Abstract
Glioblastoma multiforme (GM), the most frequent primary malignant brain tumor, is highly invasive due to the expression of proteases, including urokinase-type plasminogen activator (uPA). Here, we show the potential of our new and powerful recombinant Sendai virus (rSeV) showing uPA-specific cell-to-cell fusion activity [rSeV/dMFct14 (uPA2), named “BioKnife”] for GM treatment, an effect that was synergistically enhanced by arming BioKnife with the interferon-β (IFN-β) gene. BioKnife killed human GM cell lines efficiently in a uPA-dependent fashion, and this killing was prevented by PA inhibitor-1. Rat gliosarcoma 9L cells expressing both uPA and its functional receptor uPAR (9L-L/R) exhibited high uPA activity on the cellular surface and were highly susceptible to BioKnife. Although parent 9L cells (9L-P) were resistant to BioKnife and to BioKnife expressing IFN-β (BioKnife-IFNβ), cell–cell fusion of 9L-L/R strongly facilitated the expression of IFN-β, and in turn, IFN-β significantly accelerated the fusion activity of BioKnife. A similar synergy was seen in a rat orthotopic brain GM model with 9L-L/R in vivo; therefore, these results suggest that BioKnife-IFNβ may have significant potential to improve the survival of GM patients in a clinical setting.
Introduction
Glioblastoma multiforme (GM), the most frequent primary malignant brain tumor, is highly invasive and intractable, and the median survival of patients bearing GM is merely 1 year with the current standard treatments, including surgery and radiotherapy.1 A recent clinical trial revealed that temozolomide, an oral alkylating agent, achieved significant improvement of the median survival of patients of GM to 14.6 months;2 however, another study reported that 73.5% of patients still die within <2 years and 72.2% experience recurrence.3 The highly invasive nature of GM is a cause of great frustration among surgeons and is responsible for the high rate of local recurrence; extended resection for complete removal of tumor cells is usually difficult without injuring surrounding brain tissues, and therefore, the surgical margin is frequently regarded as positive.
There is thus an urgent need for new approaches in GM treatment. Virus-based therapeutics for glioblastoma have been evaluated in clinical studies over the last decade, including a packaging cell-mediated local production of retroviruses expressing herpes simplex virus thymidine kinase.4 Even though these viruses have been shown to increase oncolytic activity and tumor specificity, no significant therapeutic effects have been reported in the clinical trials to date;5 therefore, researchers are focusing their current efforts on improving new viruses that target GM6,7,8,9 including a unique oncolytic adenovirus that targets mutant variant III of the epithelial growth factor receptor (EGFRvIII), which is a glioblastoma-specific mutant. In our effort to achieve an efficient virus-based therapeutics, we previously demonstrated a cancer vaccine regimen combined with intratumor injection of F/M-gene-deleted nontransmissible recombinant Sendai virus (rSeV) expressing the interleukin-2 gene in a rat orthotopic model.10 This was the first experimental protocol to successfully eliminate an established rat 9L gliosarcoma in the brain, and suggested that rSeV would be safe and useful for the clinical practice of glioblastoma therapy. However, because of the highly malignant nature of the 9L tumor, the tumor elimination ratio was modest (–30%) even by this regimen, and there is thus need of a more powerful tool before moving to clinical study.
In order to develop a new therapeutic modality, we returned to the current understanding of the molecular mechanisms involved in the invasive nature of GM, and have recently focused on the protease system. It has been reported that GM frequently expresses urokinase-type plasminogen activator (uPA) and that its invasiveness is closely related to uPA activity.11 uPA can be activated by a high-affinity receptor, uPAR, on the cell surface, after which uPA activates matrix metalloproteases and plasmin, resulting in the degradation of extracellular matrix. Also, activation of these uPA-related proteins induces several intracellular signaling pathways via growth factor receptors, thereby facilitating cell adhesion, migration, and proliferation. A number of reports have demonstrated the close relationship between increased levels of local uPA and poor prognosis in cancer patients, including patients bearing GM.12 Because uPA activity is specifically upregulated in cancerous tissue but not in normal tissue, and uPA activity is observed on the cancer cell surface, it seems reasonable that a therapeutic modality targeting uPA could selectively kill cancer cells without significant damage to the surrounding normal tissue.
To realize this concept, we recently developed novel oncolytic viruses based on a type of rSeV that selectively shows matrix metalloprotease- or uPA-specific cell-killing activity via cell–cell fusion,13,14 namely, “oncolytic rSeV.” These new viruses were developed by several major genetic modifications: (i) deletion of the gene encoding matrix (M) protein, which resulted in the loss of budding of secondary viral particles and accumulation of hemagglutinin/neuraminidase and F (fusion) proteins on the cell surface; (ii) replacement of trypsin-susceptible amino acid sequences of the F-gene with targeted protease-specific ones; and (iii) truncation of the cytoplasmic domain of the F-gene. As a result, our recent study demonstrated that uPA-targeted oncolytic rSeV [rSeV/dMFct14(uPA2): named “BioKnife”] showed optimal performance and was applicable to various types of human malignancies.14
Therefore, we here examined the therapeutic potential of BioKnife for treating human glioma and a rat orthotopic model of highly malignant 9L gliosarcoma. Our results indicate that BioKnife may be useful for GM treatment. In addition, we revealed that BioKnife armed with the interferon-β (IFN-β) gene exhibited pronounced killing of GM in vitro and in vivo.
Results
uPA activity–specific cell fusion and killing using BioKnife in human GM cells
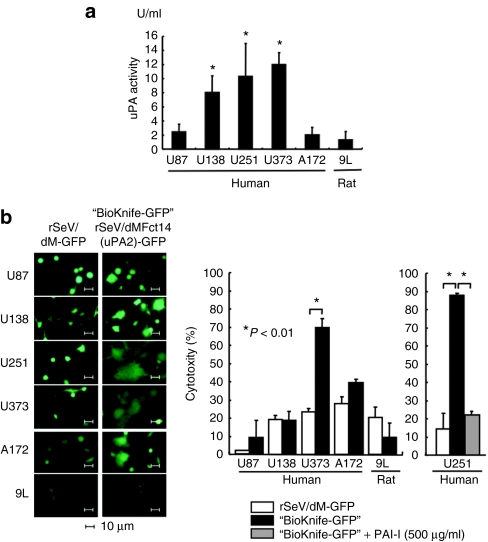
In the initial stage of this study, we tested our hypothesis, namely, that uPA activity-dependent cell fusion/killing could be accomplished using infection of the BioKnife vector into five independent lines of human GM cells (U87, U138, U251, U373, and A172) and into a rat 9L gliosarcoma. As shown in Figure 1a, a catalytic uPA activity assay revealed that 3 (U138, U251, and U373) of the six cell lines expressed a significant amount of active uPA in culture media. To assess the cytotoxic activity of BioKnife [rSeV/dMFct14(uPA2)] on these cells, a Water-Soluble Tetrazolium (WST) assay was performed. As expected, BioKnife expressing green fluorescent protein (BioKnife-GFP) efficiently killed GM cells that expressed high levels of uPA (U373 and U251; Figure 1b, left panels and two graphs). Because modest cytotoxicity due to the M-gene-deleted control recombinant virus (rSeV/dM-GFP, see Supplementary Figure S1) was found in U138, A172, and 9L cells, no significant increase in the cytotoxic effect of BioKnife-GFP was detected in these cells. Addition of a sufficient amount of PAI-1, a specific inhibitor for uPA, into the culture medium of U251 cells completely prevented the BioKnife-dependent cytotoxicity (right graph), confirming the uPA-specific killing of U251 cells.
Figure 1.
Expression of urokinase-type plasminogen activator (uPA) by human and rat brain tumor cells and their susceptibility to uPA-targeted rSeV/dMFct14(uPA2), namely, BioKnife (*P < 0.01). (a) Expression of uPA by human glioblastoma multiforme cells (U87, U138, U251, U373, and A172) and rat gliosarcoma (9L) in culture medium. Four days after changing to fresh medium, the culture supernatant was subjected to a colorimetric catalytic assay (n =6 wells/group). (b) Susceptibility of brain tumor cells to BioKnife-GFP or control vector (rSeV/dM-GFP) at an multiplicity of infection of 1.25. Four days after virus inoculation, cells were observed under a fluorescent microscope (panels) and the culture supernatant was subjected to cytotoxicity assay (graphs). In the case of U251, a group with PAI-1, a specific inhibitor of uPA, was included. Note that syncytium formation was observed in wells with U138, U251, U373, or A172 cells (panels), but not in wells with U87 or 9L cells. Significant cytotoxicity was observed against U251 and U373 cells, but not against A172 cells (left graph). Addition of PAI-1 almost completely prevented the BioKnife-dependent cytotoxicity (right graph; n = 6 wells/group).
Second, we assessed the time course of the cytotoxicity of BioKnife-GFP and the infection efficiencies based on the rSeV-dM-GFP-positive cell ratio determined by fluorescence-activated cell-sorting analyses on these five human GM cell lines and the rat gliosarcoma cell line 9L. As shown in Supplementary Figure S2, even when a small amount of viruses was used (multiplicity of infection = 1.25), the five human cell lines were relatively susceptible to rSeV/dM infection, usually showing an rSeV-dM-GFP-positive cell ratio of over 70%; however, significant and strong cell death was seen only in the cell lines U251 and U373 (see Figure 1b for representative results). 9L cells, in contrast, were highly resistant both to rSeV/dM infection and BioKnife-mediated cell death. Together with the data shown in Figure 1, these results suggest that uPA activity may predict the cytotoxic activity of BioKnife-GFP, and that the infection efficiency is not always important for BioKnife-mediated cell death.
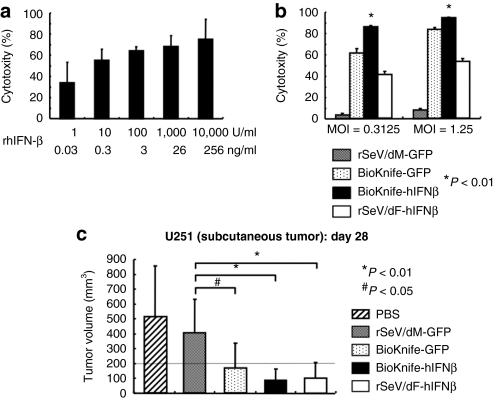
Next, we focused on U251 cells, which was the only of the human GM cell lines tested that could develop subcutaneous xenograft tumors on immunodeficient mice (data not shown). Also, the effect of BioKnife expressing the human IFN-β gene (hIFN-β) was examined, because hIFN-β has been proven effective for the treatment of experimental and clinical glioma.15,16 As shown in Figure 2a, recombinant hIFN-β protein (rhIFN-β) demonstrated a dose-dependent cytotoxic effect on U251, with over 75% cytotoxicity at 10,000 U/ml (equivalent to 256 ng/ml). A direct comparison study revealed that BioKnife-hIFNβ had a significantly greater cell-killing effect on U251 than any of the other viruses tested (all P < 0.01; Figure 2b). When U251 cells were implanted on the subcutis of the right flank of nu/nu mice, however, the superiority of the antitumor effect of BioKnife-hIFNβ was not apparent, because all the therapies tested were sufficiently effective in this model (Figure 2c).
Figure 2.
Sole or combined effect of hIFN-β and BioKnife on U251 cells in vitro and in vivo (*P < 0.01 and #P < 0.05). (a) Dose-dependent cytotoxic effect of rhIFN-β protein on U251 cells. Four days after addition of various amounts of rhIFN-β, the culture supernatant was subjected to cytotoxicity assay (n = 4 wells/group). (b) Cytotoxicity due to BioKnife-GFP (rSeV/dM-GFP as a control) and BioKnife-hIFNβ (rSeV/dF-GFP as a control). Four days after exposure of cells to viruses at an MOI of 0.3125 or 1.25, the culture supernatant was subjected to a cytotoxicity assay (n = 6 wells/group). (c) Antitumor effect of BioKnife-GFP and BioKnife-hIFNβ on U251 tumor cells subcutaneously implanted into the right flank of nude mice. When the subcutaneous tumors grew over 200 mm3 (defined as day 0, usually 14 days after tumor inoculation), 100 µl PBS with or without each vector (1 × 107 cell infection unit/dose) was injected three times (day 0, 3, and 6) intratumorally using a 26-gauge needle. The data indicate the tumor volumes on day 28 after tumor inoculation. MOI, multiplicity of infection; PBS, phosphate-buffered saline; rhIFN-β recombinant human interferon-β protein.
uPA activity on the cellular surface is critical for optimal BioKnife activity
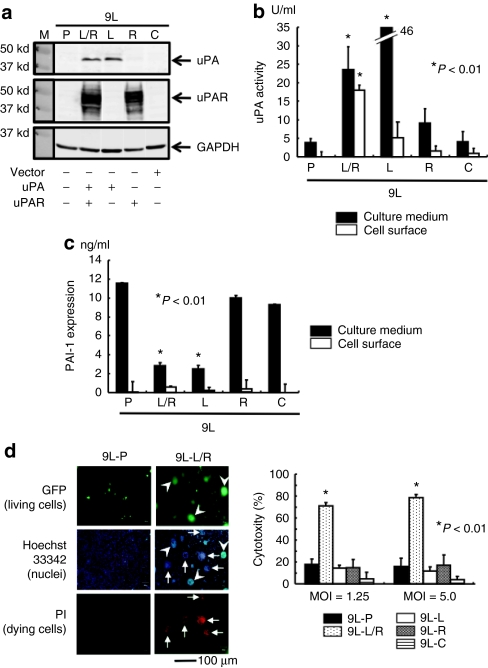
To clarify whether BioKnife shows significant synergism with IFN-β in vitro and in vivo, we generated 9L cells that were less susceptible to both BioKnife (Figure 1b) and recombinant murine IFN-β protein (rmIFN-β Figure 4a) and that stably expressed murine uPA and/or murine uPAR. As shown in Figure 3a, western blot analyses confirmed that the levels of uPA and uPAR were undetectable in the parent 9L (9L-P), and that the transfected gene-specific protein synthesis had occurred [9L-C: 9L with control plasmid; 9L-L: 9L with uPA ligand complementary DNA (cDNA); 9L-R: 9L with uPA receptor cDNA; and 9L-L/R: 9L with both uPA ligand cDNA and uPA receptor cDNA]. Interestingly, only 9L-L/R cells expressed high uPA activity in both the culture medium and on the cell surface, whereas 9L-L cells synthesized uPA only in the culture medium, suggesting that a high level of uPAR expression is required to accumulate active uPA on the cell surface (Figure 3b). In contrast, expression of the uPA-specific endogenous inhibitor PAI-1 was almost undetectable on the cell surface of all cells tested, and was significantly downregulated in the culture media of 9L-L/R and 9L-L cells (P < 0.01), suggesting a negative feedback mechanism of uPA or catalytic degradation of PAI-1 by uPA (Figure 3c).
Figure 3.
Accumulation of uPA on the cell surface is critical for the biological action of BioKnife (*P < 0.01). (a) Representative western blot analyses confirming the protein synthesis stably transfected with murine uPA and/or uPAR cDNA (9L-P: parent 9L; 9L-C: 9L with control plasmid; 9L-L: 9L with uPA ligand cDNA; 9L-R: 9L with uPA receptor cDNA; and 9L-L/R: 9L with both). Three independent experiments were performed and showed similar results. (b) Expression of uPA activity in the culture medium and on the cell surface from each transfectant (5 × 105 cells). Forty-eight hours after cell seeding, the supernatant was subjected to the colorimetric uPA activity assay. Both 9L-L/R and 9L-L expressed a high level of uPA in the culture media; however, significant accumulation of uPA activity on the cell surface was seen only in 9L-L/R cells. (c) Expression of PAI-1 in the culture medium and on the cell surface from each transfectant. The supernatant obtained in b was subjected to enzyme-linked immunosorbent assay for PAI-1. (d) Biological activity of BioKnife-GFP forming syncytium (panels, MOI = 1.25) and leading cells to death (graph, MOI = 1.25 and 5.0) on 9L-P or 9L-L/R cells (4 days after virus inoculation). Panels demonstrate the typical fluorescent microscopic findings detecting living cells (expressing foreign GFP genes) and dying cells (positive for PI). Nuclear staining was done using Hoechst 33342. No syncytium formation was found in parent 9L cells (9L-P, three left-hand panels). Note that GFP-expressing multinuclear cells (arrowheads) were negative for PI, and inversely, PI-positive dying cells (arrows) were negative for GFP (three right-hand panels). Two independent experiments were performed and showed similar results. Bar = 100 µm. Graphs represent cytotoxic assay for each transfectant. Only 9L-L/R cells demonstrated a significant increase of susceptibility to BioKnife-GFP (n = 6/group). cDNA, complementary DNA; GFP, green fluorescent protein; MOI, multiplicity of infection; PI, propidium iodide; uPA, urokinase-type plasminogen activator.
Using these cells, we next tested the biological action of BioKnife. As shown in Figure 3d, BioKnife-GFP showed optimized cytotoxicity on 9L-L/R cells (Figure 3d, graph; P < 0.01). Interestingly, Hoechst 33342 staining (middle two panels) revealed that 9L-L/R cells forming a syncytium contained both living cells strongly expressing the GFP transgene (green, upper two panels, arrowheads) and dying cells stained by propidium iodide (red, lower two panels, arrowheads).
Taken together, these findings indicate that BioKnife shows its optimized biological action—namely, the killing of tumor cells via induction of syncytium formation—when there is a sufficient amount of uPA activity on the cell surface.
Pronounced synergy of BioKnife and the IFN-β transgene for killing of 9L-L/R cells in vitro
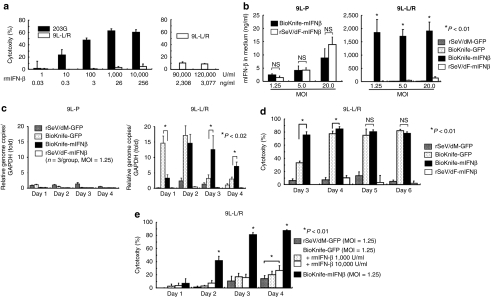
Next, we examined the synergy between BioKnife and IFN-β for GM therapy using 9L-L/R cells. We here used the mIFN-β gene and recombinant protein (rmIFN-β), because the species-specificity of IFN-β between rodents and humans is widely known. Because our preliminary study showed that the 9L-L/R cell-killing activity of BioKnife expressing the mIFN-β gene (BioKnife-mIFNβ) was superior to that of BioKnife-GFP at 3 days after virus inoculation (data not shown; representative data are shown in Figure 4d), we first examined the susceptibility of 9L-L/R cells to BioKnife-mIFNβ.
Figure 4.
Drastic synergy induced by BioKnife and mIFN-β gene transfer for killing 9L-L/R cells (*P < 0.01). (a) Direct cytotoxic effect of recombinant murine IFN-β (rmIFN-β) on mouse glioma 203G and 9L-L/R cells. Four days after treatment with various amounts of rmIFN-β, a cytotoxicity assay was performed. Note that 203G cells were susceptible to the rmIFN-β protein and dose-dependent cytotoxicity was observed, whereas 9L-L/R cells were highly resistant to rmIFN-β protein even at a concentration of over 90,000 U/ml (equivalent to 2,308 ng/ml). Each group contains n = 4. (b) Cell fusion via BioKnife enhances its mIFN-β transgene expression on 9L-L/R (susceptible to cell fusion, right graph), but not on 9L-P (resistant to cell fusion, left graph). Four days after exposure to each virus at each MOI, the mIFN-β protein level was measured by specific enzyme-linked immunosorbent assay. No significant difference in expression levels was found between rSeV/dF-mIFNβ (nonfusogenic) and BioKnife-mIFNβ (fusogenic) using 9L-P cells (left graph). Note that over 2-logs higher mIFN-β protein levels were found with BioKnife-mIFNβ than with rSeV/dF-mIFNβ at any MOI in 9L-L/R cells (right graph). Significant, but very low levels of mIFN-β protein were detected when using rSeV/dM-GFP and BioKnife-GFP. Each group contains n = 4. (c) Cell fusion via BioKnife potentiates viral genome copies on 9L-L/R (susceptible to cell fusion, right graph), but not on 9L-P (resistant to cell fusion, left graph). After exposure to each virus at an MOI of 1.25 on each time point, viral genome copies were quantified by real-time reverse transcriptase-PCR targeting of the N gene, their common sequence. Each data point was standardized by simultaneous amplification of GAPDH (the genome copies/GAPDH on day 1 in 9L-P cells treated with rSeV/dM-GFP = 1). Strong and early amplification of genome copies of BioKnife-GFP was found in 9L-L/R cells, and declined 3 days after infection. In contrast, delayed and sustained increase of genome copies was found when using BioKnife-mIFNβ. Each group contains n = 3. (d) Arming with the mIFN-β gene accelerates the cytotoxicity of BioKnife to mIFN-β-insensitive 9L-L/R cells. At various time points after exposure to each virus at an MOI of 1.25, a cytotoxicity assay was performed. Note that BioKnife-GFP showed peak cytotoxicity from day 4, whereas BioKnife-mIFNβ had already reached its peak cytotoxicity on day 3. Each group contains n = 6. (e) Acceleration of cell fusion and cytotoxicity due to BioKnife is efficiently induced by mIFN-β gene transfer, but not by rmIFN-β protein. At various time points after exposure to each virus at an MOI of 1.25, a cytotoxicity assay was performed. Note that a modest but significant acceleration of cytotoxicity via BioKnife-GFP was found only in the presence of a high concentration of rmIFN-β (10,000 U/ml) on day 4. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MOI, multiplicity of infection; NS, nonsignificant; rmIFN-β, recombinant murine interferon-β.
Recombinant mIFN-β protein (rmIFN-β) showed significant cytotoxicity to 203G murine glioma cells in a dose-dependent manner; however, 9L-L/R cells were highly resistant to rmIFN-β, even when a huge amount of rmIFN-β (~10% of cytotoxicity at 120,000 U/ml, equivalent to 3,077 ng/ml) was used (Figure 4a). In addition, inoculation of the conventional nontransmissible vector rSeV/dF-mIFNβ or BioKnife-mIFNβ to parent 9L cells that were resistant to BioKnife-mediated cell fusion produced a similar amount of mIFN-β protein in a dose-dependent manner (Figure 4b, left graph). In contrast, we were surprised to observe that the infection of BioKnife-sensitive 9L-L/R cells with BioKnife-mIFNβ dramatically enhanced mIFN-β expression (over 2-logs higher than the expression levels obtained using the others; Figure 4b, right graph), indicating that cell fusion markedly accelerated the expression of the transgene.
To seek the possible mechanism underlying the enhanced expression of the mIFN-β transgene, we next assessed the time course of the genome copy numbers of viruses by quantitative real-time reverse transcriptase-PCR (RT-PCR) targeting the N gene. In the case of the SeV, it is well known that genome replication reflects its transcription well. As shown in Figure 4c, cell fusion via BioKnife potentiates the number of viral genome copies on 9L-L/R (susceptive to cell fusion, right graph), but not on 9L-P (resistant to cell fusion, left graph). Interestingly, strong and early amplification of genome copies of BioKnife-GFP was found in 9L-L/R cells, and declined 3 days after infection. In contrast, a delayed and sustained increase of genome copies was found when using BioKnife-mIFNβ. It was suggested, therefore, that this sustained increase of genome copies and its transcription might have resulted in the enhanced IFN-β transgene expression shown in Figure 4b.
Next, we questioned the reverse effect—in other words, whether mIFN-β might accelerate cell fusion and killing even in mIFN-β-insensitive 9L-L/R cells. To examine this hypothesis, we checked the cytotoxicity over a time course. Interestingly, the cytotoxicity induced by BioKnife-GFP peaked on days 4–5, whereas cells treated with BioKnife-mIFNβ already demonstrated a nearly peak cell death on day 3 (Figure 4d). Then, we asked whether the transgene expression of mIFN-β might accelerate the cell fusion/killing and whether rmIFN-β might have an effect similar to that seen using the transgene. As shown in Figure 4e, a pronounced acceleration of cytotoxicity on 9L-L/R was observed when using BioKnife-mIFNβ. In contrast, a modest enhancement of cell-killing by BioKnife-GFP was seen under a higher concentration of rmIFN-β on day 4, indicating the drastic improvement in the biological activity of BioKnife conferred by transgene expression of the mIFN-β gene.
Pronounced synergy of BioKnife and the IFN-β transgene for killing of 9L-L/R cells in vivo
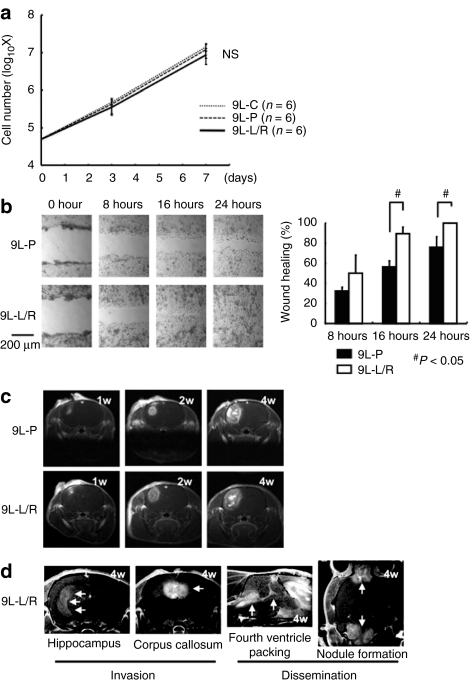
Before the in vivo study assessing the therapeutic potentials of BioKnife, we investigated the effect of forced uPA/uPAR gene expression on the growth and behavior of 9L cells in vitro and in vivo. As shown in Figure 5a, the growth activity of 9L-L/R in vitro was identical to those of 9L-P and 9L-C. In contrast, the in vitro wound-healing assay revealed the significantly enhanced migratory activity of 9L-L/R cells (Figure 5b). Magnetic resonance imaging (MRI) also demonstrated that the local growth of 9L-L/R tumors in the rat brain in vivo was roughly identical to that seen with 9L-P tumors (Figure 5c; 9L-P: n = 12 animals; 9L-L/R: n = 15 animals); however, five animals that received 9L-L/R cells showed the invasive growth of tumors to the hippocampus and/or corpus callosum, as well as the tumor dissemination to the fourth ventricle packing and/or nodule formation at the distant central nervous system (Figure 5d), indicating that forced expression of uPA/uPAR resulted in the increased malignant potential of 9L tumors.
Figure 5.
Overexpression of urokinase-type plasminogen activator (uPA)/uPA facilitates the invasive potential of 9L gliosarcoma cells. (a) Growth curve of 9L-L/R cell in vitro. No significant difference was found between 9L-P and 9L-C. (b) Wound-healing assay of 9L-P and 9L-L/R cells for assessing cell migration activity in vitro. A single linear wound was made with a 200 µl pipette, and each wound area was calculated 8, 16, and 24 hours later. Left panels are the representative phase-contrast images. Facilitated wound repair was found in 9L-L/R cells, indicating that the overexpression of uPA/uPA accelerates cell migration activity. (c,d) Time course of the growth and invasiveness of 9L-P (n = 12 animals) or 9L-L/R (n = 15 animals) cells in rat brains assessed by sequential assessment with magnetic resonance imaging (MRI). (c) All rats inoculated with 9L-P cells and 10 rats inoculated with 9L-L/R cells showed local growth around the injection site with neither detectable invasion nor dissemination. (d) A typical MR image showing invasion and dissemination. Five rats receiving 9L-L/R cells showed invasive cell transfer that included: (i) direct progression along neural fibers of the hippocampus and/or corpus callosum; (ii) cerebrospinal fluid dissemination surrounding the cerebral ventricle and/or forming nodules on the brain surface. NS, nonsignificant.
Pronounced synergy of BioKnife and the IFN-β transgene for killing of 9L-L/R cells in vivo
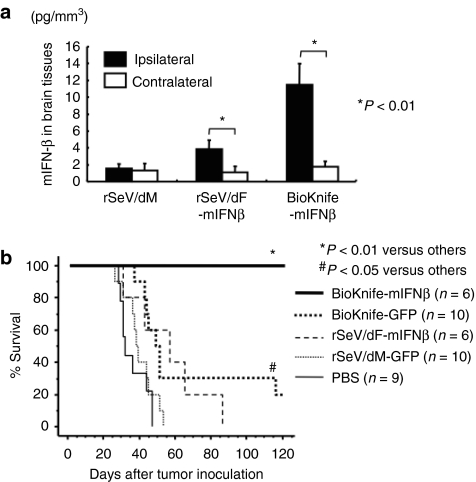
Finally, the therapeutic potential of BioKnife-mIFNβ on an orthotopic rat model of GM was investigated using 9L-L/R cells. We first assessed the expression level of transgene mIFN-β in vivo in 9L-L/R tumor-bearing rat brain tissue. Two days after injection of vector at 1 × 107 cell infection unit (CIU)/dose, the brain tissue was subjected to specific enzyme-linked immunosorbent assay (ELISA). As shown in Figure 6a, strong enhancement of transgene expression was found when using BioKnife-mIFNβ as compared to that seen using rSeV/dF-mIFNβ at the ipsilateral site, and the findings were similar to those seen in the in vitro experiment (Figure 4b).
Figure 6.
Impact of BioKnife expressing the mIFN-β gene for the treatment of an orthotopic rat brain tumor model with 9L-L/R (*P < 0.01 and #P < 0.05). (a) mIFN-β expression in ipsilateral (tumor bearing) and contralateral (no tumor) brain tissues 2 days after a single intratumor injection of BioKnife-mIFNβ or control vectors [rSeV/dM and rSeV/dF-mIFNβ 1 × 107 cell infection unit (CIU)/dose]. Each group contains n = 8 animals. (b) Therapeutic effect of BioKnife-mIFNβ on an orthotopic rat brain tumor model. On days 1, 4, and 7 after tumor injection, each virus (2 × 107 CIU) in 20 µl PBS was injected, and the cell survival was observed. The data are the sum of two independent experiments. mIFN-β, murine interferon-β PBS, phosphate-buffered saline.
The therapeutic benefit on the survival of tumor-bearing rats is shown in Figure 6b; all control rats that were treated with phosphate-buffered saline (PBS) or nonfusogenic rSeV/dM-GFP were dead within 46 or 52 days after tumor inoculation, respectively. Gene transfer of mIFN-β via rSeV/dF significantly improved the survival of rats with IFN-β-insensitive 9L-L/R (P < 0.05), suggesting that this therapeutic effect might have been due to the well-known immune-mediated antitumor effect of IFN-β, but not due to its direct cytotoxicity on tumor cells. Similarly, the survival of tumor-bearing rats that received BioKnife-GFP was also significantly prolonged compared to those in the PBS or rSeV/dM-GFP group (P < 0.05, respectively). Compared to these efficacies, BioKnife-mIFNβ showed a dramatic enhancement of the therapeutic potential of BioKnife: namely, no dead animals were observed during the trial. These surviving rats exhibited no residual tumor cells in their brain tissue either macroscopically or microscopically (data not shown).
These results provided clear evidence of a pronounced synergistic effect between the fusogenic activity of BioKnife and the IFN-β transgene in vitro and in vivo.
Discussion
Here, we investigated the potential and mechanism of our recently developed uPA-targeted oncolytic virus, BioKnife, to treat a rat model of GM. Key observations obtained in this study were as follows: (i) among 9L cells that were stably transformed with the uPA and/or uPAR gene, accumulation of uPA activity on the cellular surface was crucial for the optimal biological functioning of BioKnife; (ii) there was a dramatic synergy between cell fusion/cytotoxicity and IFN-β transgene expression via BioKnife—namely, we observed an acceleration of cell fusion and a dramatic increase of transgene expression; and (iii) the synergistic effects of BioKnife-IFNβ were effective for treating a rat orthotopic brain tumor model with 9L-L/R, even if the tumor itself was resistant to IFN-β therapy alone. These findings indicate the potential utility of BioKnife expressing the IFN-β gene and provide important information related to the biological parameters predicting the efficacy of BioKnife in future clinical trials.
During the course of this study, we had some unexpected findings; however, some of these anomalies were resolved in subsequent experiments. First, the expression level of uPA did not always predict the cytotoxic activity of BioKnife (e.g., U138, Figure 1), and this question was resolved by an experiment using 9L, which showed that the cell surface accumulation of uPA, but not its release, was critical for the biological activity of BioKnife (Figure 3). To the best of our knowledge, this is the first demonstration that the expression of uPAR is critical for facilitating the accumulation of active uPA on the cell surface. Second, although BioKnife-GFP appeared to be capable of facilitating syncytium formation of U138 and U172 (Figure 1b, panels), the observed cytotoxicity did not always support the notion that such syncytium formation occurred (Figure 1b, graphs). This contradictory data could be explained by the data in Figure 3d: namely, syncytium formation did not always correlate with dying cells, at least at this time point.
Next, we should discuss why PAI-1 expression in the culture media from 9L-L/R and 9L-L was downregulated, as it was previously shown that uPA enhances the expression of PAI-1.17 A possible explanation of this paradox is that the uPA/PAI-1 complex may mask epitopes recognized by the ELISA system, because RT-PCR analysis could not detect a significant reduction of PAI-1 mRNA in these cells (data not shown).
A most important question remains—namely, by what mechanism is the dramatic synergy between BioKnife and the IFN-β transgene realized? It has been widely accepted based on previous studies18,19 that type-I IFNs inhibit cell–cell fusion and viral replication of paramyxoviruses, including SeV. However, our data obtained by recombinant viruses suggest that this rule may not apply in all situations; for example, the recombinant virus yield during the amplification process of BioKnife encoding IFN-β using LLC-MK2 cells is significantly increased more than several-fold compared with that of BioKnife expressing other transgenes (unpublished results). Interestingly, the cell–cell fusion of epithelial and dendritic cells induced by measles virus, a member of Paramyxoviridae, was shown to be enhanced by type-I IFNs,20 suggesting that acceleration of the cell–cell fusion induced by IFN-β might be cell- and/or species-specific. In this study, we also found that BioKnife-mediated cell–cell fusion strongly enhanced IFN-β transgene expression, which reached over 2-logs higher than that seen when using nonfusogenic virus. Because the transgene of BioKnife was expressed by its own viral expression machinery, these findings theoretically indicate that cell–cell fusion facilitates viral protein synthesis; however, to the best of our knowledge, there has been no published data that could explain these findings. Therefore, further studies will be needed to investigate the molecular mechanisms of such synergies.
Finally, because in vivo experiments using the orthotopic GM model in this study demonstrated a dramatic therapeutic effect, the clinical relevance of our BioKnife-IFNβ should be discussed. First, almost all surgical specimens of GM were shown to express a high level not only of biologically active uPA but also uPAR,21,22 suggesting that a majority of GM patients would be good candidates for this therapeutics. The current model may be clinically relevant to residual and disseminated tumor cells rather than solid tumors, because virus injection was started 1 day after tumor inoculation; therefore, the adjuvant use of BioKnife-IFNβ after mass reduction via, for example, surgery and/or stereotactic radiotherapy would be reasonable. Although it may be possible that such a protease-targeting strategy would show limited efficacy, because urokinase is variably expressed at different sites in the parenchyma of growing tumors, these theoretical considerations based on this study warrant further investigations and encouraged us to move this system to the clinic.
In conclusion, we here demonstrated the efficient control of a rat orthotopic model of GM via the marked synergistic actions of IFN-β gene transfer and BioKnife, an oncolytic rSeV targeting uPA. These findings support the therapeutic potential of BioKnife-IFNβ for intractable GM in a clinical setting.
Materials and Methods
Cells, reagents, ELISA, plasmid vectors. The human brain tumor cell lines (U87, U138, U251, U373, and A172) and the rat cell line 9L were purchased from American Type Culture Collection (Rockville, MD) or the European Collection of Cell Cultures (England, UK). These cell lines were maintained in complete medium (RPMI 1640 for 203G and Dulbecco's modified Eagle's medium for the others) supplemented with 10% fetal bovine serum, penicillin, streptomycin, and 1% sodium pyruvate (Flow Laboratories, Mclean, VA) under a humidified atmosphere containing 5% CO2 at 37 °C. ELISA was performed using commercially available kits for rat PAI-1 (Hyphen Biomed, Andresy, France) and for mIFN-β (PBL Biomedical Laboratories, Piscataway, NJ). Full-length murine uPA and uPAR cDNAs were cloned from total RNA of Lewis lung cancer cells (American Type Culture Collection: CRL-1642) by RT-PCR, and human uPA and uPAR cDNAs were cloned from total RNA of human prostate tumor cells: PC3 (American Type Culture Collection: CRL-1435) by RT-PCR. The sequences of PCR primers with BamHI and NheI sites were the following: murine uPA, forward 5′-TCTAGCTAGCCGCCATGAAAGTCTGGCTGG CGAGC-3′/reverse 5′-CCGCGGATCCTCAGAAGGCCAGACCTTTCTC-3′ murine uPAR, forward 5′-TCTAGCTAGCCGCCATGGGACTCCCAAGGCGGCTG-3′/reverse 5′-CCGCGGATCCTCAGGTCCAGAGGAGGACGCC-3′). Amplified cDNA fragments were digested with BamHI and NheI and inserted into a pcDNA3.1 vector (Invitrogen, Carlsbad, CA). The full sequences of cDNAs were determined by direct sequencing.
Construction and recovery of BioKnife vectors. rSeVs and rSeV armed with IFN-β were constructed as described previously,13,14,23 and all schematic structures of viruses used in this study are demonstrated in the Supplementary Figure S1. In brief, the parent plasmid pSeV18+/dM-GFP, in which the GFP had been substituted for the deleted M gene,24 was digested with SalI and NheI, and the F-gene fragment (9,634 bp) was subcloned into LITMUS 38 (New England Biolabs, Beverly, MA). Site-directed mutagenesis was performed using a Quick-Change Mutagenesis Kit (Stratagene, La Jolla, CA), and the mutated F-gene was returned to the pSeV18+/dF-GFP backbone. mIFN-β and hIFN-β cDNAs were previously cloned25 and were inserted into the pSeV18+/Fct14(uPA2)dM-GFP plasmid. Recovery and amplification of the SeV vector were performed essentially as described before. Briefly, LLC-MK2 cells were transfected with a plasmid mixture containing each plasmid—pSeV18+/Fct14 (uPA2)dM-GFP, pGEM-NP, pGEM-P, and pGEM-L—in 110 µl of Superfect reagent (Qiagen, Tokyo, Japan). The transfected cells were maintained for 3 hours, washed three times, and incubated for 60 hours in minimum essential medium containing araC. The cells were collected and lysed by three cycles of freezing and thawing. The lysate solution was incubated on the F/M-expressing LLC-MK2 cells in a 24-well plate. Twenty-four hours later, the cells were washed and incubated in minimum essential medium containing araC and 7.5 µg/ml trypsin plus 10 ng/ml urokinase (Cosmobio, Tokyo, Japan). The virus yield was expressed in CIU, as previously described.13,14 Expression of mIFN-β or hIFN-β was determined by specific ELISA.
Virus titration. The virus titers were expressed as CIU, which were estimated by infecting confluent LLC-MK2 cells in a 6-well plate with diluted solution as previously described.13,14 In brief, LLC-MK2 cells were inoculated in duplicate with a series of dilutions of virus, then incubated for 1 hour and washed twice with PBS. Two days after infection, cells were fixed in methanol, incubated with anti-SeV primary antibodies, and then incubated with fluorescein isothiocyanate–labeled goat anti-rabbit IgG(H+L) (Invitrogen). Immunofluorescent-positive cell were counted and CIU/ml were calculated.
uPA activity assay and ELISA for PAI-1 and mIFN-β. Cells (1.5 × 104) in 60 µl medium were seeded in a 96-well dish, and the supernatants were collected after 48 hours. The uPA activity or PAI-1 expression in the culture medium and on the cell surface of denucleated cells that were washed three times with a sufficient amount of PBS was measured using a colorimetric uPA Activity Assay Kit (Chemicon International, Temecula, CA) or ELISA kit for rat PAI-1, according to the manufacturer's instruction.
Cytotoxicity assay. Cells (5 × 103) were seeded in a 96-well dish with 100 µl medium containing various concentrations of recombinant hIFN-β (PBL Biomedical Laboratories), mIFN-β (Chemicon International) proteins, or various amounts of viruses. At each time point, cytotoxicity was measured using a Premix WST-1 Cell Proliferation Assay System (Takara Bio, Otsu, Japan). Defining the cell viability of a negative control well as 100%, the cytotoxicity of each well was calculated by subtracting the corresponding viability from 100%.
uPA- and/or uPAR-expressing stable transfectants of 9L. 9L cells (3 × 105) were seeded in 6-well dishes with 2 ml medium/well, and the following day, the plasmid vectors were transfected using TransIT-LT1 Reagent (Mirus Bio, Madison, WI) according to the manufacturer's instructions. Gene-transferred 9L cells (9L-P: parent 9L; 9L-C: 9L with control plasmid; 9L-L: 9L with uPA ligand cDNA; 9L-R: 9L with uPA receptor cDNA; and 9L-L/R: 9L with both uPA ligand cDNA and uPA receptor cDNA) were maintained under 200 µg/ml of antibiotic G-418, and the clones surviving from single cells were selected twice.
Western blotting. Samples were lysed in lysis buffer containing 50 mmol/l Tris–HCl (pH 6.8) and 10% sodium dodecyl sulfate, and the protein concentration for each sample was determined by using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA). Anti-mouse uPA (ab20789, rabbit, 1:1,000; Abcam, Cambridge, UK), anti-mouse uPAR (AF534, goat, 1:1,000; R&D Systems, Abingdon, UK), and anti-glyceraldehyde 3-phosphate dehydrogenase (IMG-5143A, rabbit, 1:1,000; IMGENEX, San Diego, CA) were used as primary antibodies. The secondary antibodies, anti-rabbit IgG (65-6120, goat, 1:10,000; Invitrogen) or anti-goat IgG (561-71271, rabbit, 1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA), were applied for 30 minutes. Specific bands were visualized using an ECL Plus Western Blotting Detection System (GE Healthcare, Little Charfont, UK).
Quantitative real-time RT-PCR assessing rSeV genome copies. Total cellular RNA was extracted from cultured cells with an ISOGEN system (Wako Pure Chemicals, Tokyo, Japan) according to the manufacturer's instructions, then treated with RNase-free DNAase I (Boehringer Mannheim, Mannheim, Germany). A volume of 25 ng of total RNA was subjected to real-time RT-PCR. Real-time monitoring of the amplification and quantification of genome copies of rSeV were done using a model 7000 Sequence Detection System (Applied Biosystems, Tokyo, Japan) by the TaqMan method according to the manufacturer's instructions. The oligonucleotide sequences of PCR primers and TaqMan probes targeting the N gene (forward: 5′-CAATGCCGACATCGACCTAGA-3′ reverse: 5′-CGTGCCCATCTTTCACCACTA-3′ TaqMan Probe: FAM-ACAAAAGCCCATGCGGACCAGGAC-TAMRA), a common sequence for all vector constructs used in this study, were purchased from Applied Biosystems. Three independent experiments were performed and the obtained data were statistically analyzed. The genome copies were standardized by the glyceraldehyde 3-phosphate dehydrogenase level in each sample, and expressed as a relative fold increase compared with the control level.
Wound-healing assay. A single linear wound was made with a 200 µl pipette tip in confluent cultures of 9L-P or 9L-L/R cells and washed gently with PBS to remove cellular debris. The cells were transferred to fresh medium and incubated at 37 °C. After 8, 16, and 24 hours, the cells were photographed and the area of each wound was calculated.
Animals. Female 7- to 8-week-old Balb/c nu/nu mice and male Fisher 344 rats were obtained from the Shizuoka Laboratory Animal Center. Animals were kept under specific pathogen-free and humane conditions in the animal care facility of Chiba University's Inohana campus. The animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee and by the Biosafety Committee for Recombinant DNA experiments of Chiba University. These experiments were also done in accordance with the recommendations for the proper care and use of laboratory animals and according to The Law (no. 105) and Notification (no. 6) of the Japanese Government.
Subcutaneous tumor model of human U251 cells in nu/nu mice: A density of 1 × 107 U251 cells in 50 µl PBS plus 50 µl Matrigel (BD Biosciences, Bedford, MA) were subcutaneously injected into the right flank of nude mice. The tumor volume was calculated every 3 or 4 days, until day 28 after tumor inoculation, using the following formula: volume = a2b/2, where a is the shortest diameter and b is the longest. When the subcutaneous tumors grew over 200 mm3 (defined as day 0, usually 14 days after tumor inoculation), the mice were divided into treatment or control groups, and 100 µl PBS with or without each vector (1 × 107 CIU/dose) was injected 3 times (day 0, 3, and 6) intratumorally using a 26-gauge needle.
Orthotopic GM model of 9L-L/R cells in rats: A density of 5 × 105 9L-L/R cells with 10 µl PBS were injected into rat brains using a microinjector (Harvard Apparatus, South Natick, MA) over 5 minutes, as previously described.10 Briefly, rats were anesthetized with 50 mg/kg pentobarbital and placed in a stereotactic apparatus. A burr hole was made at an appropriate location (1 mm posterior to the bregma and 3 mm right to the midline). A 25-gauge needle was inserted at a point 3 mm ventral from the dura. On days 1, 4, and 7 after tumor injection, each SeV vector (2 × 107 CIU) with 20 µl PBS was injected in the same way. In addition, to evaluate the invasive activity of 9L-L/R, 1 × 105 of 9L-P or 9L-L/R cells with 10 µl PBS were injected into other rat brain in the same way and examined every 7 days with MRI. The invasive activity was measured at 4 weeks after tumor inoculation. The rats were observed daily until severe paresis, ataxia periophthalmic encrustations, or >20% weight loss developed.
MRI. All rats were anesthetized with 2.0% isoflurane (Abbott Japan, Tokyo, Japan) and injected with 0.4 ml of Gd-DTPA (Meglumine Gadopentetate, 0.002 ml/g, 50 mmol/l, Bayer, Leverkusen, Germany) intravenously 15 minutes before MRI measurement. All MRI experiments were performed on a 7.0-T MRI scanner (Magnet: Kobelco and JASTEC, Kobe, Japan; Console: Bruker Biospin, Ettlingen, Germany) with a volume coil for transmission (Bruker Biospin) and two-channel phased array coil for reception (Rapid Biomedical, Rimpar, Germany). Multislice T1-weighted MR images covering the entire brain (T1WI; multislice spin echo, TR/TE = 400/9.57 ms, slice thickness = 1.0 mm, slice gap = 0, number of slices = 16, matrix = 256 × 256, field of view = 25.6 × 25.6 mm2, average = 4) were acquired. The slice orientation was transaxial for all scans. Two independent neurosurgeons evaluated the MRIs to determine whether either “invasion” or “dissemination” were present. “Invasion” was defined as direct progression along neural fiber tracts and “dissemination” was defined as distant tumor growth separated from the original injection site by cerebrospinal fluid. When either invasion or dissemination was clearly observed in the transaxial plane, images in the horizontal and sagittal planes were also acquired. Image reconstruction and analysis were performed using ParaVision (Bruker Biospin).
Statistical analysis. All data were expressed as the means ± SD. The data were examined statistically using one-way analysis of variance with Scheffe adjustment. When the number of evaluated groups was small, the data were subjected to the Kruskal–Wallis or the Mann–Whitney U test. The survival curves were determined using the Kaplan–Meier's method. The log-rank test was used for comparison. A probability value of P < 0.05 was considered statistically significant. Statistical analyses were determined using StatView software (SAS Institute, Cary, NC).
SUPPLEMENTARY MATERIAL Figure S1. The structures of BioKnife and its relatives used in this study. Figure S2. Cytotoxic activities of BioKnife-GFP on various human glioblastomas and rat 9L gliosarcoma.
Acknowledgments
We thank Akihiro Tagawa, Takumi Kanaya, Hiroshi Ban, and Takashi Hironaka for their excellent technical assistance in vector construction and large-scale production; Sayaka Shibata for technical assistance of animal preparation. KN International Ltd. assisted in the revision of the language in this article. This work was supported in part by a grant from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to Y.Y.). Y.Y. is a member of the Scientific Advisory Board of DNAVEC Corporation.
Supplementary Material
The structures of BioKnife and its relatives used in this study.
Cytotoxic activities of BioKnife-GFP on various human glioblastomas and rat 9L gliosarcoma.
REFERENCES
- DeAngelis LM. Brain tumors. N Engl J Med. 2001;344:114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. National Cancer Institute of Canada Clinical Trials Group et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Brandes AA, Tosoni A, Franceschi E, Sotti G, Frezza G, Amistà P, et al. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation With MGMT promoter methylation status. J Clin Oncol. 2009;27:1275–1279. doi: 10.1200/JCO.2008.19.4969. [DOI] [PubMed] [Google Scholar]
- Rainov NG. A phase III clinical evaluation of herpes simplex virus type 1 thymidine kinase and ganciclovir gene therapy as an adjuvant to surgical resection and radiation in adults with previously untreated glioblastoma multiforme. Hum Gene Ther. 2000;11:2389–2401. doi: 10.1089/104303400750038499. [DOI] [PubMed] [Google Scholar]
- Liu TC, Galanis E., and, Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat Clin Pract Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Iwado E, Kondo Y, Aoki H, Hayashi Y, Georgescu MM, et al. Autophagy-inducing agents augment the antitumor effect of telerase-selve oncolytic adenovirus OBP-405 on glioblastoma cells. Gene Ther. 2008;15:1233–1239. doi: 10.1038/gt.2008.98. [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT, Jr, Zaupa C, Aghi M, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sarkaria JN, Petell CA, Paraskevakou G, Zollman PJ, Schroeder M, et al. Combination of measles virus virotherapy and radiation therapy has synergistic activity in the treatment of glioblastoma multiforme. Clin Cancer Res. 2007;13:7155–7165. doi: 10.1158/1078-0432.CCR-07-1306. [DOI] [PubMed] [Google Scholar]
- Piao Y, Jiang H, Alemany R, Krasnykh V, Marini FC, Xu J, et al. Oncolytic adenovirus retargeted to Δ-EGFR induces selective antiglioma activity. Cancer Gene Ther. 2009;16:256–265. doi: 10.1038/cgt.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwadate Y, Inoue M, Saegusa T, Tokusumi Y, Kinoh H, Hasegawa M, et al. Recombinant Sendai virus vector induces complete remission of established brain tumors through efficient interleukin-2 gene transfer in vaccinated rats. Clin Cancer Res. 2005;11:3821–3827. doi: 10.1158/1078-0432.CCR-04-1485. [DOI] [PubMed] [Google Scholar]
- Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fei Z, Bu X, Zhen H, Zhang Z, Gu J, et al. Expression and significance of urokinase type plasminogen activator gene in human brain gliomas. J Surg Oncol. 2000;74:90–94. doi: 10.1002/1096-9098(200006)74:2<90::AID-JSO2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Kinoh H, Inoue M, Washizawa K, Yamamoto T, Fujikawa S, Tokusumi Y, et al. Generation of a recombinant Sendai virus that is selectively activated and lyses human tumor cells expressing matrix metalloproteinases. Gene Ther. 2004;11:1137–1145. doi: 10.1038/sj.gt.3302272. [DOI] [PubMed] [Google Scholar]
- Kinoh H, Inoue M, Komaru A, Ueda Y, Hasegawa M., and, Yonemitsu Y. Generation of optimized and urokinase-targeted oncolytic Sendai virus vectors applicable for various human malignancies. Gene Ther. 2009;16:392–403. doi: 10.1038/gt.2008.167. [DOI] [PubMed] [Google Scholar]
- Mizuno M., and, Yoshida J. Effect of human interferon beta gene transfer upon human glioma, transplanted into nude mouse brain, involves induced natural killer cells. Cancer Immunol Immunother. 1998;47:227–232. doi: 10.1007/s002620050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman H, Berkey BA, Maor MH, Groves MD, Schultz CJ, Vermeulen S, Radiation Therapy Oncology Group et al. Phase II Radiation Therapy Oncology Group trial of conventional radiation therapy followed by treatment with recombinant interferon-beta for supratentorial glioblastoma: results of RTOG 9710. Int J Radiat Oncol Biol Phys. 2006;66:818–824. doi: 10.1016/j.ijrobp.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Shetty S, Bdeir K, Cines DB., and, Idell S. Induction of plasminogen activator inhibitor-1 by urokinase in lung epithelial cells. J Biol Chem. 2003;278:18124–18131. doi: 10.1074/jbc.M207445200. [DOI] [PubMed] [Google Scholar]
- Tomita Y., and, Kuwata T. Suppressive effects of interferon on cell fusion by Sendai virus. J Gen Virol. 1981;55 Pt 2:289–295. doi: 10.1099/0022-1317-55-2-289. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Cheung HC., and, Hunter E. Interferon inhibits Sendai virus-induced cell fusion: an effect on cell membrane fluidity. Proc Natl Acad Sci USA. 1982;79:835–839. doi: 10.1073/pnas.79.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschke F, Plumet S, Duhen T, Azocar O, Druelle J, Laine D, et al. Cell-cell fusion induced by measles virus amplifies the type I interferon response. J Virol. 2007;81:12859–12871. doi: 10.1128/JVI.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sawaya R, Mohanam S, Rao VH, Bruner JM, Nicolson GL, et al. Expression and localization of urokinase-type plasminogen activator receptor in human gliomas. Cancer Res. 1994;54:5016–5020. [PubMed] [Google Scholar]
- Landau BJ, Kwaan HC, Verrusio EN., and, Brem SS. Elevated levels of urokinase-type plasminogen activator and plasminogen activator inhibitor type-1 in malignant human brain tumors. Cancer Res. 1994;54:1105–1108. [PubMed] [Google Scholar]
- Li HO, Zhu YF, Asakawa M, Kuma H, Hirata T, Ueda Y, et al. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J Virol. 2000;74:6564–6569. doi: 10.1128/jvi.74.14.6564-6569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Tokusumi Y, Ban H, Kanaya T, Shirakura M, Tokusumi T, et al. A new Sendai virus vector deficient in the matrix gene does not form virus particles and shows extensive cell-to-cell spreading. J Virol. 2003;77:6419–6429. doi: 10.1128/JVI.77.11.6419-6429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Okano S, Yonemitsu Y, Onimaru M, Sata S, Nagata-Takeshita H, et al. Induction of efficient antitumor immunity using dendritic cells activated by recombinant Sendai virus and its modulation by exogenous IFN-β gene. J Immunol. 2006;177:3564–3576. doi: 10.4049/jimmunol.177.6.3564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The structures of BioKnife and its relatives used in this study.
Cytotoxic activities of BioKnife-GFP on various human glioblastomas and rat 9L gliosarcoma.