Abstract
A zebrafish heart can fully regenerate after amputation of up to 20% of its ventricle. During this process, newly formed coronary blood vessels revascularize the regenerating tissue. The formation of coronary blood vessels during zebrafish heart regeneration likely recapitulates embryonic coronary vessel development, which involves the activation and proliferation of the epicardium, followed by an epithelial-to-mesenchymal transition. The molecular and cellular mechanisms underlying these processes are not well understood. We examined the role of PDGF signaling in explant-derived primary cultured epicardial cells in vitro and in regenerating zebrafish hearts in vivo. We observed that mural and mesenchymal cell markers, including pdgfrβ, are up-regulated in the regenerating hearts. Using a primary culture of epicardial cells derived from heart explants, we found that PDGF signaling is essential for epicardial cell proliferation. PDGF also induces stress fibers and loss of cell-cell contacts of epicardial cells in explant culture. This effect is mediated by Rho-associated protein kinase. Inhibition of PDGF signaling in vivo impairs epicardial cell proliferation, expression of mesenchymal and mural cell markers, and coronary blood vessel formation. Our data suggest that PDGF signaling plays important roles in epicardial function and coronary vessel formation during heart regeneration in zebrafish.
Keywords: epicardium, mesenchymal cells, mural cells, zebrafish heart regeneration
Coronary heart disease is among the leading causes of disability and mortality in the United States and worldwide (1). Scars form in injured human hearts, which results in decreased cardiac performance and the eventual development of heart failure (2). In contrast to humans, zebrafish and newts have remarkable regenerative abilities (3, 4). After 20% resection of the ventricle, zebrafish fully regenerate lost heart tissue (3, 4). During this process, newly formed coronary blood vessels vascularize the regenerating myocardium (5, 6). Expression of the embryonic epicardial markers tbx18 and raldh2 is induced in the epicardium of adult regenerating hearts (5, 6), suggesting that an embryonic gene expression program in the epicardium is activated in response to injury. This activation starts throughout the entire ventricle and gradually becomes localized to the apex. The activated epicardium proliferates from 3 to 7 d postamputation (dpa) (5). A previous study suggested that the activated epicardium undergoes an epithelial-to-mesenchymal transition (EMT) and subsequently contributes to newly formed coronary blood vessels (5). The lineages of the different cell types in blood vessels formed during zebrafish heart regeneration have not yet been conclusively determined.
Zebrafish heart regeneration, at least in part, likely recapitulates embryonic heart development. EMT is a key step during heart development in mice and chicks, wherein the epicardium forms epicardium-derived cells (EPDCs), which then differentiate into fibroblasts, smooth muscle cells (7–9), and potential cardiomyocytes (10, 11). The molecular aspects of zebrafish heart regeneration are poorly understood. Whether EMT actually occurs during zebrafish heart regeneration has not been determined. We previously identified PDGFs as promising candidates involved in heart regeneration (12). In this study, we find that EMT and mural cell markers are up-regulated during zebrafish heart regeneration. Using an in vitro explant culture that we established for adult zebrafish hearts, we demonstrate that PDGF signaling regulates proliferation of epicardial cells. PDGF also induces stress fibers and loss of cell-cell contacts of epicardial cells in explant culture. This effect is mediated by Rho-associated protein kinase (ROCK). We further show that PDGF signaling is required for epicardial cell proliferation, expression of mesenchymal and mural cell markers, and coronary blood vessel formation in vivo. Our studies demonstrate that PDGF plays important roles in zebrafish heart regeneration.
Results
EMT Markers Are Up-Regulated During Zebrafish Heart Regeneration.
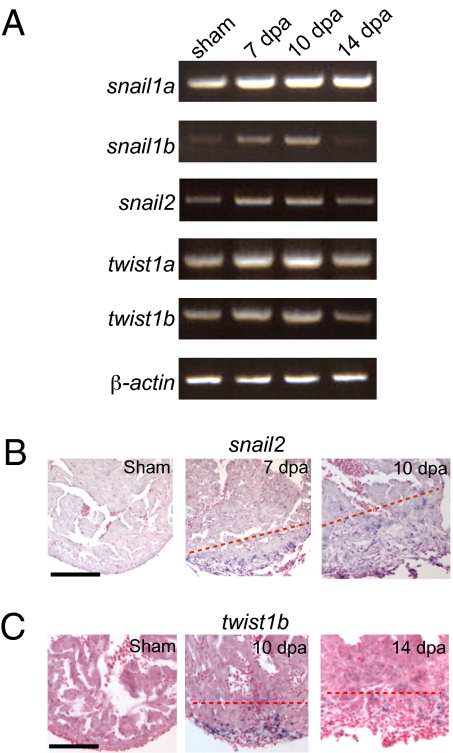
EMT has not been clearly demonstrated during zebrafish heart regeneration. To determine whether EMT of the epicardium actually occurs during heart regeneration, we examined the expression of the snail and twist genes, which are markers for EMT (13). RT-PCR using mRNA isolated from the regenerating hearts showed that expression of these markers was up-regulated (Fig. 1A). We confirmed the expression pattern of snail2 and twist1b by in situ hybridization (ISH). There was no snail2 expression in sham-operated hearts. At 7 and 10 dpa, snail2 was expressed in the wound and the fibrin clot (Fig. 1B). Similarly, we observed up-regulation of twist1b that peaked at 10 dpa (Fig. 1C). These data suggest that EMT might occur during zebrafish heart regeneration.
Fig. 1.
EMT markers are up-regulated during zebrafish heart regeneration in vivo. (A) RT-PCR analysis of EMT markers. Expression of snail1a, snail1b, snail2, twist1a, and twist1b was examined in sham and 7-, 10-, and 14-dpa hearts by semiquantitative RT-PCR. RT-PCR of β-actin is included as a loading control. (B) snail2 ISH in sham and 7- and 10-dpa hearts. (Scale bar = 100 μm.) (C) twist1b ISH in sham and 10- and 14-dpa hearts. The dashed line marks the approximate position of the amputation plane. (Scale bar = 100 μm.)
Mural Cells Are Present in Coronary Vessels During Zebrafish Heart Regeneration.
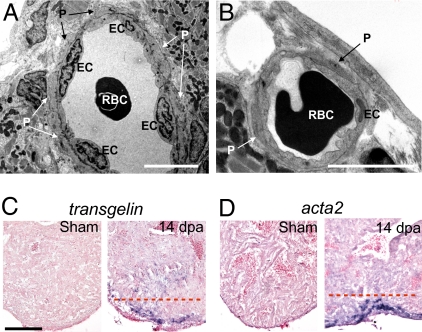
Zebrafish coronary blood vessels do not have smooth muscle cells. Instead, blood vessels are surrounded by pericyte-like mural cells (14). If epicardial cells undergo EMT during zebrafish heart regeneration, they most likely contribute to mural cells and fibroblasts of the coronary vessels. To determine whether endothelial cells in the wound and regenerating area are surrounded by mural cells and whether they are functional, we first examined blood vessels in clots from regenerating hearts by transmission EM. Although newly formed blood vessels were very primitive and consisted of only two to three endothelial cells, red blood cells could be readily observed within the lumen, suggesting that these blood vessels were functional. We observed mural cells associated with the endothelial cells in some of the blood vessels in the regenerating area (Fig. 2 A and B).
Fig. 2.
Mural cells are present in coronary vessels in the wound during zebrafish heart regeneration. (A and B) Transmission EM images of coronary vessels in the wound of 14-dpa regenerating hearts. EC, endothelial cells. P, pericytes. RBC, red blood cells. (Scale bar = 5 μm.) ISH using digoxigenin-labeled antisense probes against tagln (C) and acta2 (D) in sham-operated and 14-dpa regenerating hearts. The dashed red line marks the approximate position of the amputation plane. (Scale bar = 100 μm.)
We hypothesized that expression of mural cell markers should increase if mural cells form or are recruited to the wound during heart regeneration. We used ISH to examine the expression of mural cell markers such as sm22αb/transgelin (tagln) and α-smooth muscle actin (αSMA)/acta2 (15, 16). In 14-dpa hearts, tagln expression was strongly up-regulated at the wound site in the fibrin clot, whereas tagln expression was not detected in sham-operated control hearts (Fig. 2C). Similarly, acta2 expression was up-regulated at the wound site at 14 dpa (Fig. 2D). In addition to mural cells, these markers mark fibroblasts and mesenchymal cells. Our data suggest that mural cells, mesenchymal cells, and/or fibroblasts form or are recruited to the wound during heart regeneration.
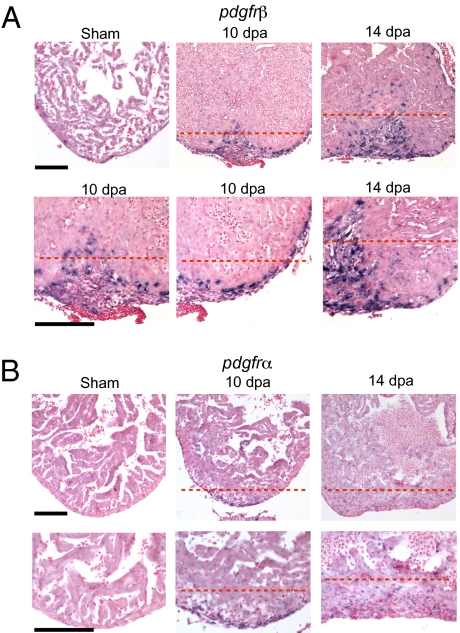
The PDGF receptor β (PDGFRβ) marks pericytes and perivascular mesenchymal cells in other organisms (17–19). Expression of the zebrafish homolog (pdgfrβ) (accession no. HM439112) (20) is strongly induced in the epicardium and subepicardium and in the fibrin clot formed at the wound site at 6, 10, and 14 dpa (Fig. 3A and Fig. S1). Compared with the strong expression of pdgfrβ, expression of the fibroblast marker pdgfrα is relatively weak (Fig. 3B). The induced expression of tagln, acta2, and pdgfrβ suggests that mural and/or mesenchymal cells but not fibroblasts form or are recruited to the wound after injury.
Fig. 3.
pdgfrβ is up-regulated during zebrafish heart regeneration. ISH using digoxigenin-labeled antisense probes against pdgfrβ (A) and pdgfrα (B) in sham-operated and 10- and 14-dpa regenerating hearts. The lower row of images in A and B contain enlarged images of the upper row. The enlarged images are also slightly panned to show the epicardium. The dashed red line marks the approximate position of the amputation plane. (Scale bar = 100 μm.)
Establishment of an Adult Zebrafish Epicardium/EPDC Explant Culture.
Interestingly, pdgfrβ is expressed in the epicardium of developing embryonic mouse hearts and is required for coronary artery formation (21, 22). The expression of pdgfrβ in the wound and the epicardium (Fig. 3) suggested that PDGF signaling might play a role in epicardial function and coronary vessel formation during heart regeneration. pdgfb, which codes for PDGF-B, is also expressed at the wound site (12). This suggested that locally expressed PDGF-B, likely via CD41-positive thrombocytes in the wound site (Fig. S2), may exert a paracrine effect on pdgfrβ-expressing epicardial cells.
To study the role of PDGF signaling on epicardial cells at the cellular level, we established an explant culture of adult zebrafish epicardial cells. To mimic the in vivo conditions under which the epicardial cells migrate into the fibrin clot, we cultured explanted hearts on a fibrin gel (details are provided in Materials and Methods). When sham-operated hearts were cultured, a monolayer of epithelium-like cells migrated out of explants within 3 d (Fig. S3). Nearly all the cells had strong nuclear capsulin/epicardin/transcription factor 21 (hereafter referred to as epicardin) staining (Fig. S3) and a typical epithelial phenotype characterized by close cell-cell contact marked by subcortical actin bundles and expression of the tight junction marker ZO1 (23, 24) (Fig. S3). Cells at the periphery of this monolayer typically did not detach. The phenotype of the epicardial cells from sham-operated hearts was similar to that of cells obtained from chick and quail embryonic epicardial explants (25, 26). These data suggest that these migrating cells are likely epicardial cells or EPDCs.
PDGF Induces Stress Fibers and Loss of Epithelial Phenotypes in Epicardial Cells in Vitro.
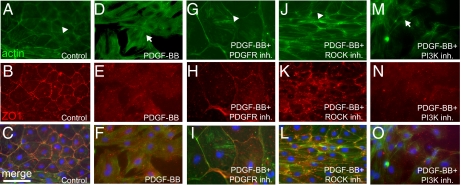
We used this explant culture to determine the effects of PDGF signaling on epicardial cells. Treatment of the epicardial cell monolayer with recombinant PDGF-BB induced the cells to detach from the monolayer and form mesenchymal-like cell types in sham-operated heart explants (Fig. S4 B and D). Compared to controls treated with BSA (Fig. 4 A–C), PDGF-BB caused a reorganization of subcortical actin into stress fibers (Fig. 4 D and F) and the loss of cell-cell contacts marked by ZO1 expression (Fig. 4 E and F) in cells at the periphery of the monolayer. These changes in gene expression and cytoskeletal structure induced by PDGF-BB were blocked by a selective PDGFR inhibitor (PDGFR inh.V; Calbiochem), which inhibits PDGF signaling in zebrafish embryos (20, 27) (Fig. 4 G–I).
Fig. 4.
PDGF signaling is sufficient to induce stress fibers and loss of cell-cell contacts in vitro. Sham-operated hearts were cultured in the presence of 0.5% serum for 3 d and then treated with 0.5% BSA as a control (n = 12 hearts) (A–C), PDGF-BB (n = 4 hearts) (D–F), PDGF-BB and PDGFR inhibitor (Inh.; 2 μM, n = 4 hearts) (G–I), PDGF-BB and ROCK inhibitor (2 μM, n = 4 hearts) (J–L), or PDGF-BB and PI3K inhibitor (2 μM, n = 4 hearts) (M–O) for 1 d (i.e., from day 3 to day 4 in culture) (actin, green; DAPI: blue; ZO1, red). Actin localized to the subcortical region is marked by arrowheads, and stress fibers are marked by arrows. Each individual explant was imaged in at least three different fields of view, and a representative image is shown. (Scale bar = 50 μm.)
Rho-associated protein kinase ROCK mediates PDGF-induced EMT in quail embryo proepicardium explants (28). ROCK also mediates EMT of neural crest cells in developing zebrafish embryos (29). To determine whether PDGF function on zebrafish epicardial cells also depends on ROCK, we tested the effect of blocking ROCK activity in our explant culture. We found that treating the cells with the selective ROCK inhibitor Y-27632 (Tocris) completely blocked PDGF-BB–induced stress fibers and loss of cell-cell contacts (Fig. 4 J–L). PI3K is another downstream effector of PDGF signaling (30). Compared with the ROCK inhibitor, the PI3K inhibitor LY-294002 (Calbiochem) did not have a strong effect (Fig. 4 M–O) but inhibited cell proliferation instead (see below). These results suggest that PDGF-BB induces a loss of epithelial phenotypes in cultured zebrafish epicardial cells and that this function requires signaling via ROCK.
PDGF Signaling Is Required for Epicardial Proliferation.
We further tested whether PDGF regulates the proliferation of epicardial cells. In vitro, ∼61% of cultured epicardial cells from 4-dpa regenerating heart explants retained their proliferative capacity and underwent DNA synthesis after treatment with the vehicle control DMSO (Fig. S5). This is consistent with the time frame of epicardial proliferation in vivo (between 3 and 7 dpa). We observed a dose-dependent inhibitory effect of PDGFR inhibitor on BrdU incorporation of epicardial cells (Fig. S5A). Furthermore, the PI3K inhibitor significantly blocked DNA synthesis, whereas the ROCK inhibitor had little effect (Fig. S5 B and C). These results suggest that PDGFR signaling is required for epicardial cell DNA synthesis in vitro and that PI3K but not ROCK is required for epicardial proliferation.
To determine whether PDGFR signaling is required for epicardial cell proliferation in vivo, we injected zebrafish with BrdU and treated the regenerating fish with PDGFR inhibitor from 2–7 dpa. Approximately 45% of epicardial cells incorporated BrdU when the fish were treated with the vehicle control DMSO (n = 11; Fig. S6 A and B), whereas only 14% of epicardial cells were BrdU-positive when zebrafish were treated with PDGFR inhibitor (n = 11; Fig. S6 A and B). These results suggest that PDGFR signaling is required for epicardial cell proliferation in vivo.
PDGF Signaling Is Required for Coronary Blood Vessel Formation During Zebrafish Heart Regeneration.
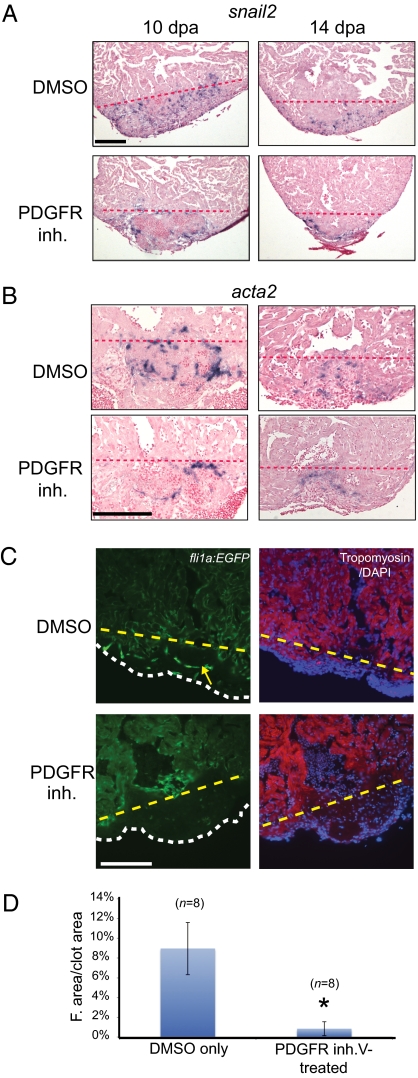
To determine the role of PDGF signaling during zebrafish heart regeneration in vivo, we treated regenerating hearts with PDGFR inhibitor from 2–14 dpa (i.e., when the EMT potentially occurs) and examined the expression of EMT and mural cell markers. At 10 dpa, snail2 expression was found in the wound and fibrin clot in DMSO-treated control fish (n = 6), similar to Fig. 1. The expression of snail2 gradually decreased by 14 dpa (Fig. 5A). By contrast, PDGFR inhibitor-treated hearts had reduced snail2 expression with residual snail2 expression mainly found outside the fibrin clot area (n = 5) (Fig. 5A). Similarly, we observed decreased expression of acta2 in the PDGFR inhibitor-treated 10-dpa hearts (Fig. 5B). These results suggest that PDGF signaling is required for expression of EMT and mural cell markers.
Fig. 5.
PDGF signaling is required for coronary blood vessel formation during zebrafish heart regeneration. (A) Treatment of wild-type fish with DMSO or PDGFR inhibitor (inh.) from 2–14 dpa. snail2 ISH was performed using 10- and 14-dpa hearts. (Scale bar = 100 μm.) (B) Treatment of wild-type fish with DMSO or PDGFR inhibitor from 2–14 dpa. acta2 ISH was performed using 10- and 14-dpa hearts. (Scale bar = 100 μm.) (C) Treatment of fli1a:EGFP fish with PDGFR inhibitor from 2–14 dpa. Hearts were collected and processed at 14 dpa (DAPI staining, blue; tropomyosin staining, red). An example of blood vessel formation (green) in the regenerating hearts is marked by the yellow arrow. The dashed lines mark the approximate position of the amputation plane in A–C. The white dashed line indicates the outline of the fibrin clot in the regenerating hearts. (Scale bar = 100 μm.) (D) Quantification of blood vessel formation. The total fluorescent (F.) area was measured relative to total clot area (n = 8 hearts for DMSO and PDGFR inhibitor treatment). *P < 0.0001.
To determine the role of PDGF signaling on blood vessel formation during zebrafish heart regeneration in vivo, we examined hearts from 14-dpa zebrafish harboring a fli1a:EGFP transgene, which marks endothelial cells (31). In DMSO-treated control fish (n = 8), blood vessels appeared in the clot and invaded the regenerating myocardium. By contrast, PDGFR inhibitor-treated hearts lacked blood vessels in the wound (n = 8) (Fig. 5 C and D). The lack of blood vessels might indicate that no endothelial cells were formed, or it might indicate that endothelial cells were formed but did not survive to 14 dpa. To distinguish between these two possibilities, we examined the amputated hearts at earlier time points. Endothelial cells first appeared in the wound as early as 7 dpa in DMSO-treated control hearts. By contrast, endothelial cells were not observed in 7- or 10-dpa PDGFR inhibitor-treated hearts, suggesting that endothelial cells do not form after PDGFR inhibition (Fig. S7). These results suggest that blood vessel formation during zebrafish heart regeneration requires PDGF signaling. However, our results cannot absolutely rule out the possibility that PDGFR inhibitor might also act with reduced efficacy on other tyrosine kinase receptors in vivo.
Discussion
Neovascularization is a promising strategy for treating ischemic heart disease (32). Formation of coronary blood vessels occurs naturally during zebrafish heart regeneration (5). To develop neovascularization further as a therapeutical strategy in mammals, it is imperative to understand how zebrafish form coronary blood vessels during heart regeneration, which likely recapitulates embryonic heart development (5). Neovascularization during heart regeneration might be similar to coronary vessel formation during heart development. This study establishes that PDGF signaling plays a central role in reactivating developmental neovasculogenesis in adult zebrafish after heart injury.
Endothelial cells in the regenerating area might arise from epicardial EMT or wound angiogenesis; however, our data do not allow us to distinguish between these two possibilities. Currently, we also cannot determine whether PDGF signaling acts directly or indirectly on endothelial cells. It is possible that PDGF signaling regulates mural cells, which can affect endothelial cell formation or angiogenesis. Epicardial cells might also undergo EMT and contribute to mural cell formation during zebrafish heart regeneration. Our results showed that PDGF signaling is required for mesenchymal and mural cell formation during heart regeneration in vivo. Interestingly, epicardium-specific pdgfrβ knock-outs in mice show abnormal clustering of endothelial cells, absence of epicardial-derived smooth muscle cells, and defective coronary artery formation (22), indicating an essential role for PDGF signaling in coronary vessel development. Therefore, despite the fact that pericytes rather than smooth muscle cells surround the endothelial cells of blood vessels in zebrafish (14), molecular events in the developing mouse heart are very similar to what we observed during zebrafish heart regeneration. For instance, some tbx18-positive cells have been shown to express pdgfrβ in embryonic mouse hearts, indicative of the epicardial contribution of pdgfrβ-marked vascular support cells (10). However, we cannot rule out the possibility that some acta2- and/or pdgfrβ-positive cells are mesenchymal cells that are not derived from epicardium but are recruited to the wound.
Zebrafish heart regeneration involves multiple tissue types, including the epicardium and myocardium. It is therefore important to delineate the separate roles of the myocardium and epicardium. Previous studies of heart regeneration that used the hsp70 promoter to drive transgene expression could not address tissue specificity (5), making it difficult to distinguish between cell-autonomous vs. non–cell-autonomous effects. In principle, zebrafish harboring epicardium-specific inducible transgenes expressing a dominant-negative form of pdgfrβ can be used to address these questions via a loss-of-function genetic approach. However, compared with other model organisms, temporal-spatial control of transgene expression has not been well established in adult zebrafish. The lack of an in vitro culture system has also hampered mechanistic studies of cellular processes. Therefore, we have developed in vitro primary cultures of cardiomyocytes (12) and epicardial cells (this study) to address the individual roles of these tissue types during heart regeneration. Our unique in vitro approach complements the strengths of in vivo studies.
We previously showed that PDGF signaling is required for cardiomyocyte proliferation during zebrafish heart regeneration. Although we demonstrated that PDGF-BB can induce DNA synthesis of primary cultured zebrafish cardiomyocytes (12), we cannot rule out an indirect effect from the epicardium, which can potentially secrete growth factors that stimulate proliferation of cardiomyocytes. The functions we observed for PDGF signaling last until 14 dpa. We observed expression of pdgfb across this time period but only a very transient expression of pdgfa (12). In addition, we observed much stronger expression of pdgfrβ than pdgfrα. Therefore, PDGF-B is most likely the major ligand that signals through PDGFRβ during heart regeneration.
Our results provide evidence for a crucial role for PDGF signaling in the epicardium during zebrafish heart regeneration. Interestingly, controlled sequential release of PDGF-BB together with VEGF-A was shown to induce mature blood vessels and improve cardiac function in cardiac ischemia in mice (33). Carefully manipulating the delivery timing and method of PDGF and/or its downstream signaling effectors can potentially serve as a therapeutical option for heart repair.
Materials and Methods
Zebrafish Husbandry, Heart Amputation, and Inhibitor Treatment.
Details are provided in SI Text. For testing the effects of PDGFR inhibitor (PDGFR inh. V), hearts of wild-type fish were amputated as described (12). Fish were then treated in water containing PDGFR inhibitor (0.25 μM). DMSO was used as a control. In vivo BrdU labeling was done as described (12). Blood vessel formation was quantified by measuring the area of fli1a:EGFP fluorescence signaling using ImageJ (National Institutes of Health).
RT-PCR.
Primer sequences for snail1a, snail1b, snail2, twist1a, and twist1b RT-PCR are listed SI Text.
ISH.
Whole-mount ISH and section ISH were performed essentially as described (5, 12).
Fibrin Gel Heart Explant Culture.
Heart ventricles of sham-operated control and regenerating fish were dissected away from heart tissue such as the atrium and heart valves. The ventricles were washed in PBS to remove blood and placed on top of a fibrin gel with the epicardium contacting the fibrin gel, which was prepared as previously described (34). For all culture conditions, 0.5% serum (FBS) was used, with no additional supplements to maintain fibrin gel integrity. The explant cultures from 4-dpa regenerating hearts were derived from the apex half of ventricles. PDGF-BB or various inhibitors were added directly to the culture media at the indicated concentrations. Immunohistochemistry and BrdU labeling were performed using standard procedures (details are provided in SI Text).
EM.
EM was performed according to standard procedures (details are provided in SI Text).
Immunostaining of Thrombocytes.
Details of thrombocyte immunostaining are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Dr. K. Poss for protocols and advice; Dr. S. Childs (University of Calgary) for αSMA (acta2) and sm22αb (tagln) in situ probes; Dr. G. Crump (University of Southern California) for twist1b probe; Dr. M. Nieto (Instituto de Neurociencias CSIC-UMH) for the snail2 probe; Drs. W. Shi and L. Perin for help with microscopy; and Drs. V. Kaartinen, D. Kohn, H. Sucov, and D. Warburton for critical reading of the manuscript. This work was supported by American Heart Association Grant 0730214N (to C.-L.L.); National Heart, Lung, and Blood Institute Grant R01HL096121 (to C.-L.L.); the Wright Foundation (to C.-L.L.), a Chinese American Faculty Association Faculty Development grant (to C.-L.L.); a Research Career Development Award from the Saban Research Institute (to C.-L.L.); National Institute of General Medical Sciences Grant R01GM055081 (to T.-L.T.); and California Institute for Regenerative Medicine (CIRM) postdoctoral fellowships (to J.K. and K.M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0915016107/-/DCSupplemental.
References
- 1.Lloyd-Jones D, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Executive summary: Heart disease and stroke statistics—2010 update: A report from the American Heart Association. Circulation. 2010;121:948–954. doi: 10.1161/CIRCULATIONAHA.109.192666. [DOI] [PubMed] [Google Scholar]
- 2.Holmes JW, Borg TK, Covell JW. Structure and mechanics of healing myocardial infarcts. Annu Rev Biomed Eng. 2005;7:223–253. doi: 10.1146/annurev.bioeng.7.060804.100453. [DOI] [PubMed] [Google Scholar]
- 3.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 4.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 6.Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Olivey HE, Compton LA, Barnett JV. Coronary vessel development: The epicardium delivers. Trends Cardiovasc Med. 2004;14:247–251. doi: 10.1016/j.tcm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 9.Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ Res. 2002;91:761–768. doi: 10.1161/01.res.0000038961.53759.3c. [DOI] [PubMed] [Google Scholar]
- 10.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu N, Yost HJ, Clark EB. Cardiac morphology and blood pressure in the adult zebrafish. Anat Rec. 2001;264:1–12. doi: 10.1002/ar.1111. [DOI] [PubMed] [Google Scholar]
- 15.Santoro MM, Pesce G, Stainier DY. Characterization of vascular mural cells during zebrafish development. Mech Dev. 2009;126:638–649. doi: 10.1016/j.mod.2009.06.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgijevic S, et al. Spatiotemporal expression of smooth muscle markers in developing zebrafish gut. Dev Dyn. 2007;236:1623–1632. doi: 10.1002/dvdy.21165. [DOI] [PubMed] [Google Scholar]
- 17.Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512–523. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- 18.Hellström M, Kalén M, Lindahl P, Abramsson A, Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl P, et al. Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Development. 1998;125:3313–3322. doi: 10.1242/dev.125.17.3313. [DOI] [PubMed] [Google Scholar]
- 20.Wiens KM, et al. Platelet-derived growth factor receptor beta is critical for zebrafish intersegmental vessel formation. PLoS ONE. 2010;5:e11324. doi: 10.1371/journal.pone.0011324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van den Akker NM, et al. PDGF-B signaling is important for murine cardiac development: Its role in developing atrioventricular valves, coronaries, and cardiac innervation. Dev Dyn. 2008;237:494–503. doi: 10.1002/dvdy.21436. [DOI] [PubMed] [Google Scholar]
- 22.Mellgren AM, et al. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 24.Sridurongrit S, Larsson J, Schwartz R, Ruiz-Lozano P, Kaartinen V. Signaling via the Tgf-beta type I receptor Alk5 in heart development. Dev Biol. 2008;322:208–218. doi: 10.1016/j.ydbio.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol. 2001;234:204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- 26.Landerholm TE, et al. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development. 1999;126:2053–2062. doi: 10.1242/dev.126.10.2053. [DOI] [PubMed] [Google Scholar]
- 27.Eberhart JK, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, et al. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev Biol. 2001;240:404–418. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- 29.Berndt JD, Clay MR, Langenberg T, Halloran MC. Rho-kinase and myosin II affect dynamic neural crest cell behaviors during epithelial to mesenchymal transition in vivo. Dev Biol. 2008;324:236–244. doi: 10.1016/j.ydbio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tallquist M, Kazlauskas A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004;15:205–213. doi: 10.1016/j.cytogfr.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 32.Molin D, Post MJ. Therapeutic angiogenesis in the heart: Protect and serve. Curr Opin Pharmacol. 2007;7:158–163. doi: 10.1016/j.coph.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Hao X, et al. Angiogenic effects of sequential release of VEGF-A165 and PDGF-BB with alginate hydrogels after myocardial infarction. Cardiovasc Res. 2007;75:178–185. doi: 10.1016/j.cardiores.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 34.Tuan TL, Grinnell F. Fibronectin and fibrinolysis are not required for fibrin gel contraction by human skin fibroblasts. J Cell Physiol. 1989;140:577–583. doi: 10.1002/jcp.1041400324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.