pfu-SecY gene clusters in the case of complete truncation (A), or point mutations (B) in the cytoplasmic C terminus of Pfu-SecY. The growth patterns at permissive and nonpermissive temperatures are shown. Pfu-Y and Ec-Y segments indicate the presence of wild-type full-length proteins; Δ segments indicate the presence of C-terminal deletion mutants in which 15 residues (454–468) are deleted from Pfu-SecY or 20 residues (424–443) are deleted from E. coli-SecY. Point mutants in the full-length Pfu protein are labeled E457A, F459A, R463A, K464A, R463A/K464A, and F459P/A461P. (C) Model of the ribosome/translocon interaction based on our structure aligned and the Sec61-translocon cryo-EM structure (12). At the ribosomal tunnel exit site, the C-terminal helix of SecY (red) packs against ribosomal RNAs (yellow) and proteins (blue). Δ indicates the position of the deletion. The sequence of Pfu-SecY is shown; the area shaded in yellow indicates the C-terminal truncation. Membrane boundaries are depicted as brown lines.
Two conclusions arise. First, an archaeal translocon can function in bacteria despite the significant differences in the membrane composition and structure between Bacteria and Archaea. Second, the C-terminal end of SecY is essential to the receptor process of protein targeting and cell viability. Because there is no known equivalent to the bacterial motor ATPase SecA in Archaea, the lethal phenotype observed for the C-terminal deletion mutants suggests a disruption of the interaction between the translocon and the SRP or the ribosomal machinery (which is conserved between Bacteria and Archaea), perhaps by preventing or interrupting signal sequence docking or nascent chain progression through the channel.
Both Structure and Sequence of the C-Terminal End of SecY Seem to Be Essential.
Point mutants along the C-terminal helix of Pfu-SecY were generated and tested for their ability to rescue the E. coli secY24 strain (Fig. 4B). Although single mutants E457A, F459A, R463A, and K464A are viable, the double mutants R463A/K464A or F459P/A461P fail to rescue. All SecY/61α sequences harbor basic amino acids at their C termini but without any strict sequence consensus (Fig. S1). The R463A/K464A double mutation removes the only cluster of basic amino acids present in the C terminus of Pfu-SecY; thus, the basic cluster is likely to be involved in anchoring the channel to the ribosome. Homology modeling suggests that these basic amino acids are vicinal to bases A535/A536 in the 25S ribosomal RNA (eukaryotic nomenclature) (Fig. 4C). The double mutation F459P/A461P introduces a triple proline P459-P460-P461 sequence in the middle of the cytoplasmic C-terminal helix of Pfu-SecY, disrupting its α-helical conformation. We propose that this C-terminal helix acts as a sensor of and a general guiding surface for the incoming nascent chain and relays conformational changes to the lateral gate.
General Model for the Priming of the Translocon for Protein Insertion.
We suggest a model for the path followed by the nascent chain in the initial steps of translocation (Fig. S5 and Movie S3). Our structural and functional analyses reveal an unexpected role for the C-terminal helix of SecY in an essential step of the translocation process and suggest that this helix is likely to interact with the nascent chains or signal sequences. The conformational changes observed show that the cytoplasmic vestibule widens and the lateral gate opens without significant rearrangement of the central plug. These observations suggest an effect on an early step of the translocation reaction, whereby the nascent chain or the signal sequence is docked against the cytoplasmic loops and C-terminal helix.
Mutagenesis in vivo in yeast showed that cytoplasmic loops CL2 and CL3 are involved in ribosome binding and cotranslational translocation, respectively (12, 18). The E. coli SecY-ribosome cryo-EM structure (19) shows that these loops come into contact with ribosomal RNAs located near the ribosomal exit tunnel. The most recent cryo-EM structure of the yeast Sec61-ribosome nascent chain complex (12), while revealing the role of cytoplasmic loops CL2 and CL3 in ribosome binding, did not involve the C terminus. The subnanometer resolution and the use of SecY structures disordered at their C termini as docking models probably obscured this level of detail. Superposition of our complex on this structure confirms that cytoplasmic loops CL2 and CL3 are well-accommodated and positioned at the tunnel exit site of the RNC, and it also indicates that the newly observed C-terminal cytoplasmic helix of Pfu-SecY projects itself at the exit of the ribosomal tunnel and fits remarkably well against ribosomal protein L26 (eukaryotic nomenclature) and helix H4 of the 25S ribosomal RNA (Fig. 4C). The incoming nascent chain is likely to be sandwiched between these three cytoplasmic elements of the channel. Molecular dynamics simulations of the SecY-ribosome association also suggest an interaction between the C terminus of SecY and the ribosome at a similar position (20).
Docking of the nascent chain mimic seems sufficient to trigger a lateral opening of the SecY subunit by the release of the clamping effect that SecE exerts on SecY; alternatively, the energetics of lateral gate opening must be small in our detergent environment. As this clamp is released, the substrate can progress farther inside the widening cytoplasmic vestibule. The channel plug is maintained but not the ring that loosens to accommodate the incoming nascent chain; this supports the idea that a substantial length of the folded (or at least, partially folded) nascent chain could be inserted inside the cytoplasmic vestibule before the vertical translocation seal is released through dislocation of the plug. The primed state of the translocon that we have trapped here could be essential to the fate of the substrate in terms of topology of insertion into the membrane. Although the ring opens to accommodate a substrate as it enters the cytoplasmic vestibule, the plug still maintains a central seal, preventing ion and solute leakage.
The crevice generated by opening of the lateral gate cannot only accommodate the signal sequence but is also likely to allow its interaction with host phospholipids, and membrane permeability is not compromised. This suggests that the lateral gate and the plug are, to a certain extent, independent of each other. Although the lateral gate opens as a substrate enters the cytoplasmic vestibule, the plug has to be actively displaced by the nascent chain as it progresses through the channel.
The closed state of the translocon is stabilized by hydrogen bonds between highly conserved residues; perturbations caused by mutations at various locations are rapidly relayed to the plug (21) and facilitate its displacement through increased hydration. Our structure also supports the notion that the channel, more precisely, its lateral gate, is exquisitely sensitive to the presence of a nascent chain in the vestibule. It also underlines the importance of early lateral gate opening, independent of plug displacement, during membrane protein biogenesis. This is supported by the observation that disulfide bridges introduced between lateral gate TMs 2 and 7 impair protein translocation because they restrict lateral gate opening (22). It is possible that, whatever the fate of the translocated protein (e.g., membrane-inserted versus secreted), the lateral gate is continuously opened while the nascent chain threads through the channel; lipids could constantly be in contact with the nascent chain, whether it is folded or not, and thus, influence the process. Studies of the folding of secretory proteins or TM sequences within the ribosomal tunnel have shown that, although secretory proteins are in an extended conformation, TM sequences adopt and maintain a compact conformation (23). More recently, α-helical nascent polypeptide chains have been visualized within distinct regions of the ribosome exit tunnel using cryo-EM (24). Membrane lipids and ribosomal recognition of the nascent chain folding may, thus, control the function of the translocation channel.
Signal sequences orient themselves in the channel on the basis of their flanking charges. In vitro translation of model proteins has been used to decipher a molecular code for TM helix recognition by the translocon (25, 26) and to reveal the effects of TM segment lengths and sequences on membrane insertion efficiency. Hydrophobic N-terminal signals initially insert head first before inversion of their orientation to translocate their C-terminal end. The rate of inversion is reduced with increasing hydrophobicity of the signal because of an increased affinity of the initial bound state at the translocon (27). Such inversion generally places the more positive end of the translocated segment on the cytosolic side of the membrane and is generalized as the positive-inside rule. Attempts to model a nascent chain (Fig. S6) in our opened and primed channel suggest that the size of the vestibule can allow for the reorientation of TM segments in a relatively protected environment and acquisition of tertiary structure.
On entry into the channel, signal sequences have been shown to be in contact with lipids, the plug helix, and the TM7 part of the lateral gate (28–30). Molecular dynamics simulations suggest that plug deletions would allow translocation and lateral gate opening more easily than the native closed Sec61 (31–33). This agrees with the observation that deletion of the plug negates the requirement of a signal sequence for channel opening (15). Mutational analyses of yeast Sec61 show the role of the Sec61a subunit in signal sequence orientation according to the positive-inside rule (34–36). Assignment of topology is partially determined by electrostatic interaction between charged residues flanking the signal core and charged residues located at the plug and the C-terminal end of TM8 in the channel; these residues are likely to contribute to an electrostatic clamp. The study of the Sec61-mediated biogenesis of aquaporin-4, an all α-helical integral polytopic protein (37, 38), showed that each TM interacted with and moved through Sec61α in a highly ordered and sequential manner and suggested the presence of a single primary binding site. Our structure may reveal the modality of such interactions at this primary binding site.
The present Pfu-SecYE structure reveals (i) the major conformational changes undergone by the channel as it accommodates a substrate mimic, (ii) a potential nascent chain binding and docking surface involving the C-terminal helix of SecY and adapted to the ribosomal tunnel exit, (iii) a clear lateral exit route to mediate the insertion of TM segments of membrane proteins and the establishment of their topology, and (iv) the intrinsic dynamics of the lateral gate of the translocon.
Conclusions.
Our Pfu-SecYE structure reveals a monomeric channel undergoing structural rearrangements to accommodate an α-helical peptide aligned along the translocation path and docked against the three main cytoplasmic loops present in all SecY sequences. The channel seems primed for sideways insertion of the individual TM segments of an integral membrane protein into the lateral plane of the membrane. Functional analysis of mutants and deletion of 15 amino acids from the C terminus in vivo support the involvement and importance of the C-terminal helix of the SecY subunit at an essential stage of membrane protein biogenesis. Based on the structure, we propose a general mechanism for nascent chain docking to and guidance along the protein-conducting channel and lateral insertion in the membrane plane. The knowledge of the structure of a channel containing trapped nascent chain intermediates is required to further elucidate the mechanisms that govern protein insertion and folding in membranes. The structures of all of the proteins constituting the SecY and SRP machineries (39, 40) from the archaeon P. furiosus are now available, providing us with a robust model system to further investigate the molecular interactions between the SecY and the SRP components and provide mechanistic insights about the SRP-mediated interactions between the protein translation and translocation machineries.
Materials and Methods
Materials and methods describing the protein expression, purification, and crystallization, the structure resolution, and the in vivo complementation tests are available in SI Materials and Methods. The SecYEb complex from P. furiosus was expressed in E. coli and purified in the detergent n-octyl-β-D-glucoside using selective heat precipitation, Ni-affinity chromatography, and gel-filtration chromatography. In vivo complementation tests were performed using the secY24 thermo-sensitive mutant E. coli strain. Data were collected at the beamline 8.3.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory). The structure was solved combining molecular replacement and seleno-MAD methods at 3.5-Å resolution. Refinement was completed with a final native dataset, collected to 2.9-Å Bragg spacings, after performing ellipsoidal truncation (dmin = 3.3, 4.1, and 2.9 Å along a*, b*, and c*, respectively) and anisotropic scaling. The model is complete except for poorly ordered segments at the N terminus and in cytoplasmic loops TM2-TM3, TM6-TM7, and TM8-TM9 and periplasmic loop TM3-TM4.
Supplementary Material
Acknowledgments
We thank Thomas Terwilliger, William Studier, Pavel Afonine, Nathaniel Echols, and Mike Sawaya for advice. We are grateful to James Holton and George Meigs for support at beamline 8.3.1 at the Advanced Light Source in Berkeley. We are grateful to Patricia Greene for help with the manuscript. Strain DHB7302 was provided by Tom Rapoport (Harvard Medical School, Boston). This work was funded by National Institutes of Health Grants GM60641 (to P.F.E.; no longer active) and GM94625 (to the Center for Structure of Membrane Proteins).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3MP7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012556107/-/DCSupplemental.
References
- 1.White SH, von Heijne G. The machinery of membrane protein assembly. Curr Opin Struct Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 3.Meyer TH, et al. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol. 1999;285:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- 4.Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr Opin Struct Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Rusch SL, Kendall DA. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry. 2007;46:9665–9673. doi: 10.1021/bi7010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Laan M, Nouwen N, Driessen AJ. SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. J Biol Chem. 2004;279:1659–1664. doi: 10.1074/jbc.M306527200. [DOI] [PubMed] [Google Scholar]
- 7.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 8.Tsukazaki T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erlandson KJ, et al. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker T, et al. Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome. New York: Science; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinch LN, Saier MH, Jr., Grishin NV. Sec61β—a component of the archaeal protein secretory system. Trends Biochem Sci. 2002;27:170–171. doi: 10.1016/s0968-0004(01)02055-2. [DOI] [PubMed] [Google Scholar]
- 14.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 15.Li W, et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Saparov SM, et al. Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell. 2007;26:501–509. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Junne T, Schwede T, Goder V, Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol Biol Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ménétret JF, et al. Ribosome binding of a single copy of the SecY complex: Implications for protein translocation. Mol Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 20.Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondar AN, del Val C, Freites JA, Tobias DJ, White SH. Dynamics of SecY translocons with translocation-defective mutations. Structure. 2010;18:847–857. doi: 10.1016/j.str.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 24.Bhushan S, et al. α-helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 25.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 26.Hessa T, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 27.Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 28.Higy M, Gander S, Spiess M. Probing the environment of signal-anchor sequences during topogenesis in the endoplasmic reticulum. Biochemistry. 2005;44:2039–2047. doi: 10.1021/bi047976z. [DOI] [PubMed] [Google Scholar]
- 29.Martoglio B, Dobberstein B. Protein insertion into the membrane of the endoplasmic reticulum: The architecture of the translocation site. Cold Spring Harb Symp Quant Biol. 1995;60:41–45. doi: 10.1101/sqb.1995.060.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 31.Gumbart J, Schulten K. Molecular dynamics studies of the archaeal translocon. Biophys J. 2006;90:2356–2367. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gumbart J, Schulten K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 33.Gumbart J, Schulten K. The roles of pore ring and plug in the SecY protein-conducting channel. J Gen Physiol. 2008;132:709–719. doi: 10.1085/jgp.200810062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goder V, Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goder V, Junne T, Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- 37.Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- 38.Daniel CJ, Conti B, Johnson AE, Skach WR. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J Biol Chem. 2008;283:20864–20873. doi: 10.1074/jbc.M803517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egea PF, Napetschnig J, Walter P, Stroud RM. Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus. PLoS ONE. 2008;3:e3528. doi: 10.1371/journal.pone.0003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egea PF, et al. Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: Implications for the targeting step at the membrane. PLoS ONE. 2008;3:e3619. doi: 10.1371/journal.pone.0003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.