Abstract
Nitrification plays a central role in the global nitrogen cycle and is responsible for significant losses of nitrogen fertilizer, atmospheric pollution by the greenhouse gas nitrous oxide, and nitrate pollution of groundwaters. Ammonia oxidation, the first step in nitrification, was thought to be performed by autotrophic bacteria until the recent discovery of archaeal ammonia oxidizers. Autotrophic archaeal ammonia oxidizers have been cultivated from marine and thermal spring environments, but the relative importance of bacteria and archaea in soil nitrification is unclear and it is believed that soil archaeal ammonia oxidizers may use organic carbon, rather than growing autotrophically. In this soil microcosm study, stable isotope probing was used to demonstrate incorporation of 13C-enriched carbon dioxide into the genomes of thaumarchaea possessing two functional genes: amoA, encoding a subunit of ammonia monooxygenase that catalyses the first step in ammonia oxidation; and hcd, a key gene in the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle, which has been found so far only in archaea. Nitrification was accompanied by increases in archaeal amoA gene abundance and changes in amoA gene diversity, but no change was observed in bacterial amoA genes. Archaeal, but not bacterial, amoA genes were also detected in 13C-labeled DNA, demonstrating inorganic CO2 fixation by archaeal, but not bacterial, ammonia oxidizers. Autotrophic archaeal ammonia oxidation was further supported by coordinate increases in amoA and hcd gene abundance in 13C-labeled DNA. The results therefore provide direct evidence for a role for archaea in soil ammonia oxidation and demonstrate autotrophic growth of ammonia oxidizing archaea in soil.
Keywords: ammonia oxidizers, nitrification, soil, autotrophy, stable isotope probing
Ammonia oxidation, the first and rate-limiting step in the nitrification process, is an important component of the terrestrial nitrogen cycle. Nitrification in agroecosystems leads to loss of a significant proportion of applied ammonia-based fertilizers (annual input of approximately 100 Tg y−1) (1), at an estimated annual cost of $15.9 billion (2). Losses occur through leaching of nitrate or its conversion to gaseous dinitrogen or nitrogen oxides, by both nitrifiers and classical denitrifiers. Ammonia oxidizers therefore contribute significantly to nitrate pollution of groundwaters and pollution of the atmosphere by the greenhouse gas nitrous oxide, which will become the dominant ozone-depleting gas in the 21st century (3). These environmental impacts add to nitrogen transformations and emissions from the oxidation of ammonia generated naturally in soil through decomposition of organic matter, and have led to renewed interest in the application of inhibitors of ammonia oxidation to reduce fertilizer loss and nitrous oxide production (4). Despite its global importance, there is uncertainty and controversy regarding the organisms oxidizing ammonia in terrestrial systems. Traditionally, the process was thought to be dominated by autotrophic ammonia oxidizers belonging to the betaproteobacteria, with small contributions in some soils from heterotrophic bacteria and fungi at low specific rates (5). Bacterial ammonia oxidizers gain energy through oxidation of ammonia to nitrite and carbon by fixation of inorganic carbon. They are readily enriched from most soils and the abundances and cell activities obtained for cultivated strains appear sufficient to support measured soil nitrification rates.
This traditional view of soil ammonia oxidation was challenged by the analysis of a soil metagenome fragment containing both a thaumarchaeal group 1.1b 16S rRNA gene and sequences encoding potential homologues of AmoA and AmoB subunits of the bacterial ammonia monooxygenase (6). This is the key functional enzyme in bacterial ammonia oxidizers, oxidizing ammonia to hydroxylamine, before its conversion to nitrite. Archaeal amoA genes are ubiquitous in soils, frequently outnumbering bacterial amoA genes (7–9), and the cultivation of Nitrosopumilus maritimus, a marine autotrophic ammonia oxidizing archaeon (10), and organisms from hot spring environments (11, 12), demonstrated the potential for autotrophic ammonia oxidation in group 1.1a, 1.1b, and a thermophilic thaumarchaeal lineage. Genome analysis of N. maritimus (13) and Cenarchaeum symbiosum (14) identified components of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle (15, 16) for carbon dioxide fixation. A key enzyme of the pathway, 4-hydroxybutyryl-CoA dehydratase, has not, to our knowledge, been identified in obligate archaeal heterotrophs (17) and could act as a marker for autotrophic thaumarchaea. To date, however, no autotrophic archaeal ammonia oxidizer has been isolated from soil, but there is evidence that ammonia oxidation by archaea may exceed that by bacteria in some soils (18–20). In these soils, nitrification is associated with higher archaeal amoA abundance, changes in abundance and relative abundance of archaeal, but not bacterial, 16S rRNA and amoA phylotypes, and greater archaeal transcriptional activity (19, 20). In contrast, growth of ammonia-oxidizing bacteria, and not archaea, correlates with nitrification kinetics in other soils (21, 22). Despite autotrophic growth of cultivated archaeal ammonia oxidizers, there is no direct evidence for archaeal autotrophy in soil, and some studies indicate heterotrophic and/or mixotrophic growth. For example, genome analysis of C. symbiosum (14) suggests the capacity for both heterotrophic and autotrophic modes of growth and isotopic analysis of marine thaumarchaeal lipids (23) indicates assimilation of some organic carbon.
Stable isotope probing (SIP) provides direct assessment of autotrophy, by incubation of samples with 13C-CO2 and molecular analysis of 13C-labeled nucleic acids. SIP has demonstrated autotrophy by bacterial ammonia oxidizers in estuarine sediments (24) and in soil (21). The latter study detected bacterial but not archaeal amoA and 16S rRNA genes in 13C-labeled DNA during nitrification in the presence of 13C-CO2. Assimilation in bacterial DNA was not observed when nitrification was inhibited by acetylene, but, interestingly, archaeal gene abundance increased during acetylene inhibition of nitrification, providing evidence for heterotrophic growth. In this study we combined SIP with analysis of bacterial and archaeal amoA genes and thaumarchaeal hcd genes to assess whether inorganic carbon fixation by archaea and bacteria is associated with ammonia oxidation in a soil in which archaea appear to play a greater role in nitrification than bacteria.
Results
Nitrification in Soil Microcosms.
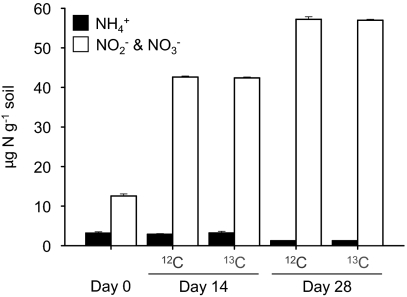
Nitrification was monitored in soil microcosms established with a headspace containing 5% (vol/vol) 12C- or 13C-CO2 and sampled destructively after incubation for 14 and 28 d. High rates of nitrification (1.1 μg NO3−-N g−1 soil d−1) have previously been observed in this soil without ammonia amendment (19) as a result of ammonia released during mineralization of organic material. Ammonia concentration was low throughout the incubation period, decreasing from an initial value of 3.2 (±0.33) μg NH4+-N g−1 soil to 1.25 (±0.07) and 1.24 (±0.01) μg NH4+-N g−1 in microcosms incubated with 12C- and 13C-CO2, respectively, after incubation for 28 d (Fig. 1). Nitrification rates were high and nitrite plus nitrate concentration increased from an initial value of 12.6 μg N g−1 to 57.2 (±0.64) and 57.0 (±0.24) μg NO2−-N/NO3−-N g−1 in microcosms incubated with 12C- and 13C-CO2, respectively, after incubation for 28 d. Ammonia and nitrite plus nitrate concentrations did not differ significantly (P > 0.05) between microcosms incubated with headspaces containing 12C- and 13C-CO2 at 14 or 28 d.
Fig. 1.
Changes in ammonia and nitrite plus nitrate concentrations in soil microcosms incubated at 30 °C for 14 or 28 d with a headspace concentration of 5% (vol/vol) 12C- or 13C-CO2. Data plotted are mean values and SEs from triplicate microcosms destructively sampled at each time point.
Quantification of amoA and hcd Genes.
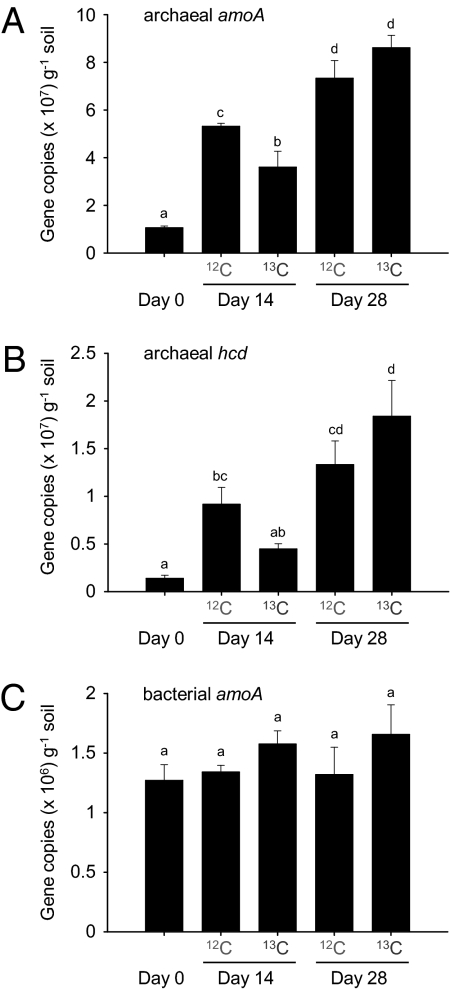
Growth of putative ammonia oxidizing archaea and bacteria during incubation was assessed by quantification of amoA genes (Fig. 2) using quantitative PCR (qPCR) assays specific for archaeal and betaproteobacterial amoA genes. Growth of putative autotrophic archaea was assessed by quantification of hcd genes (Fig. 2) using a qPCR assay specific for thaumarchaeal hcd genes. Abundance of betaproteobacterial amoA genes did not change significantly (ANOVA, log-transformed data, P = 0.46), ranging from 1.27 to 1.65 × 106 copies g−1 soil during incubation for 28 d. In contrast, archaeal amoA gene abundance increased significantly (ANOVA, log-transformed data, P < 0.001) from 1.05 to 5.31/3.60 and 7.33/8.61 × 107 copies g−1 soil in 12C/13C-CO2 microcosms after 14 and 28 d, respectively. Similarly, thaumarchaeal hcd gene abundance increased significantly (ANOVA, P = 0.002) from 0.14 to 0.92/0.45 and 1.33/1.84 × 107 copies g−1 soil in 12C/13C-CO2 microcosms after 14 and 28 d, respectively. The increase in archaeal amoA, but not hcd, gene abundance differed significantly between microcosms with headspaces containing 12C-CO2 and 13C-CO2 after incubation for 14 d, but not after 28 d. Additional soil incubations were performed to compare the effect of elevated CO2 concentrations on AOA and AOB growth (Fig. S1) and confirmed significant growth of AOA only, and no significant difference in microcosms incubated with ambient or 5% CO2 headspace concentration. Therefore no stimulation or suppression of growth could be attributed to CO2 concentration.
Fig. 2.
The abundance of archaeal amoA, archaeal hcd, and bacterial amoA genes in soil microcosms after incubation with a headspace concentration of 5% (vol/vol) 12C- or 13C-CO2 for 14 or 28 d. Data plotted are mean values and SEs from triplicate microcosms destructively sampled at each time point. The same letter above different bars indicates no significant difference (P > 0.05).
DNA Stable Isotope Analysis of Ammonia-Oxidizing Communities.
DNA SIP was used to determine which putative ammonia oxidizers were assimilating CO2. DNA extracted from 0.5 g of soil from each microcosm (15.4–17.7 μg DNA g−1 soil dry weight) was subjected to isopycnic ultracentrifugation in a CsCl gradient to separate 12C- and 13C-enriched DNA. After centrifugation for 24 h, each 5-mL tube was fractionated into aliquots of approximately 190 μL, and 20 μL from each was used to (indirectly) determine the buoyant density by measuring the refractive index. After precipitation and purification of DNA, abundances of archaeal and bacterial amoA genes in 17 fractions were determined by qPCR.
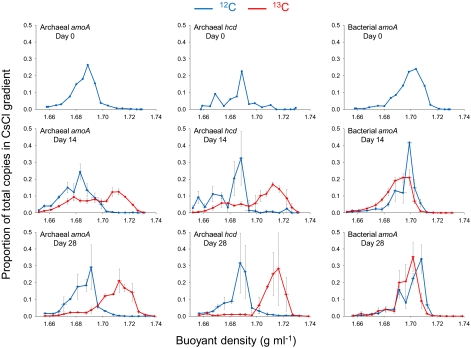
DNA from archaeal and bacterial ammonia oxidizers, determined by quantification of respective amoA genes, possessed different buoyant densities, as seen in data from microcosms sampled at day 0. Recovery of archaeal amoA genes was maximal in lighter fractions (approximately 1.69 g mL−1) than for bacterial amoA genes (approximately 1.70 g mL−1; Fig. 3), indicating a difference in the average percent guanine-cytosine (GC) content of bacterial and archaeal ammonia oxidizer communities. After incubating microcosms for 14 or 28 d, there was no apparent change in the distribution of bacterial amoA gene abundance in microcosms incubated with 12C-CO2 and 13C-CO2. There was therefore no evidence for incorporation of CO2 into the genomic DNA of bacterial ammonia oxidizers during active nitrification in this soil. In contrast, there was a large shift in the buoyant density of genomic DNA possessing archaeal amoA genes in microcosms incubated with 13C-CO2. After incubation for 14 d, the relative abundance of archaeal amoA genes was maximal in heavier CsCl fractions with a buoyant density of approximately 1.71 g mL−1 from DNA isolated from microcosms incubated with 13C-CO2, compared with a maximum at approximately 1.69 g mL−1 in microcosms incubated with 12C-CO2. After incubation for 28 d, the relative abundance of archaeal amoA genes remaining in the lighter fractions (approximately 1.69 g mL−1) decreased further, indicating continued growth of autotrophic populations, which became the dominant component of the archaeal ammonia oxidizer community.
Fig. 3.
Buoyant density distribution of archaeal and bacterial genomic DNA extracted from soil after incubation with a headspace concentration of 5% (vol/vol) 12C- or 13C-CO2. Seventeen fractions from each CsCl spin were analyzed and represented a density range from approximatel 1.66 to 1.74 g mL−1. The abundances of archaeal amoA, archaeal hcd, and bacterial amoA genes in each fraction were determined by quantitative PCR, and values converted to the proportion of total gene abundance throughout the gradient. Vertical horizontal bars represent the SE of proportional abundance from triplicate CsCl spins (representing triplicate microcosms). Horizontal error bars represent the SE of the buoyant density of fractions collected from the same point in replicate CsCl tubes. Large vertical error bars observed in adjacent samples in some density gradients indicate that the largest gene abundance was detected in each of the adjacent fractions in replicate samples. Gene distribution profiles of day 0 samples (before incubation) are mean values derived from two individual CsCl gradients only.
The relative abundance patterns of archaeal amoA and hcd genes in fractions collected from each CsCl gradient were compared to assess whether they may be derived from the same microorganisms. Recovery of archaeal hcd genes from microcosms sampled at day 0 was maximal in fractions with a buoyant density of approximately 1.69 g mL−1, as for archaeal amoA genes (Fig. 3). Similarly, the relative abundance of both archaeal amoA genes and hcd genes was highest in fractions with buoyant density of approximately 1.71 g mL−1 for microcosms incubated with 13C-CO2 and approximately 1.69 g mL−1 for microcosms incubated with 12C-CO2, after incubation for both 14 and 28 d (Fig. 3). This suggests that archaeal hcd and amoA genes were derived from the same archaea.
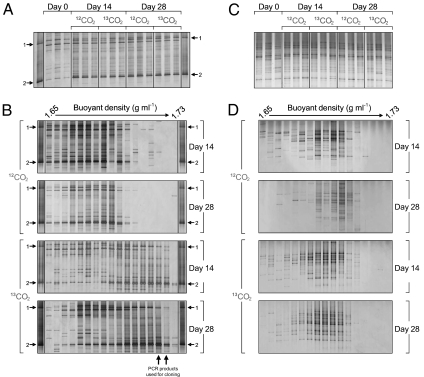
Changes in communities of archaeal and bacterial ammonia oxidizers during incubation were determined by denaturing gradient gel electrophoresis (DGGE) analysis of amplified amoA genes from individual microcosms (Fig. 4). Reproducible differences in DGGE profiles of archaeal amoA genes from microcosms incubated with 12C- or 13C-CO2 were evident after incubation for 14 and 28 d, with significant increases in the relative intensities of two amoA bands, compared with microcosms sampled at day 0 (Fig. 4A). The two amoA bands comigrated with two marker bands (bands 1 and 2, Fig. 4A), representative of sequences from archaeal amoA genes previously identified as increasing in this soil under similar incubation conditions, and belong to organisms placed within the group 1.1a lineage (Fig. S2) (19). However, no reproducible or significant differences were observed in the DGGE profiles of bacterial amoA genes after incubation for 14 or 28 d (Fig. 4C), indicating no selection of bacterial ammonia oxidizers during incubation.
Fig. 4.
DGGE analysis of amoA gene PCR products from archaeal (A and B) and bacterial (C and D) amoA-defined communities derived from soil microcosms incubated for 14 or 28 d with a headspace concentration of 5% (vol/vol) 12C- or 13C-CO2. DGGE analysis was performed on amoA genes amplified from total genomic DNA from triplicate microcosms (A and C), with each lane representing an individual microcosm. DGGE analysis was also performed on 17 fractions recovered from individual CsCl density gradients separating genomic DNA of different buoyant densities from individual microcosms incubated for 14 or 28 d for both archaeal (B) and bacterial (D) amoA-defined communities. A marker lane containing PCR products from two group 1.1a clones (1 and 2, accession numbers FJ971889 and FJ971892, respectively) was run alongside all archaeal amoA DGGE profiles and represents sequences from organisms previously identified as growing under these incubation conditions. Two arrows indicate locations of archaeal amoA PCR products from two fractions that were pooled and used to construct clone libraries (Fig. S2).
DGGE analysis of amoA PCR products derived from 17 fractions recovered from each individual CsCl spin was also performed on samples from microcosms incubated with either 12C- or 13C-CO2 after incubation for 14 and 28 d. Incorporation of CO2 into specific populations was observed for only archaeal amoA genes. Two bands increased in relative intensity in microcosms incubated for 14 and 28 d, with relative intensity increasing with increasing buoyant density only in microcosms incubated with 13C-CO2 (Fig. 4B). These bands also comigrated with marker bands 1 and 2. There was no increase in the relative intensity of bacterial amoA bands in the higher buoyant density fractions after 14 or 28 d. There was therefore no evidence for CO2 fixation by bacterial ammonia oxidizers, despite high rates of nitrification.
To confirm the phylogenetic affiliation of AOA populations growing during nitrification, archaeal amoA and 16S rRNA gene sequences were amplified from genomic DNA recovered in CsCl fractions with a buoyant density of approximately 1.725 g mL−1. These products were screened by DGGE, sequenced, and compared with reference sequences, including those already recovered from this soil after similar incubations (Figs. S2 and S3). As observed previously, archaeal amoA sequences comigrating with marker bands 1 and 2 were placed within two distinct lineages within group 1.1a thaumarchaea. The dominant archaeal 16S rRNA gene sequence increasing in relative intensity was also placed within this lineage and was presumably derived from the same organisms.
Discussion
This study provides direct evidence for autotrophic activity and autotrophic growth of thaumarchaea in soil. It also provides compelling evidence that archaeal autotrophic activity is directly linked to ammonia oxidation, i.e., that ammonia oxidation in this soil is caused by the activity and growth of autotrophic archaeal ammonia oxidizers. First, SIP demonstrated incorporation of 13C-labeled CO2 into archaeal amoA genes, the key functional gene in autotrophic ammonia oxidation, during active nitrification, but no incorporation into bacterial amoA genes. Second, CO2 fixation was accompanied by significant increases in the abundance of archaeal amoA genes, but no detectable increase in the abundance of equivalent functional genes in bacteria. Third, increases in abundance of archaeal amoA genes paralleled increases in abundance of hcd genes.
Previous studies have suggested a major role for archaea in ammonia oxidation in the agricultural soil used in this study. Archaeal amoA gene and gene transcript abundances are higher than those for bacteria, particularly in low-pH plots, suggesting greater potential and transcriptional activity, respectively (9). Archaeal communities also appear to be more dynamic during nitrification, with changes in the relative abundance of archaeal amoA gene and gene transcript phylotypes and selection at different pH values and temperatures (9, 20). In addition, both archaeal growth and nitrification are inhibited by acetylene in this soil (19). These data are consistent with the lack of response of bacterial ammonia oxidizers during observed active nitrification. The abundance and diversity (i.e., DGGE profiles) of bacterial amoA genes did not change during nitrification and there was no evidence for bacterial assimilation of CO2, a prerequisite for growth of bacterial ammonia oxidizers. In contrast, archaeal amoA genes increased in abundance. DGGE analysis also indicated changes in putative archaeal ammonia oxidizer communities, through changes in DGGE profiles and, in particular, increases in relative intensity of two DGGE bands previously observed (19) to increase during nitrification in this soil, but not after inhibition of nitrification by acetylene.
Although previous studies have provided evidence for archaeal ammonia oxidation in many soils, it has not been possible to determine whether putative archaeal ammonia oxidizers grew autotrophically or heterotrophically. CO2-SIP provides direct evidence for CO2 assimilation, and has provided evidence for autotrophic ammonia oxidation by bacteria in laboratory cultures (20), estuarine sediments (24), and soil (21). The last of these studies found assimilation of 13C-CO2 into bacterial amoA genes during nitrification and, in fact, found increases in abundance of archaeal amoA genes even when nitrification was inhibited by acetylene. Here we found the reverse situation, with assimilation of CO2 into archaeal, but not bacterial, amoA genes. amoA phylotypes associated with active nitrification dominated amoA DGGE profiles representative of higher buoyant densities and genomic DNA enriched with 13C, further strengthening evidence for association between archaeal autotrophy and ammonia oxidation. Interpretation of SIP experiments can be complicated by potential utilization of labeled substrates by secondary utilizers. This is less critical when studying autotrophs, which will reassimilate 13C-CO2 released through respiration of primary and secondary utilizers, and by targeting of specific functional genes. We cannot, however, rule out the possibility of secondary utilization of labeled organic carbon, released by autotrophic ammonia oxidizers, by heterotrophic amoA- or hcd-containing organisms. In addition, the lack of response of putative bacterial ammonia oxidizers could result from biases associated with molecular analysis or incomplete coverage of primers.
Growth of thaumarchaeal putative autotrophs and putative ammonia oxidizers was determined, independently, by analysis of hcd and amoA gene abundance. Abundance of thaumarchaeal hcd genes increased during incubation, indicating growth of thaumarchaeal autotrophs (Fig. 2). Distribution of thaumarchaeal hcd genes in SIP fractions (Fig. 3) confirmed that putative thaumarchaeal autotrophs assimilate inorganic carbon during growth. Carbon assimilation in these thaumarchaeal autotrophs might proceed through one of two autotrophic pathways characterized by the hcd gene product, i.e., the 3-hydroxypropionate/4-hydroxybutyrate cycle (16) and the dicarboxylate/4-hydroxybutyrate cycle (25, 26). Evidence for the former pathway in archaeal ammonia oxidizers is based on presence of the set of genes characteristic for this pathway in the genomes of N. maritimus (13) and C. symbiosum (14). Thaumarchaeal hcd genes detected in this study are therefore likely to originate from archaeal ammonia oxidizers. The similar distribution of thaumarchaeal hcd and amoA genes in SIP fractions indicates no major variation in the percent GC content of genomes hosting these genes, further suggesting their origin in the same organisms. There is therefore strong evidence for autotrophic growth of archaeal soil ammonia oxidizers, assimilating carbon through the 3-hydroxypropionate/4-hydroxybutyrate cycle. Although the relative increases in amoA and hcd gene abundances were very similar, there was a discrepancy between the actual abundances measured. Thaumarchaeal amoA genes were between 5.5 and eight times more abundant at each sampling point (Fig. 2). Although this could reflect differences in gene copy number per genome, it is more likely that this was a result of differences in efficiencies in the primers used to target thaumarchaeal hcd genes. As these primers were designed from a limited number of available reference sequences, they may not possess 100% sequence identity with target sequences amplified in this soil, thus underestimating thaumarchaeal hcd gene abundance. Sequence analysis of cloned PCR products from the qPCR assay confirmed the specificity of this assay (Fig. S4).
These findings contrast with those of Jia and Conrad (21), who report autotrophy by bacterial, rather than archaeal, ammonia oxidizers. Although there are several differences between these two studies (e.g., soil type, management history, incubation temperature), the most significant difference is the source of ammonia fueling nitrification. Ammonia in our system was derived from mineralization of soil organic matter and ammonia concentrations were consistently low, whereas Jia and Conrad (21) amended soil with 100 μg N-NH4+ g−1 soil at weekly intervals. The majority of archaeal ammonia oxidizer phylotypes in both soils could be placed in the group 1.1b “soil lineage” but group 1.1a phylotypes grew autotrophically in the Craibstone soil. Growth of archaeal ammonia oxidizers belonging to this lineage may have been encouraged by relatively low ammonia concentration, as demonstrated recently for the group 1.1a marine organism N. maritimus (27). This is also consistent with other studies that indicate that archaeal ammonia oxidizers may dominate oxidation of ammonia produced by mineralization of organic matter (19, 28). The data do not, however, clarify the role of group 1.1b populations, which dominated the amoA-containing lineage in this and many other soils, but did not appear to respond during soil incubations. Evidence for growth of (presumably) group 1.1b populations in soil microcosms, even during acetylene inhibition of nitrification (21), suggests that at least some of these organisms may have a mixotrophic or heterotrophic metabolism and that group 1.1a and some or all group 1.1b phylotypes differ fundamentally in their physiological and metabolic characteristics.
Materials and Methods
Construction and Sampling of Soil Microcosms.
Soil microcosms were constructed as described by Offre and colleagues (19). Briefly, microcosms consisted of 144-mL serum bottles containing 10 g sieved soil (mesh size, 3.35 mm) collected from the upper 10 cm of an agricultural plot (Scottish Agricultural College, Craibstone, Aberdeen, Scotland; grid reference NJ872104). This soil is a podzol with a sandy loam texture, is the dominant agricultural soil type in Scotland, and is a common agricultural soil in Europe. Soil pH had been maintained at approximately 7.5 since 1961, and its characteristics were described by Kemp and colleagues (29). Microcosms were sealed with rubber stoppers and aluminum caps and the headspace was adjusted to 5% (vol/vol) of either 12C-CO2 (BOC) or 13C-CO2 (99% atom enriched; CK-Gas) by injection through the rubber septum. Microcosms were opened every 3 d to allow air exchange, and maintain aerobic conditions, before resealing and reestablishing carbon dioxide concentrations. Microcosms were incubated in the dark at 30 °C, with triplicate 12C- and 13C-CO2 microcosms sampled destructively at 0, 14, and 28 d, and soil was frozen immediately at −80 °C.
Ammonia and combined nitrite plus nitrate concentrations were determined colorimetrically by flow injection analysis (FIA Star 5010 Analyzer; Tecator) (30) of 1 M KCl extracts (4 g soil in 20 mL 1M KCl). Nucleic acids were extracted from 0.5-g soil samples as described by Griffiths and colleagues (31), modified by Nicol et al. (32). Extracted nucleic acids were used for quantification, DGGE analysis of amoA and hcd genes, and SIP.
Quantification of amoA and hcd Genes.
The bacterial amoA gene assay used primers amoA1F and amoA2R (33) and a dilution series (101 to 106 amoA gene copies) of Nitrosospira multiformis ATCC25196 genomic DNA. The archaeal amoA gene assay used primers Arch-amoAF and Arch-amoAR (34) and a dilution series (103 to 108 amoA gene copies) of PCR products containing complete amo genes derived from N. maritimus genomic DNA (3-kb amplicon) or 54d9 fosmid DNA (2-kb amplicon) for CsCl gradient fractions and total soil DNA, respectively. Reactions (25 μL) contained 12.5 μL 2× SYBR Premix Ex Taq (TaKaRa), 0.2 mg mL−1 BSA, 200 nM of each primer, and 1 μL of quantified DNA template. Amplification conditions were: 95 °C for 30 min, followed by 35 cycles of 10 s at 95 °C, 30 s at 55 °C or 53 °C for bacterial or archaeal amoA genes, respectively, 1 min at 72 °C, and plate read at 85 °C or 83 °C for bacterial and archaeal amoA genes, respectively. Amplification efficiencies were 96.6% to 109.5% (r2 = 0.991–0.997) and 85.1% to 94.4% (r2 = 0.995–0.999) for bacterial and archaeal amoA genes, respectively.
Putative thaumarchaeal autotrophs were quantified by real-time PCR amplification of hcd genes using primers designed against conserved regions of sequence from seven thaumarchaeal sequences including C. symbiosum, N. maritimus, and metagenomic fragments recovered from the Global Ocean Survey (35) but biased against bacterial and hyperthermophilic crenarchaeal hcd sequences (36). Primers hcd-120F-SCM1 (5′-AGCCTGTAGACCACCCAATG-3′) and hcd-1367R-SCM1 (5′-TATTCTTTGGGCCTGTGGAG-3′) were used to amplify a 1,286-bp product of N. maritimus SCM1, a dilution series of which (102 to 107 hcd gene copies) was used as a template for a qPCR assay using primers hcd-911F (5′-AGCTATGTBTGCAARACAGG-3′) and hcd-1267R (5′-CTCATTCTGTTTTCHACATC-3′). Fragments of 395 bp were amplified with efficiency of 93.7% to 116.2% and r2 values of 0.985 to 0.998. Reactions (25 μL) contained 12.5 μL of 2× Quantifast SYBR Green PCR Master Mix (Qiagen), 0.2 mg mL−1 BSA, 1 μM of each primer, and 2 μL of extracted DNA as template. Amplification conditions were 95 °C for 15 min, followed by 35 cycles of 10 s at 95 °C, 45 s at 60 °C, plate read at 73 °C, and a final extension step of 10 min at 60 °C.
All reactions were performed using a MyIQ thermocycler (Bio-Rad) and the specificity of amplification products was determined by melting curve analysis at the end of each PCR and agarose gel electrophoresis.
DGGE Analysis of amoA Genes.
DGGE was performed with a DCode Universal Mutation Detection System (Bio-Rad) as described previously (37). Archaeal amoA genes were amplified using primers CrenamoA23f and CrenamoA616r (20) and bacterial amoA genes were amplified using primers amoA 1F-GC and amoA 2R-GG (38). Amplification was performed in a 50-μL reaction volume containing 25 μL of twofold Premix Ex Taq mix (TaKaRa), 200 nM each primer and 2 μL DNA template. Archaeal amoA genes were amplified at 95 °C for 5 min; followed by 10 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; followed by 25 cycles of 92 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; followed by 72 °C at 10 min. Bacterial amoA genes were amplified using the same conditions except a touchdown program was used, with annealing temperature from 62 °C to 57 °C during the first 10 cycles and 57 °C during the following 30 cycles. PCR products were analyzed with 8% (wt/vol) polyacrylamide gels containing a 15% to 55% or 45% to 65% linear gradient of denaturant for archaeal and bacterial amoA gene assays, respectively. DGGE gels were silver-stained (37) and scanned with a GT9600 scanner with transparency unit (Epson).
SIP.
Extracted DNA from 12C- and 13C-CO2 incubations was subjected to isopycnic density gradient centrifugation, based on the method described by Freitag and colleagues (24). Briefly, a CsCl solution was prepared in TE buffer (pH 8) by adding 1.27 g CsCl mL−1 TE. The density was determined indirectly with an ATAGO-R-5000 hand refractometer (UNI-IT) and adjusted to a refractive index of 1.399 (1.696 g mL−1 buoyant density) by adding small amounts of CsCl or TE buffer. DNA (0.5 μg) was mixed with 1.5 μL ethidium bromide (1 mg mL−1) and 200 μL CsCl in TE buffer. After thorough mixing, the suspension was added to 4.8 mL CsCl in TE buffer in 5.1-mL quick-seal polyallomer tubes (Beckman Coulter). Suspensions were then centrifuged in a VTI65.2 vertical rotor (Beckman Coulter) at 184,388 × g (45,000 rpm) for 24 h at 20 °C.
CsCl density gradients were fractionated into equal volumes using a fraction recovery system (Beckman Coulter) and an LKB 8Romma 2112Redirac fraction collector (Pharmacia). CsCl solution was displaced by supplying water to the top of the polyallomer tube using a Gilson MiniPlus3 peristaltic pump (Anachem) and 190-μL aliquots (n = 24) were collected from the base. For each individual fraction, 20 μL CsCl was used to determine the refractive index and buoyant density. DNA was recovered by overnight precipitation in two volumes of PEG 6000 in 1.6 M NaCl at 4 °C, washing with 70% ethanol and dissolving the DNA pellet in 20 μL of sterile water.
Statistical Analysis.
Differences in ammonia and nitrite plus nitrate concentrations and qPCR data were compared using one-way ANOVA followed by an S-N-K test using SPSS software, version 13.0 (SPSS).
Supplementary Material
Acknowledgments
We thank Lawrence Maurice and the Scottish Agricultural College Craibstone Estate (Aberdeen, UK) for access to the Woodlands Field pH plots and the laboratories of Prof. David Stahl and Prof. Christa Schleper for cultures of N. maritimus and fosmid clone 54d9, respectively. This work was supported by a Royal Society of Edinburgh International Exchange grant (to J.I.P.); Natural Science Foundation of China Grants 30811130224 and 40871129 (to L.-M.Z. and J.-Z.H.); the K.C. Wong Education Foundation, Hong Kong (L.-M.Z. and J.-Z.H.); Advanced Fellowship NE/D010195/1 from the Natural Environment Research Council (to G.W.N.); Marie Curie Intra-European Fellowship PIEF-GA-2008-220639 (to P.R.O.); and Grant BB/F022646/1 from the Biotechnology and Biological Sciences Research Council (to D.T.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ168069–HQ168103 and HQ225747–HQ225751).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004947107/-/DCSupplemental.
References
- 1.Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 2.Raun WR, Johnson GV. Improving nitrogen use efficiency for cereal production. Agron J. 1999;91:357–363. [Google Scholar]
- 3.Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 4.Subbarao G, et al. Scope and strategies for regulation of nitrification in agricultural systems - challenges and opportunities. Crit Rev Plant Sci. 2006;25:303–335. [Google Scholar]
- 5.De Boer W, Kowalchuk GA. Nitrification in acid soils: Micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- 6.Treusch AH, et al. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 7.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 8.He JZ, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 9.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 10.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 11.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 13.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallam SJ, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strauss G, Fuchs G. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur J Biochem. 1993;215:633–643. doi: 10.1111/j.1432-1033.1993.tb18074.x. [DOI] [PubMed] [Google Scholar]
- 16.Zarzycki J, Brecht V, Müller M, Fuchs G. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc Natl Acad Sci USA. 2009;106:21317–21322. doi: 10.1073/pnas.0908356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 18.Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- 19.Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 20.Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- 21.Jia ZR, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 22.Di HJ, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- 23.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freitag TE, Chang L, Prosser JI. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ Microbiol. 2006;8:684–696. doi: 10.1111/j.1462-2920.2005.00947.x. [DOI] [PubMed] [Google Scholar]
- 25.Huber H, et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2008;105:7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thauer RK. Microbiology. A fifth pathway of carbon fixation. Science. 2007;318:1732–1733. doi: 10.1126/science.1152209. [DOI] [PubMed] [Google Scholar]
- 27.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 28.Gubry-Rangin C, Prosser JI, Nicol GW. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010 doi: 10.1111/j.1574-6941.2010.00971.x. in press. [DOI] [PubMed] [Google Scholar]
- 29.Kemp JS, Paterson E, Gammack SM, Cresser MS, Killham K. Leaching of genetically modified Pseudomonas fluorescens through organic soils: Influence of temperature, soil pH, and roots. Biol Fertil Soils. 1992;13:218–224. [Google Scholar]
- 30.Allen SE. Chemical Analysis of Ecological Materials. 2nd Ed. Boston: Blackwell Scientific Publications; 1989. [Google Scholar]
- 31.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66:5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicol GW, Tscherko D, Embley TM, Prosser JI. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol. 2005;7:337–347. doi: 10.1111/j.1462-2920.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- 33.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Offre PR, Nicol GW, Prosser JI. Autotrophic community profiling and quantification of putative autotrophic thaumarchaeal communities in environmental samples. Environ Microbiol Reports. 2010 doi: 10.1111/j.1758-2229.2010.00217.x. in press. [DOI] [PubMed] [Google Scholar]
- 37.Nicol GW, Tscherko D, Chang L, Hammesfahr U, Prosser JI. Crenarchaeal community assembly and microdiversity in developing soils at two sites associated with deglaciation. Environ Microbiol. 2006;8:1382–1393. doi: 10.1111/j.1462-2920.2006.01031.x. [DOI] [PubMed] [Google Scholar]
- 38.Nicolaisen MH, Ramsing NB. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J Microbiol Methods. 2002;50:189–203. doi: 10.1016/s0167-7012(02)00026-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.