Abstract
The CESA1 component of cellulose synthase is phosphorylated at sites clustered in two hypervariable regions of the protein. Mutations of the phosphorylated residues to Ala (A) or Glu (E) alter anisotropic cell expansion and cellulose synthesis in rapidly expanding roots and hypocotyls. Expression of T166E, S686E, or S688E mutants of CESA1 fully rescued the temperature sensitive cesA1-1 allele (rsw1) at a restrictive temperature whereas mutations to A at these positions caused defects in anisotropic cell expansion. However, mutations to E at residues surrounding T166 (i.e., S162, T165, and S167) caused opposite effects. Live-cell imaging of fluorescently labeled CESA showed close correlations between tissue or cell morphology and patterns of bidirectional motility of CESA complexes in the plasma membrane. In the WT, CESA complexes moved at similar velocities in both directions along microtubule tracks. By contrast, the rate of movement of CESA particles was directionally asymmetric in mutant lines that exhibited abnormal tissue or cell expansion, and the asymmetry was removed upon depolymerizing microtubules with oryzalin. This suggests that phosphorylation of CESA differentially affects a polar interaction with microtubules that may regulate the length or quantity of a subset of cellulose microfibrils and that this, in turn, alters microfibril structure in the primary cell wall resulting in or contributing to the observed defect in anisotropic cell expansion.
Keywords: Arabidopsis, cell wall, CESA protein, microtubule, regulation
In higher plants, cellulose is a composite polymer of long unbranched β-1,4-linked glucan chains that are hydrogen-bonded to form microfibrils of approximately 36 chains (1). The microfibrils are the primary load-bearing component of cell walls (2, 3), and defects in cellulose synthesis or organization (4) result in decreased anisotropy of cell wall expansion and an inability to achieve differentiated cell shape (2–4).
Cellulose is synthesized by plasma-membrane complexes that have been visualized by freeze-fracture EM as hexagonal rosettes 25 to 30 nm in diameter (5). The complexes are thought to be composed of three functional types of structurally similar CESA proteins that are believed to be the catalytic subunits. For primary wall cellulose synthesis, CESA1 and CESA3 appear to be absolutely required whereas CESA2, CESA5, CESA6, and CESA9 are at least partially redundant (6). Dynamic visualization of CESA complexes in growing Arabidopsis cells showed that labeled (YFP::CESA6) synthetic complexes appeared as discrete particles at the plasma membrane that move bidirectionally along linear paths coincident with underlying cortical microtubules (7). These observations, together with previous (8) and subsequent studies (9–11), suggest that cortical microtubule organization plays a role in guiding the orientation of cellulose microfibril deposition.
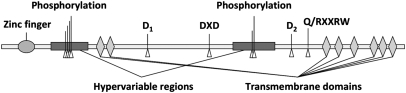
All CESA proteins are predicted to have two transmembrane helices in the N-terminal region and six in the C-terminal region (Fig. 1). Between the two regions lies a large central domain that, for the most part, is highly conserved among all CESA proteins, including three conserved aspartate residues and a Q/RXXRW motif that are believed to be important for substrate binding and catalysis. The central and N-terminal domains each contain a hypervariable region that is not found within the catalytic subunit of bacterial and cyanobacterial cellulose synthases (12). Comparison of CESA sequences from many species reveals that the hypervariable region in the central domain of CESA proteins differs between putative paralogues but is well conserved in putative orthologues, leading to this region being renamed the class-specific region (13).
Fig. 1.
Illustration of the structure of CESA1 (1081 aa). Indicated phosphorylation sites include S162, T165, T166, S167, S686, and S688.
A proteomics survey of plasma membrane phosphoproteins revealed that CESA1, CESA3, and CESA5 proteins were phosphorylated at a number of sites clustered in the two hypervariable regions (14). In this study we explored the effects of mutating the identified threonine or serine residues in CESA1 to alanine (A) or glutamate (E) residues to eliminate or mimic phosphorylation, respectively.
Results
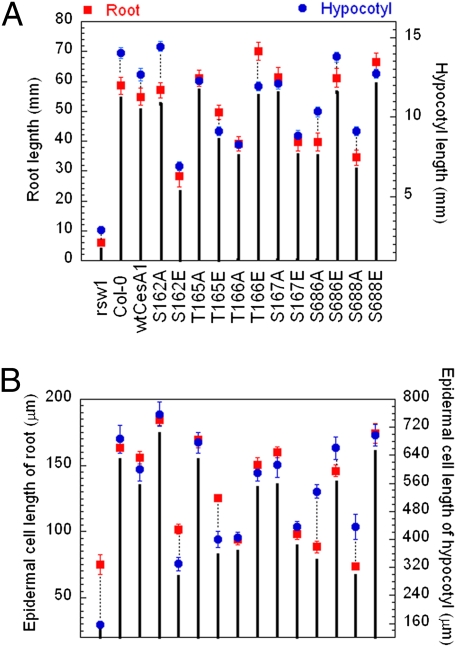
We mutagenized all known and potential phosphorylation sites in CESA1 to A and E by site-directed mutagenesis of a cDNA sequence. We refer to these as cesA1P mutations. The WT and mutated cDNAs were introduced into a temperature-sensitive cesA1 mutant, rsw1 (2), under transcriptional control of the native cesA1 promoter. For each of the 14 gene constructs, we analyzed at least three independent transgenic lines to confirm phenotypes. Tissue expansion was used as an indicator of CESA complex activity. As reported previously (2), growth of rsw1 was strongly inhibited when seedlings were grown at a restrictive temperature of 30 °C (Fig. 2A). Expression of WT CESA1 complemented the rsw1 mutation, and root elongation was restored to levels similar to those seen with Col-0 (Fig. 2A).
Fig. 2.
Lengths of primary roots and hypocotyls (A) and their epidermal cells at the mature zone (B) of Col-0, rsw1, and rsw1 transgenic lines containing WT CESA1 cDNA or cesA1P mutants. For root measurements, seedlings were grown in continuous light (200 μmol/m2/s) on vertical 0.5× MS agar plates at 30 °C for 7 d. For hypocotyl measurements, seedlings were grown on vertical 0.5× MS agar plates at 30 °C for 5 d in darkness. Mean values are shown with SEs (≥50 measurements).
Effects of Phosphorylation of the Central Domain.
In rsw1 lines expressing constructs with alanine substitutions at S686 and S688, roots and etiolated hypocotyls were significantly shorter than those in rsw1 expressing WT CESA1 (Fig. 2A). By contrast, when these residues were mutated to glutamate, a mild promotion of root length was observed for both mutants (P < 0.001 for S686E and S688E, t test) and in etiolated hypocotyl length for S686E (P < 0.001; Fig. 2A). In light-grown seedlings, the size of aerial organs of these four transgenic lines was unaltered relative to the WT control line. All together, these results suggest that phosphorylation of position 686 or 688 promotes CESA1 activity, particularly in tissues with rapid cell elongation.
Phosphorylation of the N-Terminal Domain.
Four residues were mutated in the N-terminal domain. Similar to the central domain mutations, a glutamate substitution at T166 promoted root growth, whereas an alanine substitution at this residue resulted in root stunting (a 27% increase and a 56% decrease, respectively, compared with WT transgenic line; Fig. 2A). A contrasting pattern of growth was observed for mutations at the other three residues in the N-terminal domain. The root lengths of S162E, T165E, and S167E transgenic lines were reduced relative to WT control whereas alanine mutations at these residues resulted in root lengths similar to WT. The size of aerial organs of light grown seedlings was not changed in the S162E, T165E, and S167E transgenic lines.
A similar pattern of changes in growth rate was observed for hypocotyl growth in dark-grown seedlings. Relative to the WT control, T166A caused an approximate 35% decrease in hypocotyl length (Fig. 2A). The hypocotyl length of the T166E transgenic line was, however, similar to that of the WT. Mutations of S162E, T165E, and S167E led to short hypocotyls relative to WT control or S162A, T165A, and S167A mutants, and a promotion of hypocotyl length was observed with the S162A mutant (P < 0.001). Considered together, these results suggest that phosphorylation of T166 results in a more active CESA complex whereas phosphorylation of surrounding residues (i.e., S162, T165, and S167) reduces activity.
Effects on Anisotropic Cell Expansion.
Radial swelling of the root is commonly the result of a loss in cell elongation anisotropy (15). At the nonpermissive temperature, the rsw1 mutation caused root swelling that was typically associated with root tips (Fig. S1). Root swelling was also observed in the transgenic lines that had displayed reduced root and etiolated hypocotyl lengths: T166A, S162E, S167E, S686A, and S688A (Fig. S1). However, the root tips of these transgenic lines were not swollen or abnormal (Fig. S1). Instead radial swelling was observed further up the root axis, in the elongation or differentiation zones of these transgenic lines. Radial swelling of the root axis was accompanied by both a reduction in cell length (Fig. 2B) and by swelling of individual epidermal cells in both the primary root axis and etiolated hypocotyl (Figs. S1 and S2). By contrast, the transgenic lines with normal or increased root or dark-grown hypocotyl elongation (S162A, T165A, T166E, S167A, S686E, and S688E) had relatively long epidermal cells (Fig. 2B) and cell swelling was not observed (Figs. S1 and S2). These results suggest that phosphorylation of T166, S686, or S1688 promotes anisotropic cell expansion, whereas phosphorylation of S162, T165, or S167 inhibits anisotropic cell expansion.
Effects on Cellulose Synthesis in the Primary Cell Wall.
Measurements of cellulose content showed that the transgenic lines with abnormal organ and cell expansion (S162E, T165E, T166A, S167E, S686A, and S688A) had slight but significant reductions of crystalline cellulose in their roots as compared with WT or mutant lines with normal organ and cell expansion (Table S1). However, compared with rsw1, all the transgenic lines contained relatively high levels of cellulose.
To examine directly the effects of the various mutations on cellulose synthase activity, we visualized CESA complexes in epidermal cells of dark-grown hypocotyls using a functional YFP::CESA6 fusion expressed from the native CESA6 promoter (7). Transgenic lines were created that contained both YFP::CESA6 and CESA1 transgenes in a background that was homozygous for both prc1 (7, 16) and rsw1.
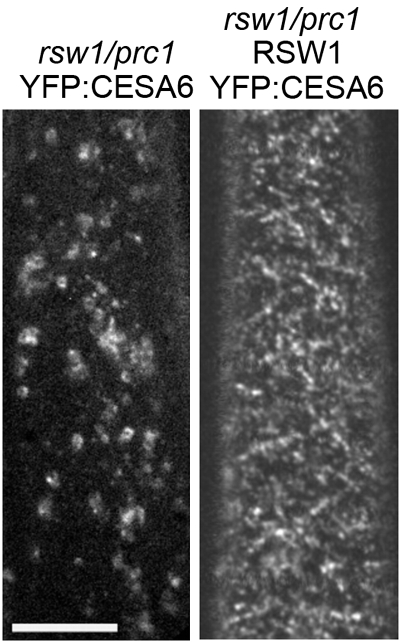
In control experiments, in which YFP::CESA6 was expressed in the rsw1/prc1 mutant, YFP::CESA6 was detected in the Golgi and plasma membrane at permissive temperature but was observed in the Golgi and other intracellular compartments only at nonpermissive temperature (Fig. 3). These observations are consistent with a previous freeze-fracture analysis of the rsw1 mutant (2), which demonstrated that exposure to restrictive temperature causes CESA rosettes to disappear from the plasma membrane. Expression of WT CESA1 under the CESA1 promoter in the rsw1/prc1/YFP::CESA6 line restored the accumulation of CESA complexes in the plasma membrane at nonpermissive temperature (Fig. 3).
Fig. 3.
Effect of temperature on the accumulation of YFP::CESA6 in Golgi and plasma membrane in an rsw1 prc1 background. Etiolated seedlings were grown at restrictive temperature (30 °C) for 2 d, then used to visualize CESA complexes at 29 °C. YFP::CESA6 localization in etiolated hypocotyl cells of rsw1/prc1-1 seedlings expressing YFP::CESA6 (Left) or both YFP::CESA6 and WT CESA1 (Right). Both transgenic lines are heterozygous for YFP::CESA6. (Scale bar: 10 μm.)
Effects of CESA1 Phosphorylation on Bidirectional Mobility of CESA Complexes.
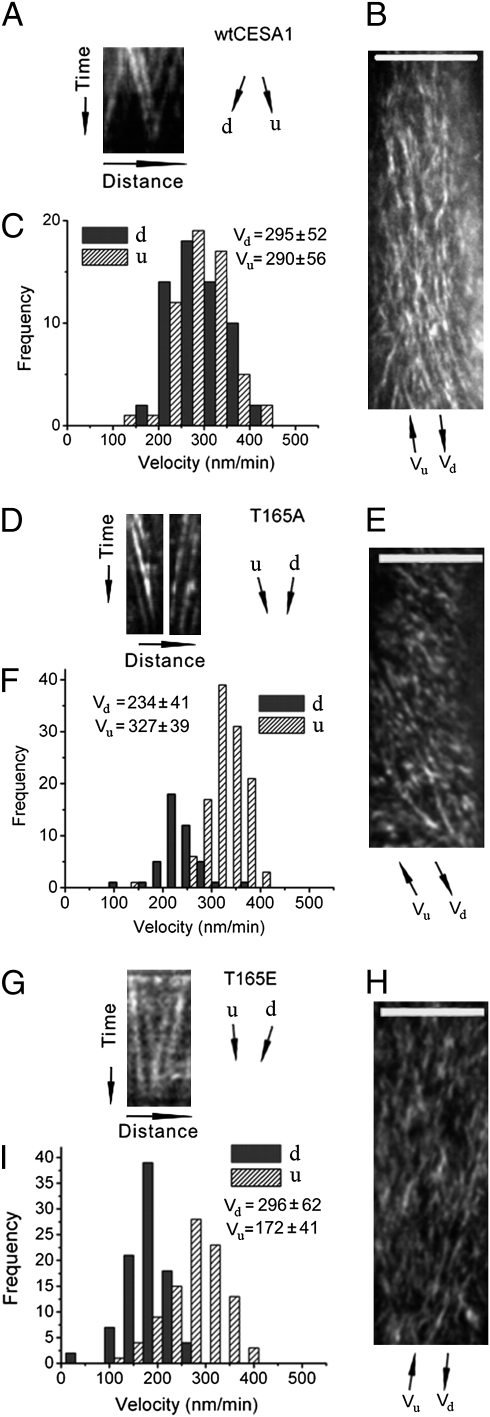
Previous measurements of movement of CESA complexes in the plasma membrane of epidermal cells in the upper hypocotyl of dark-grown seedlings revealed that WT CESA complexes move in opposite directions along linear tracks defined by cortical microtubules at similar rates (6, 7). We confirmed that CESA complexes in the WT control moved bidirectionally with similar velocities, as reflected by the similar slopes of the tracks derived from kymograph analysis of CESA complexes moving along common linear tracks (Fig. 4 A–C), and by measurement of particle movement in extended image regions using automated particle tracking analysis (Fig. S3 A–D and Table S1). The CESA particle trajectories also tended to be distributed evenly in opposite directions (50 ± 6% as measured by kymograph analysis of 2,916 particles in 11 cells from seven plants; Fig. 4 A–C and Movie S1) (6, 7). Most transgenic lines with patterns of organ and cell growth similar to WT (i.e., S162A, T166E, S167A, S686E, and S688E) also showed similar bidirectional velocities by both kymography analysis (Fig. S4) and automated particle tracking (Fig. S3 and Table S1). By contrast, all the transgenic lines with reduced organ elongation and loss of cell anisotropy (S162E, T165E, T166A, S167E, S686A, and S688A) showed a marked partitioning of particle velocities moving in opposite directions, as reflected by the different slopes of the tracks derived from kymograph analysis of CESA complexes moving along common linear tracks (Fig. 4 G–I and Fig. S4). The discrepancy was also observed by measurements of particle movement in extended image regions, i.e., labeled particles moving in one direction were, on average, significantly faster than those moving in the opposite direction (Fig. 4 G–I, Figs. S3 I–L and S4, and Table S1).
Fig. 4.
Kymograph analysis of bidirectional movement of CESA complexes in the WT, T165A, and T165E transgenic line. The data for the other mutants is presented in Fig. S4. Time-lapse confocal images of YFP::CESA6 in hypocotyl cells of etiolated seedlings grown at restrictive temperature for 2 d were used to measure CESA particle velocity at 29 °C. To analyze bidirectional movement of CESA particles, we defined the average axis of CESA particle movement in a given image series of a cell as the major axis and classified CESA particles as moving in upward (u) or downward (d) direction along the major axis, as illustrated in Fig. S4 A and B. (A, D, and G) Representative kymograph displays effects of mutations on bidirectional particle translocations, derived from kymograph analysis of CESA particles moving along common linear tracks. (B, E, and H) Average time projections of 61 frames representing 5 min. (C, F, and I) Histograms of particle velocities calculated from kymograph analysis of 100 to 200 particles from three to six cells of a single seedling. Similar results were obtained from different cells in different seedlings. (Scale bars: 10 μm in B, E, and H.)
T165A was an exception. This line had normal morphology but CESA complexes moved at different rates in opposite directions (Fig. 4 D–F, Fig. S3 E–H, and Table S1). However, in this line, a significant majority of labeled CESA particles moved in one direction along individual kymograph analysis tracks (70% and 30% as measured by kymograph analysis of 1,460 particles in nine cells from four plants; the probability of measuring these frequencies if the true frequency is 50% was P < 0.05 in nine of nine cells by binomial test; Fig. 4 D–F and Movie S2).
Altogether, the results suggest that organ and cell morphologies observed with the cesA1P mutants correlate not only with cellulose content but also with bidirectional motilities of CESA complexes in the plasma membrane of these transgenic lines, particularly with the velocity discrepancy of bidirectional movement of CESA complexes.
Effects of Oryzalin on Bidirectional Mobility of CESA Complexes.
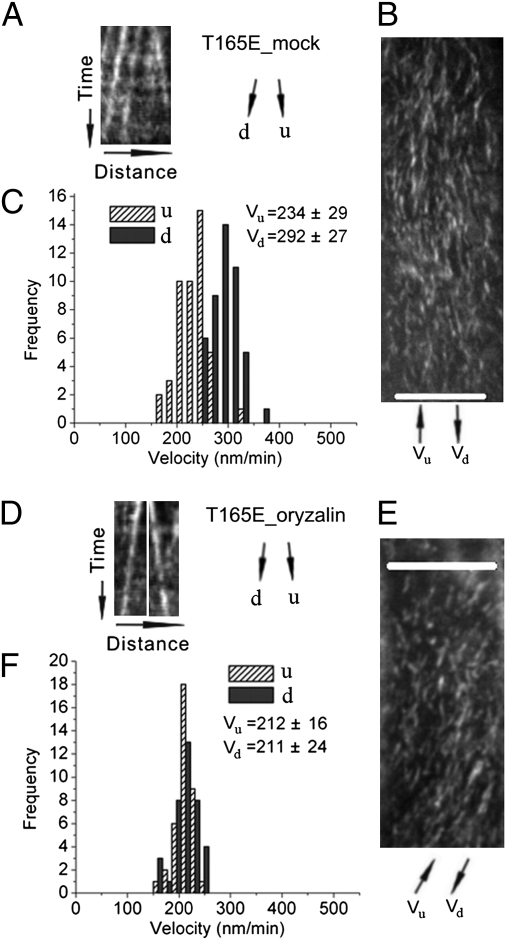
To examine whether the observed discrepancy of bidirectional movement of CESA complexes relates to interactions between CESA complexes and microtubules, we applied oryzalin, a drug that disrupts the cytoskeleton. After treatment of intact seedlings expressing the microtubule marker GFP::MAP4 at a nonpermissive temperature of 29 °C with 20 μM oryzalin for 2 h, most detectible microtubules were cleared from the cortex (Fig. S5), as also previously observed by Debolt et al. (9), at a permissive temperature. Kymograph analysis of CESA particle movement in both the T165A and T165E lines indicated that, after oryzalin treatment, CESA particle velocities no longer defined a bimodal population (Fig. 5 and Fig. S6). These results were confirmed by automated particle tracking analysis, which also indicted that CESA particle velocities after oryzalin treatment were not significantly different compared with those in untreated WT cells (Table S1).
Fig. 5.
Effects of oryzalin on bidirectional movement of CESA complexes in the T165E line. Time-lapse confocal images of YFP::CESA6 in hypocotyl cells of 2-d-old etiolated plants grown at 30 °C were used to measure CESA particle velocity. u and d represent two opposite directions of particle movement relative to the major axis (Fig. S4 A and B). (A–C) Control treatment with 0.02% methanol or DMSO at 30 °C for 2 to 3 h. (D–F) Treatment with 20 μM oryzalin at 30 °C for 2 to 3 h. (A and D) Representative kymograph displays effects of oryzalin treatment on bidirectional particle translocation. (B and E) Average time projections of 61 frames representing 5 min. (C and F) Histograms of particle velocities calculated from kymograph analysis of 100 to 200 particles from three to six cells of a single seedling. (Scale bars: 10 μm in B and E.)
We also examined effects of oryzalin on S167A, one of the transgenic lines that showed symmetrical bidirectional velocity. As observed previously with WT seedlings (9), oryzalin treatment appeared to have no significant effect on bidirectional velocities of CESA particle movement in this line compared with mock treatment (Fig. S6 and Table S1).
Discussion
Cell Expansion and Cellulose Contents in the Primary Cell Wall.
Organ morphology depends on cell placement and directional cell expansion, but the underlying mechanisms of primary cell wall expansion still remain incompletely understood (17). The primary cell wall has to be strong enough to withstand hydrostatic pressure and external loads as well as extensible enough to allow anisotropic cell expansion. It has been proposed that, in addition to the amount of cellulose in the primary cell wall, directional cell expansion depends on the orientation, length, and tensile properties of cellulose microfibrils (18).
Cellulose-deficient mutants, such as rsw1 (2) and korrigan (19, 20), show a dramatic loss of anisotropic expansion of light-grown and etiolated seedlings. Similar morphological effects can result from inhibition of cellulose synthesis by dichlorobenzonitrile (9). In the current study, cesA1P mutants that showed defects in cell expansion and axial growth also tended to have reduced cellulose content (Table S1). A simple hypothesis is that the observed defects in cell expansion were caused, at least in part, by a reduction in total cellulose content of the wall.
However, the reductions in cellulose content in cesA1P mutants with cell growth defects were less than observed in rsw1 (Table S1), and the phenotypes of these mutants differed in discrete respects from those observed in mutants such as rsw1 and korrigan or following treatment with inhibitors. In particular, rsw1 caused severe cell swelling in root tips of light-grown seedlings but the cesA1P mutant lines showed cell swelling only in the elongation and differentiation zones of primary roots (Fig. S1). Similarly, mutations, such as cob1, only influence a subset of the cell types that are affected by rsw1, and swollen cells are observed in the elongation zone but not within the meristem of cob1 roots grown under restrictive conditions (21). It has been proposed that COB1 participates directly in regulating microfibril orientation, with slightly decreased cellulose synthesis reflecting feedback from the disordered deposition (4). By analogy, it is possible the defects in microfibril assembly or deposition also contribute to the observed organ and cell morphologies of the cesA1P mutants.
Bidirectional Synthesis of Cellulose Microfibrils.
The studies reported here confirm previous observations (7) that CESA complexes move bidirectionally along linear tracks in the plasma membrane at similar rates. However, cellulose microfibrils from higher plants are in the form of cellulose I, in which individual microfibrils consist of parallel cellulose chains. This arrangement might be created if fibrils are formed from one complex or if they are formed by multiple complexes moving in one direction. Thus, the observation that complexes move bidirectionally suggests that nearby microfibrils might often be antiparallel. The cellulose I allomorph can be converted to cellulose II by swelling and recrystallizing cellulose I, a phenomenon that has long been puzzling. In cellulose II, glucan chains have an antiparallel arrangement and there is extensive intrasheet hydrogen bonding conferring the greatest thermodynamic stability on this allomorph. Previous studies on conversion of cellulose I to cellulose II (22) suggest that, whereas cellulose chains within native microfibrils are parallel, adjacent microfibrils are antiparallel, and during swelling in alkali, the native microfibrils are broken into separate sheets. When the swelling agent is removed, preferential reassociation occurs between chains of adjacent microfibrils. As the cellulose II structure is more favorable energetically than cellulose I, the net result of reassociation would be expected to be enrichment of cellulose II structures. Thus, the studies reported here and elsewhere appear to provide an explanation for the conversion from cellulose I to cellulose II.
Cell Expansion and Bidirectional Synthesis of Cellulose Microfibrils.
Measurements of bidirectional movement of CESA complexes in the plasma membrane indicate that WT complexes in rapidly growing cells move at similar velocities in opposite directions along microtubules or microtubule bundles (7). The cesA1P mutations affect the velocity of the complexes and have differential effects depending on the direction of displacement relative to the major axis of complex movement. The velocity discrepancy in bidirectional movement of CESA complexes was closely correlated with defects in organ or cell expansion observed in the mutant transgenic lines.
Wasteneys has proposed that the length of cellulose microfibrils, in addition to the orientation and tensile properties of cellulose microfibrils, may be a determinant of cell expansion anisotropy (18). Presumably, the length of microfibrils is a function of the rate of cellulose microfibril deposition and the lifetime of the CESA complexes. Taylor reported that in vitro phosphorylation of CESA7 causes the protein to be rapidly degraded via a 26s proteasome-dependent pathway and proposed that phosphorylation of CESA proteins may affect their lifetimes (23). However, the role of phosphorylation may be different for the CESA proteins that synthesize primary and secondary cell walls.
Recent images of growing cell walls by atomic force microscopy showed that fibrils of primary cell walls are likely bundles of microfibrils or cellulose microfibrils coated with a dense layer of matrix polysaccharides (24). Ding et al. (25) observed macrofibrils or large bundles of microfibrils that were found to exist only on the innermost layer of the primary cell wall, the layer adjacent to the plasma membrane. It is likely that interactions between microtubules and CESA complexes influence microfibril assembly and/or arrangement and thus direct or indirect interactions of adjacent microfibrils, which may in turn affect growth anisotropy (15, 18). This is consistent with the observation that microtubule removal by oryzalin causes rapid loss of growth anisotropy (26).
Phosphorylation of CESA1 may affect growth anisotropy through effects on microfibril arrangement or interactions of adjacent microfibrils. For example, if the velocity of CESA complexes represents microfibril synthetic rate (9), and if CESA complex stability and structure are independent of the direction and velocity of CESA complex movement, the discrepancy in the velocity of bidirectional movement of CESA complexes in some of the cesA1P mutants would be predicted to result in differences in the length of oppositely oriented microfibrils. This local difference in microfibril length could result in decreased uniformity of cell wall structure, leading to a loss of anisotropic restriction of cell expansion. By contrast, in the WT and transgenic lines in which CESA complexes move bidirectionally along microtubules at the same velocities, a more equal distribution of microfibril lengths would be produced in opposite directions and the guided synthesis and deposition of cellulose microfibrils would produce an antiparallel microfibril arrangement with a more uniform microfibril composition. The results with the T165A transgenic line were an exception to the relationship of observed discrepancy in bidirectional CESA complex velocity and defects in cell expansion. However, labeled CESA complexes in this line were also observed to show a significant bias in the percentage of complexes moving in one direction along common tracks. This pattern of movement would tend to produce a more parallel arrangement of microfibrils along these tracks, with most of the reducing ends pointing to the same direction. It is possible that this structure may have a mechanical strength more similar to the uniform antiparallel arrangement inferred in WT. However, it is not clear whether the directional bias observed in T165A is specific to dark-grown hypocotyls.
Regulation of Cellulose Synthase by Phosphorylation.
Interactions between CESA complexes and microtubules have previously been shown to influence cellulose microfibril orientation and deposition (7, 9). In view of the differential effects of cesA1P mutations on bidirectional mobility of CESA complexes, we propose that such mutations affect the interaction between CESA complexes and microtubules or modify the effects of such interactions. This idea is supported by the observation that removal of microtubules by oryzalin eliminated the discrepancy of bidirectional movement of CESA complexes in the T165A line (Table S1 and Figs. S5 and S6).
We envision that each cortical microtubule, or microtubule bundle, has lateral interactions with CESA complexes, resulting in organization of two linear arrays of CESA complexes, one on either side of the microtubule (27) (Fig. S7). This could account for the fact that clear evidence for collision of CESA complexes is not observed (27) and is consistent with freeze-etching experiments that revealed rosettes to lie alongside, but not directly on top of, cortical microtubules (28). Lateral interactions, whether direct or through a protein linker, would have inherent asymmetry with respect to each side of the microtubule as a result of microtubule polarity and confinement of CESA orientation by its integration into the plasma membrane (Fig. S7 A and B). Thus, mutations affecting these interactions and their effects on complex activity could also act asymmetrically, resulting in different complex velocities on each side of the microtubule. If there is a strong bias in the net polarity of the cortical microtubule array, such as has been observed by both Eb1-GFP (29, 30) and by high-resolution scanning EM (31), a bias in CESA activity with respect to the cell axis would result. The relative orientation of the CESA complex and tubulin could also affect delivering or distributing CESA complexes onto microtubules (32, 33), as in the case of the T165A mutant.
Phosphorylation sites in CESA1 are clustered in its two hypervariable regions (Fig. 1). These regions are not present within the catalytic subunits of bacterial and cyanobacterial cellulose synthases, suggesting that they are not required for the catalysis of cellulose chain polymerization (12). In view of the effects of phosphorylation on interaction with microtubules, it seems possible that these regions evolved to support or regulate the CESA–microtubule interactions.
In view of the relatively large number of phosphorylation sites present in CESA1, CESA3, and CESA5, it is notable that the single-site mutations described here have significant effects. Presumably, the combined effects of many phosphorylations or dephosphorylations may have profound effects that cannot be envisioned by studying the sites one at a time. It is also noted that, under in vivo conditions, phosphorylation or dephosphorylation may modify only some of the CESA1 subunits in a complex whereas mutagenesis changes all the CESA1 subunits in the complex, but at only one site. It is also worth noting that mutations at T166 had different effects on cell and organ morphology and CESA movement from mutations at the surrounding residues. Thus, a full accounting of the effects of phosphorylation on cellulose synthase activity will require additional multifactor studies and the development of additional tools.
In conclusion, results presented here show that phosphorylation of primary cell wall CESA proteins plays a critical role in regulation of anisotropic cell expansion. In particular, phosphorylation of CESA1 regulates cell elongation in rapidly expanding tissues. This appears to be achieved through regulation of bidirectional deposition of cellulose microfibrils. Further study of phosphorylation of CESA proteins and its effects on microfibril assembly, deposition, and properties, including microfibril length, tensile properties, and arrangements in cell walls will help to further understand the relationships among cellulose microfibril synthesis and deposition, cell wall structure, and cell expansion.
Materials and Methods
Construction of cesA1P Mutants.
pCesA1-gateway, a Ti plasmid containing the CESA1 promoter flanking recombination ATT sites, and a donor vector containing a CESA1 cDNA was constructed and used for site-directed mutagenesis as described in SI Materials and Methods. Sequence-verified constructs were introduced into the Arabidopsis thaliana Col-0 cesA1 mutant (rsw1) by Agrobacterium-mediated transformation.
Tissue, Cell Length, and Cellulose Measurements.
Standard methods were used for measurements of tissue and cell size as described in SI Materials and Methods. Cellulose content of freeze-dried dried material was determined as described by Updegraff (35).
Measurements of CESA Particle Velocity.
Seeds were germinated on 0.5× Murashige and Skoog (MS) agar and grown vertically in the dark for 2 d at 30 °C. Seedlings were manipulated under a red safety light to avoid the blue light response known to inhibit growth (35). Seedlings were mounted between two coverslips in water with their apexes pointing to the back of microscope. To examine oryzalin effects, seedlings were submerged in a 20-μM oryzalin solution and incubated in the dark before mounting in the same solution between coverslips. Imaging of YFP::CESA6 particles or GFP::MAP4-labeled microtubules was performed in hypocotyl cells just below the hook of etiolated seedlings. EGFP and YFP were excited at 488 nm, and fluorescence was collected through a 525/50-nm band-pass filter (Chroma Technologies). All image processing was performed by using Metamorph (Molecular Dynamics) and ImageJ (National Institutes of Health) software. The obtained sequential frames were averaged by using the Walking Average plug-in (three-frame window). To correct for tissue movement during imaging, subpixel frame alignment was performed using a nonlinear least squares algorithm [StackReg plug-in for ImageJ (36)]. As frame alignment can fail, fiducial markers in each data set were analyzed for drift and only those data sets with less than 135 nm of drift over a period of 5 min were kept for analysis. The fiducial markers used were intrinsic autofluorescent spots on cell walls. Particle velocities were measured by using two methodologies as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Andrew Carroll for help with Imaris analysis. We are grateful to Herman Höfte and Clive Lloyd for helpful critiques of the manuscript. This work was supported in part by United States Department of Energy Grants DE-FG02-03ER20133 and DE-FG02-09ER16008 and by the Carnegie Institution for Science and the Energy Biosciences Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012348107/-/DCSupplemental.
References
- 1.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 2.Arioli T, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 3.Heim DR, Skomp JR, Tschabold EE, Larrinua IM. Isoxaben inhibits the synthesis of acid insoluble cell-wall materials in Arabidopsis thaliana. Plant Physiol. 1990;93:695–700. doi: 10.1104/pp.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roudier F, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell. 2005;17:1749–1763. doi: 10.1105/tpc.105.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxena IM, Brown RM., Jr Cellulose biosynthesis: Current views and evolving concepts. Ann Bot (Lond) 2005;96:9–21. doi: 10.1093/aob/mci155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persson S, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 8.Baskin TI. On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma. 2001;215:150–171. doi: 10.1007/BF01280311. [DOI] [PubMed] [Google Scholar]
- 9.DeBolt S, et al. Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA. 2007;104:5854–5859. doi: 10.1073/pnas.0700789104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paredez AR, Persson S, Ehrhardt DW, Somerville CR. Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 2008;147:1723–1734. doi: 10.1104/pp.108.120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneda A, et al. Chemical genetic screening identifies a novel inhibitor of parallel alignment of cortical microtubules and cellulose microfibrils. Plant Cell Physiol. 2007;48:1393–1403. doi: 10.1093/pcp/pcm120. [DOI] [PubMed] [Google Scholar]
- 12.Delmer DP. Cellulose biosynthesis: Exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 13.Vergara CE, Carpita NC. β-D-glycan synthases and the CesA gene family: Lessons to be learned from the mixed-linkage (1—>3),(1—>4)β-D-glucan synthase. Plant Mol Biol. 2001;47:145–160. [PubMed] [Google Scholar]
- 14.Nühse TS, Stensballe A, Jensen ON, Peck SC. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baskin TI. Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol. 2005;21:203–222. doi: 10.1146/annurev.cellbio.20.082503.103053. [DOI] [PubMed] [Google Scholar]
- 16.Fagard M, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somerville C, et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 18.Wasteneys GO. Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol. 2004;7:651–660. doi: 10.1016/j.pbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Nicol F, et al. A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato S, et al. Role of the putative membrane-bound endo-1,4-β-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:251–263. doi: 10.1093/pcp/pce045. [DOI] [PubMed] [Google Scholar]
- 21.Schindelman G, et al. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okano T, Sarko A. Mercerization of cellulose. II. Alkali-cellulose intermediates and a possible mercerization mechanism. J Appl Polym Sci. 1985;30:325–332. [Google Scholar]
- 23.Taylor NG. Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites—a potential role in regulating protein degradation. Plant Mol Biol. 2007;64:161–171. doi: 10.1007/s11103-007-9142-2. [DOI] [PubMed] [Google Scholar]
- 24.Szymanski DB, Cosgrove DJ. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol. 2009;19:R800–R811. doi: 10.1016/j.cub.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 25.Ding SY, Himmel ME. The maize primary cell wall microfibril: A new model derived from direct visualization. J Agric Food Chem. 2006;54:597–606. doi: 10.1021/jf051851z. [DOI] [PubMed] [Google Scholar]
- 26.Baskin TI, Wilson JE, Cork A, Williamson RE. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or Taxol. Plant Cell Physiol. 1994;35:935–942. [PubMed] [Google Scholar]
- 27.Ehrhardt DW. Straighten up and fly right: Microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr Opin Cell Biol. 2008;20:107–116. doi: 10.1016/j.ceb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Giddings T, Staehelin L. In: The Cytoskeletal Basis of Plant Growth and Form. Lloyd C, editor. New York: Academic Press; 1991. pp. 85–99. [Google Scholar]
- 29.Chan J, Calder G, Fox S, Lloyd C. Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat Cell Biol. 2007;9:171–175. doi: 10.1038/ncb1533. [DOI] [PubMed] [Google Scholar]
- 30.Dixit R, Chang E, Cyr R. Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol Biol Cell. 2006;17:1298–1305. doi: 10.1091/mbc.E05-09-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton DA, Vantard M, Overall RL. Analysis of cortical arrays from Tradescantia virginiana at high resolution reveals discrete microtubule subpopulations and demonstrates that confocal images of arrays can be misleading. Plant Cell. 2008;20:982–994. doi: 10.1105/tpc.108.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowell EF, et al. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 34.Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 35.Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP. Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 2003;36:203–214. doi: 10.1046/j.1365-313x.2003.01870.x. [DOI] [PubMed] [Google Scholar]
- 36.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.