Abstract
AIM: To investigate the accuracy of serum alanine aminotransferase (ALT) in diagnosing lamivudine resistance and factors that contributed to abnormal serum ALT.
METHODS: This was a retrospective study of chronic hepatitis B patients on lamivudine therapy who were followed for 3-mo with liver function tests and hepatitis B virus (HBV) DNA measurement. Lamivudine resistance was defined as HBV DNA ≥ 1 log from nadir on at least 2 occasions, confirmed by genotyping. Serum ALT levels in patients with lamivudine resistance were compared to serum ALT levels in those without lamivudine resistance.
RESULTS: There were 111 patients with and 117 without lamivudine resistance. The area under the receiver operating characteristic of serum ALT to diagnose lamivudine resistance was 0.645 ± 0.037. Serum ALT > 42.5 U/L gave the best diagnostic accuracy with sensitivity = 61%, specificity = 60%, positive predictive value = 60%, negative predictive value = 61%, positive likelihood ratio = 1.53 and negative likelihood ratio = 0.65 for predicting lamivudine resistance, missing 39% of resistant patients. Using other serum ALT cutoffs, diagnostic accuracy was lower. By multivariate analysis, baseline abnormal serum ALT was associated with abnormal ALT during resistance (OR = 5.98, P = 0.003), and males were associated with serum ALT flares during resistance (OR = 8.9, P = 0.016).
CONCLUSION: Serum ALT is inadequate for diagnosing lamivudine resistance and has implications where viral resistance testing is suboptimal and for reimbursement of rescue therapy.
Keywords: Antiviral therapy, Chronic hepatitis B, Sensitivity, Specificity, YMDD mutants
INTRODUCTION
Chronic hepatitis B (CHB) is a global public health problem, being the most common cause of chronic viral hepatitis, and the major cause of hepatocellular carcinoma worldwide[1]. There are now five licensed oral antiviral agents for CHB: lamivudine, adefovir, entecavir, telbivudine and tenofovir. Although lamivudine is not recommended as first line therapy for CHB in the American Association for Study of Liver Disease (AASLD) CHB guidelines[2,3], and the European Association for Study of Liver (EASL) CHB guidelines[4], which recommend oral nucleosides with high genetic barrier (entecavir and tenofovir) as first line monotherapy, lamivudine is still widely used in Asia due to cost constraints. Lamivudine is an effective treatment for hepatitis B “e” antigen (HBeAg) positive patients, with HBeAg seroconversion in 16%-18% of patients compared with 4%-6% of untreated controls[5,6]. Lamivudine is also used to treat HBeAg negative Hepatitis B virus (HBV) infection, with serum HBV DNA suppressed to undetectable levels using polymerase chain reaction (PCR) assays in 60%-70% of patients after 1 year of treatment[7,8]. However, lamivudine resistance is a major concern, with genotypic resistance increasing from 14% in year 1 to 38%, 49%, 66% and 69% after 2, 3, 4 and 5 years of treatment, respectively[9]. Detection of genotypic resistance is the first event to occur in the development of viral resistance, followed by the occurrence of viral breakthrough, described as a 1 log rise in HBV DNA above the nadir (in a compliant patient), and finally clinical resistance occurs when alanine aminotransferase (ALT) is abnormal[10]. However, genotypic lamivudine resistance tests are not readily available in developing countries and are expensive. Once resistance develops, rescue therapy with add-on adefovir is usually recommended early before the onset of clinical resistance[4] as the efficacy of rescue therapy appears to be better with early rescue compared to delayed rescue, but it is not clear whether all patients will have clinical resistance if left untreated. Consequently, the utility of ALT to diagnose lamivudine resistance is unclear. Once resistance occurs, strategies for the management of viral resistance are likely to be varied depending on the availability of resources, reimbursement policies, expertise of the physicians, and available generic medications. In addition, monitoring for viral resistance with either genotypic tests or HBV DNA may be prohibitively expensive in developing countries and some patients may only be able to afford to have frequent ALT testing as a surrogate for lamivudine resistance. Thus, we aimed to determine the accuracy of ALT in diagnosing lamivudine resistance, and to determine factors associated with abnormal ALT upon development of viral resistance.
MATERIALS AND METHODS
Study design and patient population
This study was a retrospective analysis of a prospectively collected database. All patients started on lamivudine for clinical indications at the Hepatology Clinic in the National University Hospital, Singapore were enrolled into a clinical database and followed every 3 mo with liver function tests and HBV DNA measurement from December 1999 to January 2007. Patients were excluded if they had prior lamivudine therapy or had organ transplantation. Patient demographic data, baseline biochemical parameters, HBV DNA viral load and lamivudine resistance mutations were entered into the database. This study was approved by the National Healthcare Group Institutional Review Board. Waiver of consent was approved by the Institutional Review Board as patient identifiers were removed (anonymised) during data collection.
Liver function tests (LFTs) were performed using Advia Chemistry, (Advia Centaur Systems, Siemens Medical Solutions Diagnostics Pty Ltd, Bayswater, Australia). HBsAg, HBeAg and anti-HBe were tested using Roche Diagnostic kits (Roche Diagnostics GmbH, Mannheim, Germany). Up to April 2006, serum HBV DNA was measured with the Hybrid Capture II HBV DNA Test (Digene Corporation, Gaithersburg, MD, USA) with a detection range of 1.4 × 105 copies/mL (6.1 × 104 IU/mL) to 1.7 × 109 copies/mL (7.4 × 108 IU/mL). Since April 2006, HBV DNA levels were measured with the Artus HBV RG (real time) PCR kit (Qiagen Diagnostics, Hamburg, Germany), with a detection range of 100 copies/mL (1.7 × 101 IU/mL) to 1 × 109 copies/mL (1.7 × 108 IU/mL). HBV DNA results were standardized by converting to WHO IU/mL[11]. The conversion factors for the Hybrid Capture II HBV DNA Test was 2.3 copies/mL[12], and for the Artus HBV RG (real time) PCR kit was 5.8 copies/mL equivalent to 1 IU/mL[13]. Lamivudine resistance was tested at the time of virological breakthrough, and was not tested at baseline. To test for lamivudine resistance, the HBV DNA polymerase gene RT domain was amplified by PCR followed by the INNO-LiPA HBV DR v2 detection kit (INNOGENETICS N.V. Belgium). Screened mutations for lamivudine resistance included rtL80V/I, rtV/G173L, rtL180M and rtM204V/I/S.
Definition of terms
Patients with lamivudine resistance were defined as those who had virological breakthrough and the presence of mutations conferring resistance confirmed by genotyping, including rtL80V/I, rtV/G173L, rtL180M and rtM204V/I/S.
Virological breakthrough was defined as a rise in HBV DNA of 1 log from nadir on at least 2 occasions after achieving virologic response during continuous treatment. Persistent abnormal ALT was defined as ALT above the upper limit of normal (ULN) for more than 1 mo during the treatment period, regardless of prior ALT normalization. Patients with persistently elevated ALT during treatment (absence of biochemical response) and with viral resistance were also considered to have persistently abnormal ALT. ALT flare was defined as ALT more than 5 times the ULN[14]. ALT during lamivudine resistance (taken at the time point of virological breakthrough) was compared to ALT levels in those without lamivudine resistance (using the mean ALT value of all time points during treatment). The diagnosis of cirrhosis was based on liver biopsy, when available, or clinical, biochemical and ultrasonography findings.
Statistical analysis
All data was analyzed using the statistical package SPSS (version 12.0: SPSS Inc., Chicago, IL, USA). Categorical data were described in number and percentage, and tested by Fisher’s exact test for univariate analysis. Continuous data were tested for normality, described in mean (95% CI), and analyzed by independent sample t-test. Statistically significant differences between analyzed groups were defined when the P value < 0.05. All clinically important variables were included in multivariate analysis using multiple logistic regression. Variables with multi-collinearity were excluded from the model. Analyzed results were shown as point estimates and 95% CI. The sensitivity and specificity of ALT in diagnosing lamivudine resistance was tested using the area under the receiver operating characteristic (AUROC) curve. ALT levels used for this analysis were those measured at the same time point during treatment in which viral rebound was detected.
RESULTS
A total of 228 subjects were included in the analysis, 111 of whom had lamivudine resistance, and 117 had no lamivudine resistance. The majority were Chinese [n = 215 (94.3%)] males [n = 167 (73.2%)], with a median age of 48.2 years. Seventy-one (31.1%) had cirrhosis, and 119 (52.2%) had HBeAg positive CHB. At baseline, the 2 groups with and without lamivudine resistance were similar in gender (P = 0.928), race (P = 0.183), age (P = 0.13), baseline bilirubin (P = 0.899), albumin (P = 0.541), ALT (P = 0.650), aspartate aminotransferase (AST) (P = 0.891) and cirrhosis (P = 0.682). However, patients who developed lamivudine resistance were more likely to be HBeAg positive and have higher HBV DNA at baseline (Table 1), and these parameters were significant after multivariate analysis (Table 2). Of these 111 patients, 74 had abnormal ALT after the development of lamivudine resistance, with a mean duration between viral breakthrough and abnormal ALT of 11.1 + 15.1 mo.
Table 1.
Baseline characteristics of lamivudine treated patients
| Lamivudine resistance (n = 111) | Lamivudine no resistance (n = 117) | P value | |
| Male, n (%) | 81 (73.0) | 86 (73.5) | 0.928 |
| Chinese, n (%) | 107 (96.4) | 108 (92.3) | 0.183 |
| Age, yr [mean (95%CI)] | 46.5 (44-49) | 49.6 (47-52) | 0.080 |
| Cirrhosis, n (%) | 36 (32.4) | 35 (29.9) | 0.682 |
| Baseline bilirubin, μmol/L [mean (95% CI)] | 19 (15-23) | 30 (17-43) | 0.124 |
| Baseline albumin, g/L [mean (95% CI)] | 36 (34-37) | 37 (36-38) | 0.326 |
| Baseline ALT, U/L [mean (95% CI)] | 175 (131-219) | 269 (191-348) | 0.038 |
| Baseline AST, U/L [mean (95% CI)] | 132 (88-176) | 197 (132-262) | 0.100 |
| HBeAg positive, n (%) | 77 (70.0) | 42 (35.9) | < 0.001 |
| Baseline log HBV DNA, IU/mL [mean (95% CI)] | 6.5 (6.3-6.8) | 5.7 (5.4-6.1) | < 0.001 |
| Baseline abnormal ALT, n (%) | 59 (53.15) | 65 (55.55) | 0.716 |
Patients with (n = 111) and without lamivudine resistance (n = 117). ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; HBV: Hepatitis B virus; HBeAg: Hepatitis B "e" antigen.
Table 2.
Multivariate analysis of baseline characteristics between patients with and without lamivudine resistance
| Adjusted P value | Adjusted OR (95% CI) | |
| Male | 0.842 | 1.080 (0.506-2.307) |
| Chinese | 0.343 | 2.099 (0.454-9.707) |
| Mean age, yr | 0.448 | 0.989 (0.960-1.018) |
| Cirrhosis | 0.977 | 1.011 (0.479-2.133) |
| Baseline bilirubin, μmol/L | 0.457 | 0.996 (0.984-1.007) |
| Baseline albumin, g/L | 0.108 | 0.950 (0.893-1.011) |
| Baseline ALT, U/L | 0.099 | 0.997 (0.994-1.000) |
| Baseline AST, U/L | 0.521 | 1.001 (0.998-1.005) |
| HBeAg positive | 0.005 | 2.857 (1.383-5.903) |
| Baseline log HBV DNA, IU/mL | 0.014 | 1.415 (1.073-1.865) |
| Baseline abnormal ALT | 0.923 | 0.959 (0.405-2.27) |
Baseline hepatitis B virus (HBV) DNA and hepatitis B "e" antigen (HBeAg) status were independent predictors of lamivudine resistance. ALT: Alanine aminotransferase; AST: Aspartate aminotransferase.
HBV DNA and lamivudine resistance
Out of the 228 patients’ baseline HBV DNA results, only 13 were measured with the Artus HBV RG PCR kit. The rest were measured with the Hybrid Capture II HBV DNA Test. Among the 111 patients with lamivudine resistance, viral breakthrough in 86 patients was detected with the Hybrid Capture II HBV DNA Test. The other 25 patients were detected with the Artus HBV RG PCR kit. For the 117 patients with no lamivudine resistance, these patients continued to have undetectable HBV DNA which was confirmed on subsequent tests with the Artus HBV RG PCR kit during follow-up.
Accuracy of serum ALT as a diagnostic marker for lamivudine resistance
When the entire group of lamivudine resistant patients was analyzed, the AUROC was 0.645 (95% CI: 0.569-0.715, standard error: 0.037) for ALT in the diagnosis of lamivudine resistance, with ALT > 42.5 U/L giving the best diagnostic accuracy with a sensitivity of 61%, specificity of 60%, positive predictive value of 60%, negative predictive value of 61%, positive likelihood ratio of 1.53, and negative likelihood ratio of 0.65 for predicting lamivudine resistance (Figure 1 and Table 3). Using this cutoff, lamivudine resistance would be missed in 39% of resistant patients. Using other ALT cutoffs, the diagnostic accuracy for lamivudine resistance was poorer (Table 3). Based on the normal range in our hospital laboratory (ULN ≤ 70 U/L), only 39% of patients would be diagnosed with lamivudine resistance and 61% would be missed. Based on the new AASLD guidelines (2), which suggested that the ULN for ALT should be decreased to 30 U/L for men and 19 U/L for women, 85% of males and 90% of females would be diagnosed with lamivudine resistance, but a high false positive resistance rate would be observed (72% of males and 93% of females).
Figure 1.
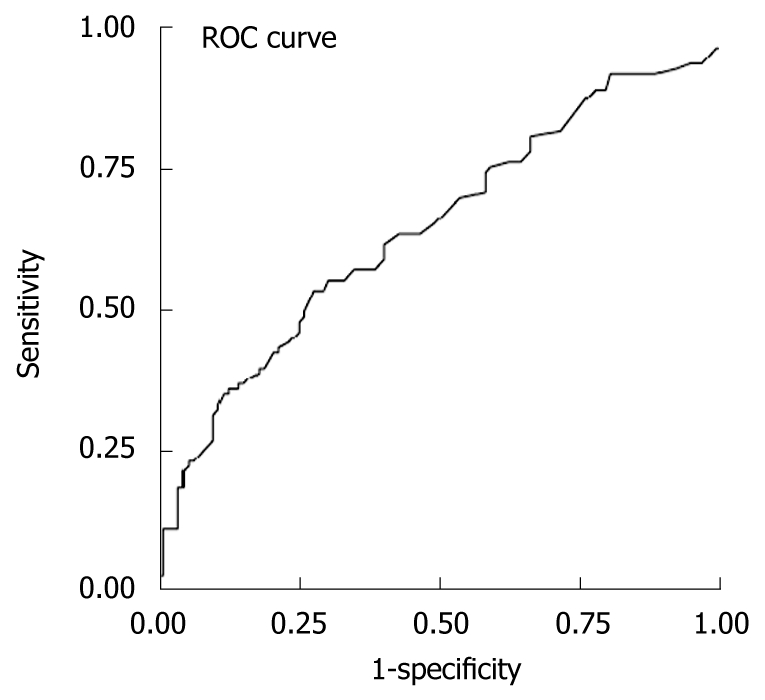
Receiver operating characteristics curve using different alanine aminotransferase levels as cutoffs. Alanine aminotransferase (ALT) cutoff of 42.5 gives the best area under the receiver operating characteristic (ROC) curve 0.645.
Table 3.
Diagnostic characteristics of various alanine aminotransferase cut-off levels for detecting lamivudine resistance
| ALT (U/L) | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | Positive likelihood ratio | Negative likelihood ratio |
| > 42.5 | 61 | 60 | 60 | 61 | 1.53 | 0.65 |
| > 70 | 39 | 81 | 67 | 58 | 2.05 | 0.75 |
| > 30 (males) | 85 | 28 | 53 | 66 | 1.18 | 0.56 |
| > 19 (females) | 90 | 7 | 50 | 40 | 0.96 | 1.40 |
The alanine aminotransferase (ALT) value of 42.5 U/L was found to be the best curve to fit receiver operating characteristics, while the ALT value of 70 U/L was the upper cutoff limit of the normal range in our laboratory, and ALT 30 U/L and 19 U/L were the cutoff values for males and females, respectively, based on the new Association for Study of Liver Disease guidelines.
A possible confounding factor is that early adefovir rescue (arbitrarily defined as adefovir treatment within 3 mo of lamivudine resistance) would result in resolution of virological breakthrough and arrest the rise in ALT. Consequently, we examined patients who had started adefovir 3 mo after the diagnosis of lamivudine resistance. Of 111 patients, 68 fulfilled this criterion, and we re-analyzed the utility of ALT in the diagnosis of lamivudine resistance using this subgroup. The results were virtually identical to those of the entire cohort of lamivudine resistant patients (n = 111), with the AUROC curve being 0.653, and ALT > 42.5 U/L giving the best sensitivity and specificity for predicting lamivudine resistance.
Abnormal ALT and the duration of lamivudine resistance
Of the 74 patients with abnormal ALT post-resistance, 27 (36%) belonged to the group that started adefovir within 3 mo of developing genotypic lamivudine resistance, indicating that abnormal ALT can occur soon after viral breakthrough.
Patients with persistently normal ALT
For the 37 patients (33.3%) with normal ALT (ULN < 70 U/L) after lamivudine resistance, the mean (SD) follow-up was 11 + 22 mo from the development of lamivudine resistance until adefovir rescue, 24% had at least 6 mo of follow-up from the development of lamivudine resistance until adefovir rescue. However, if the new AASLD guidelines were utilized, only 13 males (ALT < 30 U/L) and 5 females (ALT < 19 U/L) would have fulfilled the criteria for persistently normal ALT.
Patients with abnormal ALT
Of the patients with abnormal ALT (ULN < 70 U/L) [n = 74 (66.7%)] after the development of lamivudine resistance, 24 (21.6%) had ALT flares during lamivudine resistance. Of these 74 patients, 42 (57%) had persistently abnormal ALT, while 32 (43%) had transiently abnormal ALT during resistance. By univariate analysis, only baseline abnormal ALT was associated with abnormal ALT during resistance (P = 0.009). Gender (P = 0.111), race (P = 0.107), cirrhosis (P = 0.673), baseline HBeAg status (P = 1.00) and duration of lamivudine treatment (P = 0.42) were not associated with abnormal ALT during resistance by univariate analysis. Multivariate analysis showed that only abnormal ALT at baseline was associated with abnormal ALT during resistance (OR = 5.98, P = 0.003, 95% CI: 1.8-19.7). Gender, baseline HBV DNA, and baseline HBeAg status were not associated with abnormal ALT during resistance, by multivariate analysis. By univariate analysis, male gender was associated with ALT flares during resistance (P = 0.02). Race, age, baseline cirrhosis, albumin, ALT, AST, bilirubin, ALT flare at baseline and HBV DNA level were not associated with ALT flares during resistance. By multivariate analysis, only male gender was associated with ALT flares during resistance (OR = 8.9, P = 0.016, 95% CI: 1.5-53.3). Baseline ALT, abnormal ALT, and HBeAg status were not associated with ALT flares during resistance by multivariate analysis.
DISCUSSION
Development of viral resistance is a major concern in the treatment of CHB. This is particularly true for lamivudine, the first licensed nucleoside analogue for therapy of CHB. Lamivudine is highly efficacious but this efficacy is blunted by the rapid development of viral resistance[15]. Since the first descriptions of lamivudine resistance were published, much has been learned about the development of resistance. The first appearance of HBV resistance is the detection of genotypic mutations that confer resistance. Subsequently, viral breakthrough occurs and finally biochemical resistance (defined as the development of abnormal ALT due to viral resistance) appears[10]. However, the question of whether all patients develop biochemical resistance[16] has not been addressed. We have shown that not all patients with viral resistance develop abnormal ALT, with 33% having persistently normal ALT (ULN < 70 U/L) after the development of lamivudine resistance. This finding, however, may be confounded by the insufficient length of follow-up before adefovir rescue. Consequently, when we excluded patients who had early adefovir rescue (arbitrarily defined as adefovir rescue within 3 mo of lamivudine resistance), there were still 31% of patients with persistently normal ALT (ULN < 70 U/L) of which 71% of patients were followed for > 6 mo before adefovir treatment, indicating that length of follow-up was unlikely to be a confounder. Our study showed ALT to be an inadequate diagnostic test for viral resistance, with an AUROC of 0.645 in the best case scenario using an ALT cutoff of 42.5 U/L, which showed a sensitivity of 61%, specificity of 60%, positive predictive value of 60%, negative predictive value of 61%, positive likelihood ratio of 1.53 and negative likelihood ratio of 0.65, and with 39% of patients with lamivudine resistance being missed. Using other ALT cutoff levels, such as the normal range for our laboratory (ULN = 70 U/L) or the AASLD guidelines (males ULN = 30 U/L, females ULN = 19 U/L), did not improve diagnostic accuracy.
Although two different assays were used in our study to measure HBV DNA levels, all patients who were initially assessed to have no viral breakthrough based on the less sensitive Hybrid Capture II HBV DNA Test, were subsequently found to have undetectable HBV DNA when tested with the sensitive Artus HBV RG real time PCR kit during follow-up, with a lower limit of detection of 100 copies/mL (1.7 × 101 IU/mL). The majority of HBV DNA quantifications were carried out using a relatively insensitive method (lower limit of detection of 140 000 copies/mL), which might have led to an overestimation of the predictive value of ALT for diagnosing viral resistance, as "late" viral rebounds could result in more cases of viral resistance with abnormal ALT, thus making the poor performance of ALT even more striking.
Our study also showed that not all patients develop clinical resistance. Hence, the postulated evolution of HBV resistance starting with the development of genotypic resistance mutations, followed by viral breakthrough, then clinical resistance may not be applicable to all patients. This is particularly pertinent since the EASL guidelines for the management of CHB[4] state that rescue therapy should be instituted before the advent of clinical resistance, however, this latter finding may never be seen if ALT remains persistently normal. The most important predictor of the development of abnormal ALT upon development of lamivudine resistance was the baseline ALT value and abnormal ALT at baseline. While the ALT value may not be a very useful diagnostic marker for lamivudine resistance, there are additional implications of having a normal ALT despite the presence of viral resistance. Normal ALT values can be associated with histological damage[17] and disease progression in patients with CHB, but does this apply in cases of viral resistance? The evidence is mixed. We previously reported that patients with cirrhosis and lamivudine resistance have a high risk of mortality when untreated. In this group, ALT was normal in a substantial proportion of patients (40%)[18]. In the presence of lamivudine resistance, liver histology may still show improvement. In a pathological study of liver biopsies in patients with and without lamivudine resistance, improvement in histology was highest in those who had no evidence of genotypic resistance, and those who developed lamivudine resistance still had improvement in histology, albeit in a smaller proportion of patients[19]. Other than the implications for histological damage and disease progression, in some Asia-Pacific countries such as South Korea[20], Japan[20], Australia[20,21], and Taiwan (China)[22], an abnormal ALT during viral resistance is a requirement for reimbursement from the Government for rescue therapy with adefovir. This would mean that a significant proportion of patients with lamivudine resistance would not be able to receive rescue therapy and may run the risk of disease progression.
In conclusion, abnormal ALT does not occur in a significant proportion of patients with lamivudine resistance, consequently the predictive value of ALT in diagnosing lamivudine resistance is low, and cannot be used as a surrogate for lamivudine resistance. Although lamivudine is the cheapest oral antiviral agent available in Asia and is globally still the most widely used oral antiviral agent for CHB[23], the added cost of monitoring with expensive HBV DNA assays and viral resistance tests such as InnoLIPA, makes it economically attractive to consider cheaper options to evaluate lamivudine resistance. Unfortunately serum ALT cannot fulfill this role.
COMMENTS
Background
Chronic hepatitis B (CHB) is a global public health problem, being the most common cause of chronic viral hepatitis, and the major cause of hepatocellular carcinoma worldwide. Lamivudine is still used widely to treat CHB in Asia. However, lamivudine resistance is a major concern.
Research frontiers
The authors determined that alanine aminotransferase (ALT) is an inadequate surrogate marker for detecting lamivudine resistance.
Applications
Other than the implications for histological damage and disease progression, in some Asia-Pacific countries such as South Korea, Japan, Australia, and Taiwan, an abnormal ALT during viral resistance is a requirement for reimbursement from the Government for rescue therapy with adefovir. This would mean that a significant proportion of patients with lamivudine resistance would not be able to receive rescue therapy and may run the risk of disease progression.
Peer review
The manuscript is interesting, since lamivudine is still used in several countries and considering that hepatitis B virus (HBV) DNA quantification and detection of HBV genotypic resistance are not easily available in many of these countries.
Footnotes
Supported by (in part) Grant 02/N01 from the Health Services Development Program, Ministry of Health, Singapore
Peer reviewer: Roberto J Carvalho-Filho, MD, PhD, Hepatitis Section, Division of Gastroenterology, Federal University of Sao Paulo, Rua Botucatu, 740, 2.o andar, Vila Clementino, State of Sao Paulo, 04023-060, Brazil
S- Editor Wang JL L- Editor Webster JR E- Editor Ma WH
References
- 1.Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34 Suppl 1:S1–S3. doi: 10.1016/s1386-6532(05)00384-7. [DOI] [PubMed] [Google Scholar]
- 2.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507–539. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 3.Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 4.EASL Clinical Practice Guidelines: management of chronic hepatitis B. J Hepatol. 2009;50:227–242. doi: 10.1016/j.jhep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Wu PC, Dent JC, Barber J, et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 6.Dienstag JL, Schiff ER, Wright TL, Perrillo RP, Hann HW, Goodman Z, Crowther L, Condreay LD, Woessner M, Rubin M, et al. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256–1263. doi: 10.1056/NEJM199910213411702. [DOI] [PubMed] [Google Scholar]
- 7.Lok AS, Hussain M, Cursano C, Margotti M, Gramenzi A, Grazi GL, Jovine E, Benardi M, Andreone P. Evolution of hepatitis B virus polymerase gene mutations in hepatitis B e antigen-negative patients receiving lamivudine therapy. Hepatology. 2000;32:1145–1153. doi: 10.1053/jhep.2000.19622. [DOI] [PubMed] [Google Scholar]
- 8.Hadziyannis SJ, Papatheodoridis GV, Dimou E, Laras A, Papaioannou C. Efficacy of long-term lamivudine monotherapy in patients with hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2000;32:847–851. doi: 10.1053/jhep.2000.17915. [DOI] [PubMed] [Google Scholar]
- 9.Guan R, Lai CL, Liaw YF, Lim SG, Lee CM. Efficacy and safety of 5-years lamivudine treatment of Chinese patients with chronic hepatitis B. J Gastroenterol Hepatol. 2001;16(suppl 1):A60. doi: 10.1111/j.1440-1746.2004.03428.x. [DOI] [PubMed] [Google Scholar]
- 10.Fournier C, Zoulim F. Antiviral therapy of chronic hepatitis B: prevention of drug resistance. Clin Liver Dis. 2007;11:869–892, ix. doi: 10.1016/j.cld.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Baylis SA, Heath AB, Chudy M, Pisani G, Klotz A, Kerby S, Gerlich W. An international collaborative study to establish the 2nd World Health Organization International Standard for hepatitis B virus DNA nucleic acid amplification technology-based assays. Vox Sang. 2008;94:358–362. doi: 10.1111/j.1423-0410.2008.01023.x. [DOI] [PubMed] [Google Scholar]
- 12.Shyamala V, Arcangel P, Cottrell J, Coit D, Medina-Selby A, McCoin C, Madriaga D, Chien D, Phelps B. Assessment of the target-capture PCR hepatitis B virus (HBV) DNA quantitative assay and comparison with commercial HBV DNA quantitative assays. J Clin Microbiol. 2004;42:5199–204. doi: 10.1128/JCM.42.11.5199-5204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locarnini S, Hatzakis A, Heathcote J, Keeffe EB, Liang TJ, Mutimer D, Pawlotsky JM, Zoulim F. Management of antiviral resistance in patients with chronic hepatitis B. Antivir Ther. 2004;9:679–693. [PubMed] [Google Scholar]
- 14.Liaw YF, Leung N, Guan R, Lau GK, Merican I, McCaughan G, Gane E, Kao JH, Omata M. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2005 update. Liver Int. 2005;25:472–489. doi: 10.1111/j.1478-3231.2005.01134.x. [DOI] [PubMed] [Google Scholar]
- 15.Zoulim F. Assessment of treatment efficacy in HBV infection and disease. J Hepatol. 2006;44:S95–S99. doi: 10.1016/j.jhep.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Liaw YF, Leung N, Kao JH, Piratvisuth T, Gane E, Han KH, Guan R, Lau GK, Locarnini S; for the Chronic Hepatitis B Guideline Working Party of the Asian-Pacific Association for the Study of the Liver. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2008 update. Hepatol Int. 2008;2:263–283. doi: 10.1007/s12072-008-9080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papatheodoridis GV, Chrysanthos N, Hadziyannis E, Cholongitas E, Manesis EK. Longitudinal changes in serum HBV DNA levels and predictors of progression during the natural course of HBeAg-negative chronic hepatitis B virus infection. J Viral Hepat. 2008;15:434–441. doi: 10.1111/j.1365-2893.2007.00957.x. [DOI] [PubMed] [Google Scholar]
- 18.Dan YY, Wai CT, Lee YM, Sutedja DS, Seet BL, Lim SG. Outcome of lamivudine-resistant hepatitis B virus is generally benign except in cirrhotics. World J Gastroenterol. 2005;11:4344–4350. doi: 10.3748/wjg.v11.i28.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dienstag JL, Goldin RD, Heathcote EJ, Hann HW, Woessner M, Stephenson SL, Gardner S, Gray DF, Schiff ER. Histological outcome during long-term lamivudine therapy. Gastroenterology. 2003;124:105–117. doi: 10.1053/gast.2003.50013. [DOI] [PubMed] [Google Scholar]
- 20.Kim DJ, Kim HS, Yim HJ, Suh JI, Cheong JY, Kim IH, Tark WY, Lee YS, Lee S, Lee JY. [Problems faced by Korean patients with chronic liver disease and the role of the Korean Association for the Study of the Liver--emphases on social discrimination, insufficiency of reimbursement coverage, and deficiency of the welfare system] Korean J Hepatol. 2008;14:125–135. doi: 10.3350/kjhep.2008.14.2.125. [DOI] [PubMed] [Google Scholar]
- 21.Feller R, Strasser S, Ward J, Deakin G. Primary care management of chronic viral hepatitis. In: Bradford D, Hoy J, Matthews G, editors. HIV, Viral Hepatitis and STIs: A Guide for Primary Care. Australasian Society for HIV Medicine; 2008. pp. 111–124. [Google Scholar]
- 22. Available from: http://www.nhi.gov.tw/cgi/searcher.exe?sortfields = 2,3,4&o = 4&p = adefovir&s = 5,6,7,8. Accessed 1st June 2008.
- 23. Available from: http://www.imshealth.com/. Accessed 1st June 2008.


