There is a massive push to develop new drugs to treat viral infection. Traditional therapeutic strategies aim at viral proteins responsible for each and every step of viral replication. The main drawbacks of these approaches include an ever-increasing pool of drugs specific for a given virus and selection for drug-resistant viruses. An alternative strategy, which has recently gained popularity, targets cellular factors (not limited to viral receptors) involved in virus entry and replication (1, 2). Numerous cellular proteins aiding viral replication have recently emerged from genome-wide screens (3–6), showing the virus’ reliance on various cellular processes. Targeting less variable host factors is an attractive concept that is less prone to selecting for drug-resistant viruses. The flip side of this approach is the potential for serious side effects and the need to target a large and often nonoverlapping number of cellular factors. A study by St. Vincent et al. (7) in PNAS and the paper published earlier by another group (8) introduce an exciting paradigm that focuses on a universal cellular target, which happens to be an intricate part of all enveloped viruses. The authors (7) show that infection by enveloped viruses can be blocked by altering their membrane composition in a way that disfavors their merger with a target cell membrane.
Enveloped viruses surround their nucleocapsids with a host cell-derived lipid membrane and therefore must merge the viral and target cell membranes to initiate new infection. This step is promoted by structurally diverse fusion glycoproteins, which are activated by a specific cellular receptor (or several receptors) and/or acidic endosomal pH (9). Fusion proteins are believed to promote membrane merger by engaging the target membrane, and subsequently, refolding into a stable hairpin structure (Fig. 1A) (10, 11). Because viruses cannot directly use chemical energy released upon ATP or GTP hydrolysis, conformational energy stored in their envelope proteins seems to be the only driving force for membrane fusion (12).
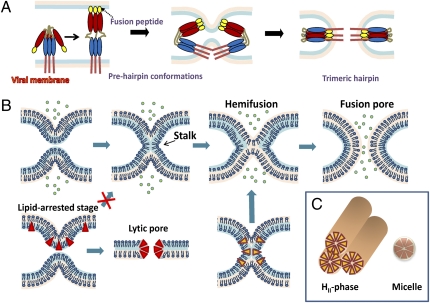
Fig. 1.
Viral protein refolding and lipid intermediates en route to membrane fusion. (A) Viral fusion protein refolding into a stable hairpin of trimers, which is coupled to lipid rearrangement through the formation of prehairpin intermediates. (B) Progression of lipid bilayer fusion through the stalk, hemifusion, and fusion pore formation. Lipophilic compounds conferring positive curvature (red triangles) stabilize prefused membranes, preventing the stalk formation and promoting the formation of lytic pores in lipid bilayers. Lipids conferring negative curvature (yellow inverted triangles) augment hemifusion. (C) Negative curvature lipids tend to form an inverted hexagonal HII-phase, whereas positive curvature lipids assemble into micelles.
Depending on the virus, the number of envelope glycoproteins could reach several hundred. The apparent surplus of these proteins reflects, in part, the importance of the fusion step and the nondeterministic nature of this process, which often fails to reach completion. There is evidence that several viral proteins must act in concert to effectively mediate fusion (13–18). Accordingly, the fusion efficiency is known to critically depend on the density of activated viral proteins (13, 19). The above considerations imply that, in general, the energy released from a single viral protein refolding may not be sufficient to destabilize lipid bilayers and promote their fusion. Thus, a synchronous activation and assembly of several fusion proteins into multimeric complexes might help overcome the energy barrier for membrane fusion.
The merger of lipid membranes involves the formation of highly curved (and thus energetically unfavorable) intermediates—stalk, hemifusion, and a fusion pore (Fig. 1B). The main contribution to the overall energy of these intermediates comes from elastic energy of bent monolayers (20), which depends on the intrinsic propensity of lipid sheets to deviate from planarity (described in terms of spontaneous curvature) (21). Lipids with larger polar head groups compared with their hydrocarbon tails confer a positive curvature by bending the membranes away from polar heads (Fig. 1 B and C). By contrast, lipids in which the cross-sectional area of the polar heads is smaller than that of the hydrophobic moiety confer a negative curvature. The merger of contacting monolayers is known to be augmented by negative curvature constituents, whereas lipids favoring the positive membrane curvature disfavor hemifusion (20). The inhibitory effect of positive curvature agents exemplified by lyso-lipids has been shown for diverse fusion reactions mediated by viral and cellular fusion proteins (22). These findings strongly imply that (i) all protein-mediated fusion reactions converge to a common lipid intermediate with a net negative curvature, most likely a hemifusion, and (ii) lipids are essential determinants of the outcome of protein-mediated fusion.
The study by St. Vincent et al. (7) introduces a class of wedge-shaped rigid amphipathic fusion inhibitors (RAFIs) that block infectivity of unrelated enveloped viruses, apparently through conferring a positive curvature to their lipid membranes. Like lyso-lipids, RAFIs seem to counteract the well-balanced action of fusion proteins through the lipid phase, without directly interacting with viral proteins. An important feature of RAFIs, as well as other compounds that target the viral membrane (8), is that these are not toxic for cells. The selective effect of RAFIs on the metabolically inactive viral membrane most likely originates from the lack of membrane repair mechanisms, which are effective in cells. As expected for compounds that alter the propensity of viral lipids to undergo fusion, RAFIs blocked infection by several enveloped viruses but did not affect the infectivity of nonenveloped viruses at much higher doses.
The overall shape of RAFIs and their ability to disfavor the transition from a lamellar to an inverted hexagonal phase (highly curved inverted lipid cylinders) (Fig. 1C) are consistent with the notion that these molecules confer a positive curvature to viral lipids, thereby antagonizing the action of viral fusion proteins. These results provide an exciting proof-of-concept for developing broad-spectrum entry inhibitors that could block fusion of virtually all enveloped viruses. Importantly, this class of drugs is unlikely to select for resistant variants, because viruses have virtually no control over their lipid composition.
Although the study by St. Vincent et al. (7) is an important milestone for future antiviral strategies, a number of questions
RAFIs blocked infection by several enveloped viruses but did not affect the infectivity of nonenveloped viruses.
remain unanswered. It is not completely clear yet whether altering the membrane curvature is the only or even the main mechanism of the RAFIs’ effect on enveloped viruses. Certain features of these compounds reported in this study might be indicative of additional modes of action. First, whereas amphipathic molecules conferring positive curvature lyse membranes by favoring the formation of lipidic pores (Fig. 1B), high concentrations of RAFIs did not seem to damage cells or lyse viruses. It is worth pointing out, however, that the results presented by St. Vincent et al. (7) argue against dissolution of the viral membrane by RAFIs but do not rule out the membrane permeabilizing effect. Second, the predominantly polar nature of amphipaths conferring positive curvature is manifested in a relatively high critical micelle concentration and the ease of their extraction from membranes on washing (13). In contrast, RAFIs seem to incorporate into viral membranes virtually irreversibly, as evidenced by their long-lasting inhibitory effect on pretreated viruses.
Further studies are needed to fully elucidate the mechanism of inhibition of viral fusion by RAFIs and related compounds. For instance, it would be interesting to determine the molar fraction of RAFIs in the viral membrane at an inhibitory concentration. This could help evaluate the corresponding change in spontaneous curvature based on the shift in the temperature of lamellar to inverted hexagonal phase transition measured in this study. Irrespective of the exact mechanism of action, the low cytotoxic effect of this class of viral fusion inhibitors might prove decisive for their future clinical applications.
Acknowledgments
The work on the mechanism of viral fusion in my laboratory is supported by National Institutes of Health Grants R01GM054787, AI053668, and R21AI079714.
Footnotes
The author declares no conflict of interest.
See companion article on page 17339.
References
- 1.Reeves JD, Piefer AJ. Emerging drug targets for antiretroviral therapy. Drugs. 2005;65:1747–1766. doi: 10.2165/00003495-200565130-00002. [DOI] [PubMed] [Google Scholar]
- 2.Kuritzkes DR. HIV-1 entry inhibitors: An overview. Curr Opin HIV AIDS. 2009;4:82–87. doi: 10.1097/COH.0b013e328322402e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 5.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 6.König R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St. Vincent MR, et al. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci USA. 2010;107:17339–17344. doi: 10.1073/pnas.1010026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf MC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci USA. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kielian M, Rey FA. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 13.Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: Restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentz J. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys J. 2000;78:227–245. doi: 10.1016/S0006-3495(00)76587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blumenthal R, Sarkar DP, Durell S, Howard DE, Morris SJ. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche S, Gaudin Y. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 2002;297:128–135. doi: 10.1006/viro.2002.1429. [DOI] [PubMed] [Google Scholar]
- 18.Markovic I, Pulyaeva H, Sokoloff A, Chernomordik LV. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J Cell Biol. 1998;143:1155–1166. doi: 10.1083/jcb.143.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markovic I, Leikina E, Zhukovsky M, Zimmerberg J, Chernomordik LV. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J Cell Biol. 2001;155:833–844. doi: 10.1083/jcb.200103005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helfrich W. Elastic properties of lipid bilayers: Theory and possible experiments. Z Naturforsch C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 22.Chernomordik LV, et al. Lysolipids reversibly inhibit Ca(2+)-, GTP- and pH-dependent fusion of biological membranes. FEBS Lett. 1993;318:71–76. doi: 10.1016/0014-5793(93)81330-3. [DOI] [PubMed] [Google Scholar]