Abstract
In this study, we compared the genomic integration efficiencies and transposition site preferences of Sleeping Beauty (SB or SB11), Tol2, and piggyBac (PB) transposon systems in primary T cells derived from peripheral blood lymphocytes (PBL) and umbilical cord blood (UCB). We found that PB demonstrated the highest efficiency of stable gene transfer in PBL-derived T cells, whereas SB11 and Tol2 mediated intermediate and lowest efficiencies, respectively. Southern hybridization analysis demonstrated that PB generated the highest number of integrants when compared to SB and Tol2 in both PBL and UCB T cells. Tol2 and PB appeared more likely to promote clonal expansion than SB, which may be in part due to the dysregulated expression of cancer-related genes near the insertion sites. Genome-wide integration analysis demonstrated that SB, Tol2, and PB integrations occurred in all the chromosomes without preference. Additionally, Tol2 and PB integration sites were mainly localized near transcriptional start sites (TSSs), CpG islands and DNaseI hypersensitive sites, whereas SB integrations were randomly distributed. These results suggest that SB may be a preferential choice of the delivery vector in T cells due to its random integration site preference and relatively high efficiency, and support continuing development of SB-mediated T-cell phase I trials.
Introduction
The use of nonviral DNA transposons as a tool for mammalian cell gene transfer has recently emerged as a viable option. The Sleeping Beauty (SB) transposon system is a reconstructed DNA transposon of the Tc1/mariner superfamily and mediates DNA transfer via a “cut-and-paste” mechanism.1 SB transposase has been shown to mediate transposition at TA-dinucleotide genomic sequences in a wide range of vertebrate cells and tissues.2,3 We have previously reported that the SB transposon system can mediate stable gene transfer in human peripheral blood lymphocytes (PBL) with a 5–20% efficiency.4,5 We also demonstrated that this system could be used for genetic modification of T cells from both PBL and umbilical cord blood (UCB) to target CD19+ B-cell malignancies in vitro and in mice.6 Our previous studies have suggested that SB-modified T cells may be useful in the treatment of refractory leukemia and lymphoma and may have general applications in adoptive cell therapy.
There are two major drawbacks for gene therapy with SB-based vectors: cargo size limitation (~5 kb) and overproduction inhibition (lower transposition at higher transposase concentration).7,8 However, PiggyBac (PB) and Tol2 transposon systems lack these disadvantages and thus may have potential uses in gene therapy including human T-cell engineering. The PB system, derived from the cabbage looper moth Trichoplusia ni,9 has been shown to mediate efficient transposition at TTAA-tetranucleotide sequences in both mammalian cells and mouse germline.10,11,12,13,14,15 Tol2, a natural medaka fish hAT gene family element,16 can efficiently integrate DNA sequences >10 kb into human cells.17 Both PB and Tol2 seem to tolerate overproduction inhibition,7,11,12,17 thus making these systems viable options for use in in vitro and in vivo gene delivery studies. In a side-by-side comparison, PB consistently exhibits the highest transposition activity compared to SB11, Tol2, and Mos118 (derived from Drosophila mauritiana) transposases when assayed in human cell lines.11 A chimeric PB transposase, but not SB11 and Tol2, fused to heterologous site-specific DNA-binding domain can be engineered, which retains the same level of transposase activity as the wild-type PB transposase.11,12,19,20,21 These unique properties could potentially allow for the engineering of PB to transfer therapeutic genes into a “safe haven” in the human genome.22
It has not been reported whether PB is a more active transposase than SB in human primary T cells. Although several studies have demonstrated that integrations mediated by both SB and PB are relatively random, these studies have used cultured human cell lines.10,12,23 Therefore, the integration profiling reported in these studies may not accurately reflect the integration profiles that are present in primary cells in which transposition has occurred. In this report, we performed a side-by-side comparison of the transposition efficiency and genomic integration profiles of PB, SB11, and Tol2 transposases in primary T cells. We show that PB provides more efficient gene transfer than SB11 and Tol2 in primary human T cells and that PB and Tol2 integration sites tend to locate near transcriptional start sites (TSSs), CpG islands, and DNaseI hyperactive sites, whereas SB integration sites are more randomly distributed in the primary human T-cell genome.
Results
PB is superior to SB11 and Tol2 in mediating stable gene transfer in PBL
SB has been shown to mediate efficient transposition in primary T cells derived from both PBL and UCB.4,5,6,24,25 However, it is not yet determined whether PB and Tol2 are active in human primary T cells or whether PB is more potent than SB in primary T cells, as has been reported in studies using other mammalian cell types.11 In order to perform a direct comparison of SB-, Tol2-, and PB-mediated transposition in T cells, each transposon and transposase vector was constructed in the identical plasmid backbone (Figure 1a). A cytomegalovirus (CMV) promoter was used to drive expression of the SB11, Tol2, and PB transposase genes (Figure 1b).
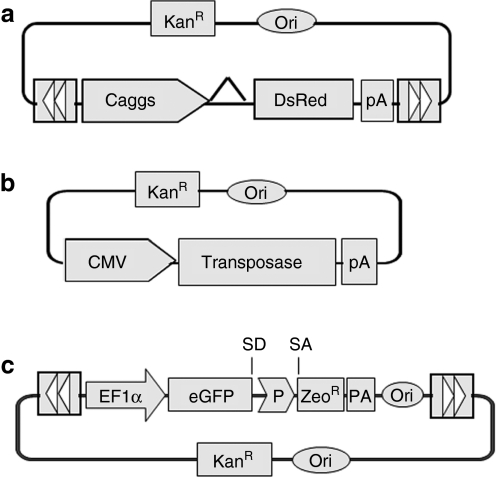
Figure 1.
SB, Tol2 and PB transposon vectors used in this study. (a) Transposon-expressing vectors. The DNA transposon vectors contain the inverted terminal repeats (indicated by arrowheads) flanking the gene of interest. The SB, Tol2, and PB transposons were pKT2-Ca-DsRed, pKmTol2-Ca-DsRed, and pKPB-Ca-DsRed, respectively. The Caggs promoter is a chimeric promoter derived from chicken β-actin and cytomegalovirus (CMV) immediate-early promoter sequences; (b) Transposase-expressing vectors. The SB, Tol2, and PB transposase driven by the CMV promoter were pK/CMV-SB11, pK/CMV-Tol2ase, and pK/CMV-PBase. pK/CMV-empty was used as filler DNA. (c) Recoverable transposon vectors used to isolate transposon insertion sites. DsRed, red fluorescent protein; EF1α, human elongation factor 1α promoter; eGFP, enhanced green fluorescence protein; P, EM7 prokaryotic promoter; KanR, kanamycin-resistance gene; Ori, origin of replication; pA, polyadenylation signal; PB, piggyBac; PBase, PB transposase; SA, splice acceptor; SB, Sleeping Beauty; SB11, SB transposase; SD, splice donor; Tol2ase, Tol2 transposase; ZeoR, zeocin-resistance gene.
We have previously reported that 5 µg of SB transposon and 10–15 µg of SB10 or SB11 transposase-expressing plasmids are optimal for nucleofection of 5 × 106 PBL.4 In this study, we further optimized these conditions using 5 µg of transposon plasmid and titrating the amount of transposase-expressing plasmid (0, 2.5, 5, 10, 15, and 20 µg) while keeping the total amount of transfected DNA constant by supplementing pK/CMV-empty plasmid. The percentage of cells positive for red fluorescent protein (DsRed+) was analyzed on days 1, 14, and 28 after transfection. Similar transfection efficiencies were achieved for all conditions with all donors; 40–60% cells were DsRed+ and cell viability was ~50–60% 1 day after transfection (Figure 2; data not shown). On days 12–14 after nucleofection, a greater proportion of DsRed+ cells was observed in PB-transfected PBL (1.38–11.21%, 15 µg transposase) than in SB- (0.68–5.73%) or Tol2-transfected (0.07–2.14%) PBL (PB versus Tol2, P < 0.001; PB versus SB, P = 0.1899). It appears that SB was more efficient than Tol2 in mediating stable DsRed expression in PBL (SB versus Tol2 on week 2, P = 0.016). The superior transposition activity of PB over SB and Tol2, and SB over Tol2 was maintained on days 29–34 (PB versus SB, P = 0.0207; PB versus Tol2, P < 0.0001; SB versus Tol2, P = 0.006) and day 43.
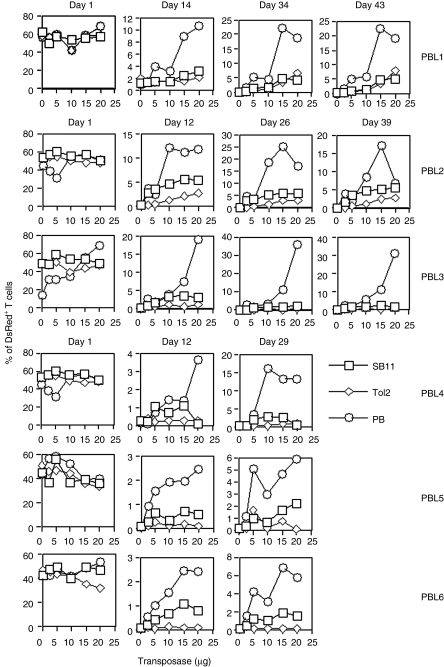
Figure 2.
Stable transgene expression in human PBL by SB, Tol2, and PB transposons. Freshly isolated PBL (5 × 106) were conucelofected with 5 µg of transposon plasmids and different amounts of transposase-encoding plasmids (0, 2.5, 5, 10, 15, and 20 µg). pK/CMV-empty plasmid was used to adjust the total amount of DNA transfected in each sample. The nucleofected cells were exposed to anti-CD3/CD28 beads overnight after transfection. DsRed expression was analyzed on days 1, 12–14, 26–34, and 39–43. The data from six individual PBL are shown. P values to compare SB versus Tol2, SB versus PB, and PB versus Tol2 on day 1 were all >0.05. On days 12–14, P values for PB versus SB, PB versus Tol2, and SB versus Tol2 were 0.189, <0.001, and 0.016. On days 26–34, P values for PB versus SB, PB versus Tol2, and SB versus Tol2 were 0.0207, <0.001, and 0.006. SB, Sleeping Beauty; PB, piggyBac; PBL, peripheral blood lymphocytes; UCB, umbilical cord blood.
To confirm that PB is the most efficient transposon in UCB T cells, mononuclear cells (UCBMNCs) purified from 12 UCB donors were individually nucleofected with SB, Tol2, and PB, and exposed to anti-CD3/CD28 beads 2–4 hours (UCB1–7) or overnight (UCB8–12) after transfection. PB appeared the most efficient transposon in UCB1–8, whereas SB was most effective in UCB10 on days 14, 20–26, 35, and 42 after transfection. Tol2 was the least efficient transposon in both PBL and UCB T cells (Supplementary Figure S1 and Figure 2). We observed that more UCB T cells died from nucleofection than PBL. Moreover, UCB T cells were less responsive to anti-CD3/CD28 bead activation following nucleofection than PBL T cells.6 Therefore, we were unable to analyze DsRed+ cell populations 1 day after transfection. We conclude that PB is the most active transposon in PBL-derived T cells 2 and 4 weeks after gene transfer (P < 0.0001).
PB generated the highest number of integrants per cell
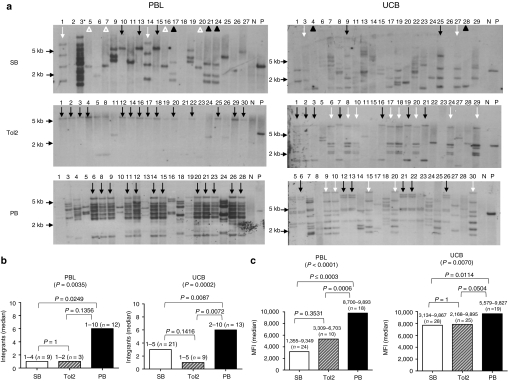
To determine whether stable DsRed expression in PB- and Tol2-transfected T cells is due to genomic integration of the transposon, Southern hybridization of nucleofected T-cell clones derived from PBL donor 6 and UCB donor 1 was performed. Approximately 168–180 DsRed+ clones were derived from SB-, Tol2-, and PB-transfected T cells under identical conditions. Of these, 22 T-cell clones from each transposon-transfected PBL and UCB donor (a total of 132 clones) were analyzed for transposon integration by Southern hybridization. Figure 3a demonstrates that each clone derived from PB- or Tol2-transfected PBL or UCB contained DsRed transgene integrated in the genomic DNA as has been previously described for SB-transfected T-cell clones.4 These data confirm that PB and Tol2 can mediate stable gene transfer in primary human T cells. Notably in Figure 3b, PB generated the highest number of integrants compared with SB and Tol2 in both PBL- (median = 6, 1, 1, respectively) and UCB- (median = 6, 3, 1, respectively) derived T-cell clones (P = 0.0035 for PBL; P = 0.0002 for UCB). Moreover, it appears that there was a strong correlation between numbers of integration and levels of DsRed expression in PB-transfected PBL and UCB T-cell clones (P < 0.0001 and P = 0.007, respectively). However, this correlation was insignificant in SB- and Tol2-transfected PBL and UCB (Figure 3c). DsRed expression levels in PB-transfected T cells were more narrow than SB- and Tol2-transfected T cells (Figure 3c).
Figure 3.
Molecular analysis of integration in both PBL- and UCB-derived T-cell clones after transfection with SB, Tol2, and PB transposons. (a) Southern hybridization of genomic DNA isolated from DsRed+ T-cell clones with a DsRed probe. PBMNCs and UCBMNCs derived from identical donors were isolated and nucleofected with 5 µg of DsRed transposon-containing vector and 5, 10, or 15 µg of transposase-expressing vector. Three weeks after nucleofection, cells were pooled and sorted for DsRed expression. DsRed+ cells were subsequently cloned by limiting dilution and used for Southern hybridization analysis. Ten µg of genomic DNA extracted from each T-cell clone (clone identification # is shown) was digested with EcoRI and BamHI over night at 37 °C. Ten micrograms of genomic DNA from mock nucleofected T cells with or without 6 pg of transposon plasmid was used as positive and negative controls, respectively. Genomic DNA fragments were separated by electrophoresis through a 0.9% agarose gel and transferred to a Hybond membrane (GE Healthcare, Piscataway, NJ). A radiolabeled probe specific for a portion of DsRed was created by random priming. Probe hybridization was performed in 0.2 mol/l NaHPO4 pH 7.2/1 mmol/l EDTA/1% bovine serum albumin/7% sodium dodecyl sulfate at 65 °C overnight. Blots were subsequently washed three times and exposed to film. Identical symbols indicate the clones with similar patterns of integrants. PB: 12 (~10 bands) out of 22 clones in PBL, 5 (~7 bands, white arrow) and 6 (~5 bands, black arrow) out of 22 in UCB; Tol2: 14 (~1 band) out of 22 PBL, 8 (~5 bands, white arrow), and 7 (~ 2 bands, black arrow) out of 22 UCB; SB: 4 (~1 3-kb band, white triangle), 3 (~3 bands, black triangle), 3 (~1 5.5-kb band, black arrow), and 2 (~4 bands, white arrow) out of 21 PBL, 2 out of 22 UCB with three different banding patterns. Asterisk in PBL-SB clone 3 indicates that this clone was actually derived from PB-transfected T cells confirmed by linker-mediated PCR. One of the two representative data are shown. (b) Median number of integrants per clone. The number above each bar indicates the minimal and maximal number of integrants. (c) Median expression of MFI per clone. The number above each bar indicates the minimal and maximal MFI. MFI, mean fluorescence intensity; N, negative control; n, number of clones analyzed; P, positive control; PBL, peripheral blood lymphocytes; UCB, umbilical cord blood.
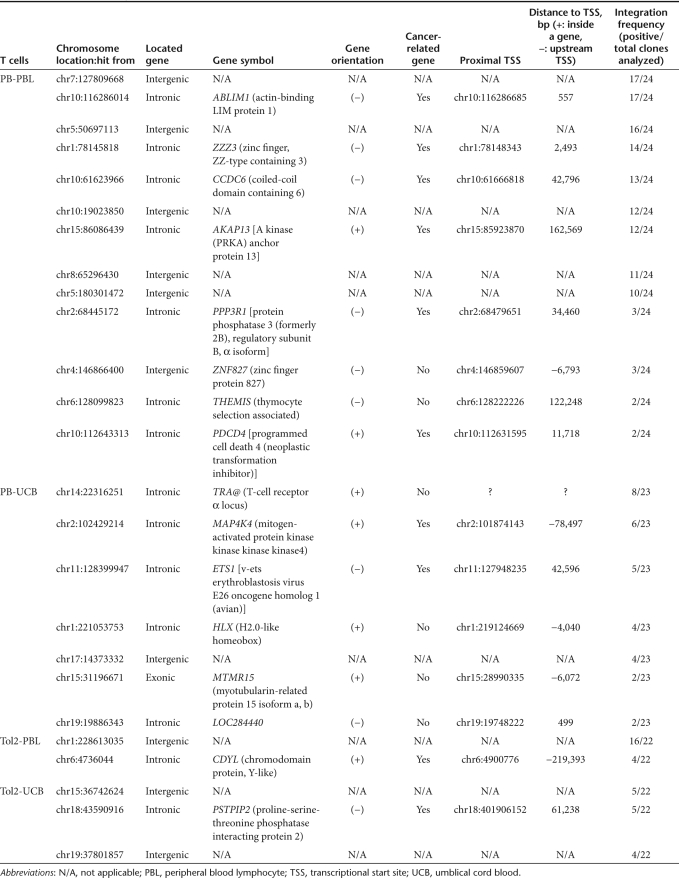
Surprisingly, Southern hybridization showed that more clones with similar band patterns were found in PB- and Tol2-transfected PBL and UCB than SB-transfected clones (Figure 3a), indicating that PB and Tol2 integration may promote T-cell clonal expansion. To further confirm transposon-mediated integration and clonality, linker-mediated PCR (described in Supplementary Materials and Methods) was performed to identify integration sites in PB- and Tol2-transfected PBL- and UCB-derived T-cell clones. Out of 25 clones, 15 (as indicated by * in Supplementary Figure S2a) derived from PB-transfected PBL had similar PCR banding patterns. However, 24 out of 25 clones were subject to integration site analysis, and 20 integrant plasmids from each T-cell clone were sequenced. Of these 24 clones, 17 had the same integration sites on chromosomes 7(128099823) and 10(116286014), 16 on chromosome 5(50697113), 14 on chromosome 1(78145818), and 10–13 on other chromosomes (Supplementary Figure S2a and Table 1). In PB-UCB, Tol2-PBL, and Tol2-UCB T-cell clones, 56–89% of PCR bands (68/~76, 24/~32, and 24/~43, respectively) were recovered and sequenced for integration site analysis. In PB-transfected UCB T-cell clones, 8 out of 23 clones had integrations on chromosome 14(22316251) (Supplementary Figure S2b). By comparison, 16 out of 22 clones from Tol2-transfected PBL had transposon integrations at the same site on chromosome 1(228613035) (Supplementary Figure S2c), and 5 out of 22 clones from Tol2-transfected UCB had transposon integrations on chromosome 15(36742624) and 18(43590916) (Supplementary Figure S2d). Table 1 summarizes the nature of these integrants in 83–96% of T-cell clones (24/25, 23/24, 20/24, and 20/24 T-cell clones derived from PB-PBL, PB-UCB, Tol2-PBL, and Tol2-UCB, respectively) including location, proximity to cancer-related gene and TSS, and integration frequency. These data further confirm PB- and Tol2-mediated transposition in primary T cells and suggest that PB and Tol2 transposons may have higher possibility than SB in promoting T-cell clonal expansion potential.
Table 1. The nature of integrants in T-cell clones.
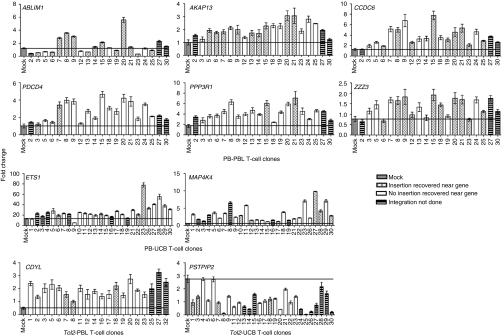
To investigate whether the cancer-related genes near the insertion sites are transcriptionally activated, quantitative reverse transcription-PCR was used to quantify the expression of all 10 cancer-related genes in transposon integrated 96 T-cell clones (20 PB-PBL, 27 PB-UCB, 20 Tol2-PBL, and 29 Tol2-UCB). Figure 4 shows that five out of six cancer-related genes (AKAP13, CCDC6, PDCD4, PPP3R1, and ZZZ3, but not ABLIM1) were upregulated in PB-PBL clones compared to those genes in pooled mock T cells (P < 0.0001). Additionally, three proto-oncogenes (ETS1, MAP4K4, and CDYL) were significantly upregulated in PB-UCB and Tol2-PBL, whereas one proto-oncogene (PSTPIP2) was downregulated in Tol2-UCB (P < 0.0001). These data suggest that PB and Tol2 integration may have altered the expression of neighboring cancer-related genes in T cells, likely causing clonal dominance.
Figure 4.
Expression of cancer-related genes near integration sites in human T-cell clones. Total RNA was extracted from 96 T-cell clones derived from PB- and Tol2-transfected T cells (PB-PBL, 20 clones; PB-UCB, 27 clones; Tol2-PBL, 20 clones, and Tol2-UCB, 29 clones). TaqMan quantitative PCR was used to quantify gene expression of each sample in duplicate and the assay was repeated once. The relative gene expression is presented as fold change (mean ± SEM, n = 4). A line is drawn to indicate the baseline of gene expression in mock T cells. P <0.0001 for both the PB-PBL and PB-UCB groups when the median of each gene (n = 80 in PB-PBL, n = 108 in PB-UCB) was divided by the mean of the mock and compared. P <0.0001 in all genes except ABLIM1 (P = 0.2790) when median of each gene (n = 80 in PB-PBL, n = 108 in PB-UCB, n = 80 in Tol2-PBL, n = 116 in Tol2-UCB) was divided by the mean of the mock with constant 1 and compared. Mock indicates pooled PBL or UCB from three separate donors. PB, piggyBac; PBL, peripheral blood lymphocytes; UCB, umbilical cord blood.
To further investigate the clonal expansion potential, flow cytometry–based T-cell receptor Vβ repertoire (described in Supplementary Materials and Methods) analysis was performed. MNCs isolated from 6 PBL and 3 UCB donors were nucleofected with SB, Tol2, and PB transposons encoding nerve growth factor receptor (NGFR) reporter gene (Supplementary Figure S3a) and their cognate transposase-expressing plasmids. Three to four weeks after nucleofection and OKT3-mediated expansion, NGFR+ cells were sorted, cultured for 2–3 months, and the T-cell receptor Vβ gene usage was analyzed in CD4+ and CD8+ T cells (Supplementary Figure S3b) and compared with mock (no DNA)-transfected cells from the same donor and cultured under the same condition. Using arbitrary tenfold above mock as the cutoff, it appeared that Tol2-mediated clonal expansion of T cells in CD4+ and/or CD8+ subsets occurred in 6 of 9 donors (PBL1, 3, 5, and UCB1, 2, 3, indicated in green arrows in Supplementary Figure S3c). PB- and SB-transfected T cells also showed some clonal expansion in 1 PBL5 (purple arrow indicated in both CD4+ and CD8+ subsets) and 1 UCB1 (black arrow indicated in CD8+ subset) of 9 donors, respectively, as indicated in Supplementary Figure S3c. No clonal expansion of either CD4+ or CD8+ T-cell subsets was detected in mock-transfected T cells from all nine donors. Supplementary Table S1 summarizes clonal expansion analysis by T-cell receptor Vβ family usage, suggesting that Tol2 mediated higher levels of clonal expansion than PB and SB.
Genome-wide integration profiling
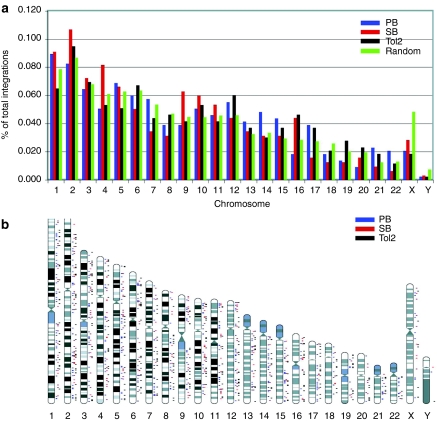
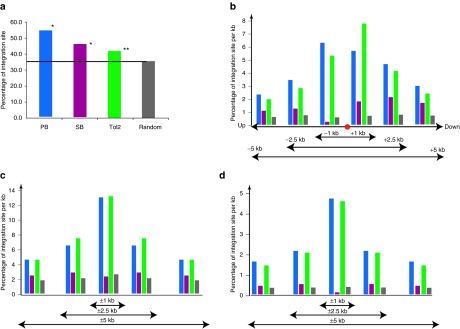
We have cloned 291 unique integration sites for SB transposon, 402 for Tol2, and 436 for PB from identical 2 PBL and 2 UBC T cells. The integration sites for all three transposons are distributed across all chromosomes and generally the number per chromosome correlates with the size of each chromosome (Figure 5a,b), although the X chromosome is less favored for all three transposons than the random data set. These results indicate that all chromosomal regions are accessible for transposon integration. Surprisingly, compared to 10,000 random integration sites generated by computer, SB, Tol2, and PB transposons showed distinct integration site preferences. All three transposons showed preferences for RefSeq genes, with PB showing the highest preference, followed by SB and Tol2 (P < 0.01, by a χ2-test comparing to random data set; Figure 6a). PB and Tol2 showed striking preferences for CpG island regions, DNaseI hypersensitive site regions, and transcription start sites of RefSeq genes (Figure 6b–d), with the bell-shaped distributions near the center of the above features; this is not observed for SB. CpG islands and DNaseI hypersensitive sites are frequently found near promoter regions and transcriptions start sites of genes.
Figure 5.
Genome-wide mapping of SB, Tol2, and PB integrations in human primary T cells. (a) Distribution of SB, Tol2, and PB integration events at the chromosome level. (b) The relative positions of SB, Tol2, and PB integration sites on each chromosome. PB, peripheral blood lymphocytes; SB, Sleeping Beauty.
Figure 6.
SB, Tol2, and PB integration site distribution. (a) Frequency of integration sites within RefSeq genes. (b) Proximity of integration sites to transcription start sites (TSSs). (c) Percentage of integration sites to CpG islands. (d) Percentage of integrations to DNaseI hypersensitive site. PB, peripheral blood lymphocytes; SB, Sleeping Beauty.
Discussion
In this study, we report several novel observations. First, the PB system is superior to SB11 and Tol2 in mediating stable gene expression in primary T cells derived from PBL. Second, PB-modified T cells from both PBL and UCB contained more integrations than SB- and Tol2-transfected PBL- and UCB-derived T cells. Third, PB and Tol2 integrations may have a higher likelihood than SB in promoting T-cell clonal expansion. Moreover, PB and Tol2 integrations significantly altered the expression of 9 out of 10 cancer-related genes near the insertion sites. Fourth, no clear chromosomal preferences were observed for SB, Tol2, or PB transposon integrations. Fifth, compared to SB, Tol2 and PB had similar integration site preferences in human T cells; Tol2 and PB transposons preferentially integrated near TSSs, CpG islands, and DNaseI hypersensitive sites whereas SB transposon integrations occurred randomly across the genome.
It is not known why PB is a more efficient transposon than SB11 and Tol2 in human PBL-derived T cells (this report), B cells (H. Guo and X. Zhou, unpublished results) and other mammalian cell lines.11 There can be several possible explanations. First, PB transposase may mediate a more efficient “cut-and-paste” process than the SB or Tol2 transposases. In support of this hypothesis, Figure 3b shows that the median number of integrants in PB-transposed T-cell clones from both PBL and UCB was higher than those in SB- or Tol2-transposed T-cell clones; second, the human genome contains ~2,000 PB-like elements.22,26 These endogenous sources of PB-like transposases could excise and remobilize a transgene flanked by exogenous PB transposon's termini. However, this hypothesis is not supported by the fact that T cells transfected with PB transposon without PB transposase did not show stable DsRed+ cells; third, because PB transposons favor integration near promoter regions, CpG islands, and DNaseI hypersensitive sites (Figure 6), PB transposons may have an increased potential for activating proto-oncogenes or disrupting tumor-suppressor genes, thereby promoting cell proliferation and clonal expansion (Figures 3a and 4; Table 1). Finally, PB may be less dependent on host factors than SB to mediate transposition in human cells due to the vast evolutionary distance between humans and moths, and therefore there may be a reduced possibility of transposition interference by host factors.22 It has been shown that SB transposition efficiency can be modulated by interaction with host proteins (e.g., high-mobility group protein 1 and Myc-interacting zinc finger protein 1).27,28
Oligoclonal expansion of transfected T cells was more pronounced in Tol2- and PB-transfected T cells than SB-transfected T cells (Figure 3a, Supplementary Figures S2a–d and S3c; Table 1). It is not known whether clonal expansion is due to transposon-mediated integrations or whether it is an experimental artifact of gene transfer and cell culture. Our gene expression analysis data support the likelihood of the former as 9 out of 10 cancer-related genes showed significant upregulation and downregulation compared to the mock T cells (Figure 4). For example, MAP4K4 and ETS1 expression in PB-UCB derived T-cell clones displayed a median 4.83- and 1.76-fold greater than the mock T cells, respectively, whereas CCDC6, PPP3R1, PDCD4, and AKAP13 expression in PB-PBL clones were 2.65-, 2.39-, 2.29- and 1.9-fold higher. CDYL expression was upregulated 2.28-fold and PSTPIP2 expression was downregulated 0.37-fold in Tol2-PBL and Tol2-UCB T-cell clones, respectively. MAP4K4 is a serine/threonine kinase and mediates Jun N-terminal kinase and tumor necrosis factor-α signaling pathways.29,30 Studies have shown that MAP4K4 is overexpressed in many types of human cancer cell lines and primary tumors compared to normal tissue.31 ETS1 has long been known to harbor oncogenic activity through the regulation of genes responsible for extracelluar matrix remodeling, migration, and invasion, as well as transforming growth factor-α–mediated proliferation.32 AKAP13, a RhoA GTPase-specific guanine exchange factor, has been shown to anchor cyclic adenosine monophosphate protein kinase A to subcellular locations and promotes oncogenesis.33,34 However, as the magnitude of dysregulated gene expression in PB- and Tol2-transfected T-cell clones is less than fivefold, the definitive role of the dysregulated genes in T-cell clonal expansion remains to be determined.
It is possible that the lower efficiency of stable gene transfer by Tol2 compared to PB and SB led to a poorer expansion and subsequent skewing of T-cell receptor Vβ repertoire. In general, transposon-transfected UCB T cells exhibit higher toxicity and poorer cell expansion than transposon-transfected PBL T cells (data not shown). In addition, clonal expansion may be correlated to multiple insertions, as in the case of PB integration. However, even clones with a single integration in Tol2-transfected T cells may potentially be subjected to clonal expansion (Figure 3a,b). Donor-dependent variance in spontaneous clonal dominance could also be possible. Clonal expansion in PB- or Tol2-transfected T cells in this study is likely due to dysregulated proto-oncogene expression near the integration sites and/or their preferential integration near regulatory gene regions based on our genome-wide integration analysis (Figures 4 and 6).
It is well established that retroviruses like murine leukemia virus have high preferences for integrating near promoter regions, whereas human immunodeficiency virus highly prefer to integrate inside active genes.35,36 Our data demonstrate that Tol2 and PB integration preferences resemble those seen using γ-retroviral vectors: both Tol2 and PB preferentially integrate near TSSs, CpG islands, and DNaseI hypersensitive sites. In contrast, SB integrations appear randomly and lack preference for these sites. Our results using human primary T cells are in agreement with published experiments performed in cell lines with SB23,37 and PB.10,12 Yant et al.23 studied 1,336 SB insertions in primary mouse liver and cultured mammalian cell lines (NIH 3T3 mouse fibroblasts and Huh-7 human hepatoma) and reported that 39.1% of SB integrations mapped within human RefSeq genes, a slightly higher frequency than computer-simulated random integrations (33%) but a much lower frequency than murine leukemia virus (50.7%) and human immunodeficiency virus (83.4%) vectors.35,36 A 11.2% of SB integrations were found near (±5 kb) TSSs compared with 21.4% of murine leukemia virus integrations.23 Wilson et al.12 mapped 575 PB integration sites in human HEK-293 and HeLa cells and reported that 48.8% of integrations were found in RefSeq annotated genes, a slightly higher frequency than that observed for SB integrations (39.1%).23 Interestingly, PB exhibits a greater bias than SB for integration at locations ±5 kb of TSSs (16.2% versus 8.5%) and ±1 kb of CpG islands (3.8% versus 2.5%). Bushman's group has characterized SB integration sites using a genomic annotation approach and reached the same conclusion, as ours, that SB integrations show no consistent preference for integration in active transcriptional units, CpG islands, near transcription start and stop features, and DNase I hypersensitive sites.37
Our study has important implications for transposon-based gene therapy. It appears that SB is a desirable choice of delivery vector in human T cells for the following reasons: (i) SB integration sites appear to be random compared to PB and Tol2; (ii) SB-transfected T cells exhibited fewer numbers of integrants per cell than PB, thus it may reduce insertional genotoxicity; (iii) SB can mediate an equivalent efficiency of stable gene transfer in UCB T cells as PB. Although it is less efficient than PB in PBL T cells, SB-transfected cells can be positively selected using an antibody to a reporter gene and further expanded in culture;6 (iv) With latest development of the hyperactive SB100X transposase,38 we demonstrate that SB100X is superior to SB11 and PB in mediating stable DsRed expression in cord blood–derived hematopoietic stem and progenitor cells.39 Thus, it would be interesting to test SB100X versus the latest generation of PB transposase40 in human T cells; and (v) stable integration of SB transposase-encoding sequences is so rare that no transposase mRNA transcript or protein can be detected in 14 SB transposed bulk T-cell populations.41 Inclusion of a suicide approach in SB-engineered T cells or use of mRNA as a source of transposase may lift this concern.
Materials and Methods
Construction of transposons and transposase-encoding plasmids. SB, miniTol2 (mTol2) and PB transposons and transposases (Figure 1a,b) were constructed under an identical plasmid backbone using standard molecular cloning techniques. Briefly, SB, mTol2, and PB transposon terminal repeat sequences were cloned from pT2/DsRed (SB-specific),4 pDB-mTol2 (Tol2-specific),17 or pXL-Bac II (PB-specific)42,43 (kindly provided by Malcolm Fraser Jr, University of Notre Dame, Notre Dame, IN) individually into a plasmid backbone containing a kanamycin-resistance gene. Subsequently, a Caggs promoter, DsRed reporter gene, and polyadenylation signal (Invitrogen, Carlsbad, CA) were inserted. All three transposases were cloned separately into another vector in which expression was under the control of the CMV promoter (Figure 1b). In the recoverable transposon system, SB, Tol2, and PB transposons were constructed with a minimum intron containing an RNA splice donor site; a splice acceptor site on either side of the prokaryotic promoter EM7 was inserted between the green fluorescent protein (GFP) and zeocin-resistant (ZeocinR) portions of a GFP-ZeocinR fusion protein, the PUC origin was followed by the bovine growth hormone polyadenylation signals within transposon cargo (Figure 1c).
Human T-cell gene transfer and generation of T-cell clones. Human T-cell gene nucleofection with DNA transposons has been previously described.4,5,6 PBL was either purchased from Memorial Blood Centers (St Paul, MN) or obtained from healthy donors with informed consent. Discarded UCB was obtained from the Duke University Cord Blood Center, St Louis Blood Center, New York Blood Center, and the American Red Cross in the Twin Cities following informed consent. MNCs from PBL and UCB were isolated by Lymphocyte Separation Medium (Mediatech Cellgro, Herndon, VA), washed, and resuspended in human T-cell Nucleofector solution (Lonza, Walkersville, MD). Nucleofections were performed using a Nucleofector device (Lonza), program U14. Briefly, 5 µg of transposon plasmid were co-transfected with 0, 2.5, 5, 10, 15, or 20 µg of transposase plasmid and a sufficient quantity of supplemental empty vector (pK/CMV-empty) so that a total of 25 µg of plasmid was always added to 5 × 106 MNCs from PBL or MNCs from UCB per sample. Following nucleofection, the cells were immediately transferred into 24-well plates containing human T-cell medium [RPMI-1640 (Invitrogen), 10% fetal calf serum (Hyclone), 2 mmol/l -glutamine, 50 U/ml penicillin, 50 µg/ml streptomycin, 25 µmol/l β-mercapto-ethanol, supplemented with rhIL-2 (50 IU/ml; Chiron, Emeryville, CA) and rhIL-7 (10 ng/ml; National Cancer Institute Biological Resources Branch, Rockville, MD)] and incubated at 37 °C/5% CO2 overnight or for 2–4 hours (UCB8–12 in Supplementary Figure S1). Cells were activated with anti-CD3/CD28 beads (provided by Bruce L. Levine, University of Pennsylvania, Philadelphia, PA) at a target-to-bead ratio of 1:3.44 Five days later, beads were removed and activated T cells were maintained in human T-cell medium supplemented with rhIL-2 (50 IU/ml) and rhIL-7 (10 ng/ml) and restimulated every 10–14 days with OKT3 (Ortho Biotech, Raritan, NJ).45 Cell samples were periodically harvested and transgene expression was assayed by fluorescence-activated cell-sorting analysis using a FACSCalibur (BD Biosciences, San Jose, CA).
T-cell clones were generated by limiting dilution from DsRed+ bulk T-cell populations after 3–4 rounds of expansion and cell sorting. T-cell clones were expanded in 24-well plates or flasks with OKT3, irradiated allogeneic MNCs from PBL, and Epstein–Barr virus–transformed B cells.45
When the recoverable transposon system was employed, 5 µg transposon plasmid and 15 µg transposase plasmid were used in the nucleofection protocol. After 5 days of activation the anti-CD3/CD28 beads were removed and zeocin (200 µg/ml; Invitrogen) was added in the T-cell medium to select ZeocinR T cells. The cells were further expanded in culture for 1–2 months using OKT3. GFP+ cells were confirmed by fluorescence-activated cell sorting.
Construction of transposition library. Genomic DNA from transposed T cells (ZeocinR and GFP+) was purified using Purege DNA Purification Kit (Qiagen, Valencia, CA). One microgram of genomic DNA was cut with BamHI for SB, EcoRI for Tol2, or XbaI for PB. The resulting genomic DNA fragments were ligated and electroporated into DH10B cells (Invitrogen). Transformed colonies were selected for ZeocinR and kanamycin-sensitive clones. Single colonies were grown in 96 deep-well culture blocks (Edge Biosystems, Gaithersburge, MD) and plasmids were purified using Perfectprep plasmid 96 Vac Kit (Qiagen). The plasmid library was confirmed by restriction digest. Plasmids containing transposon:genomic DNA fragments were sequenced using Big-Dye reagent.
Linker-mediated PCR used to recover the genomic DNA sequences flanking transposon inserts have been described previously.4,6 Linker and primer sequences are listed in Supplementary Table S2. DNA sequencing was performed at the BioMedical Genomics Center at University of Minnesota. The sequence results were subjected to BlastN analysis against the human genome using the UCSC database. Cancer-related genes were identified using the Cancer Genes database at Memorial Sloan-Kettering Cancer Center (http://cbio.mskcc.org/CancerGenes/). TSS data were extracted from the UCSC database originally from SwitchGear Genomics. The closest TSS on the same strand with the mapped region was used.
Southern hybridization analysis. Southern hybridization was performed as previously described.4 A radiolabeled probe specific for a portion of DsRed was used.
Quantification of gene expression by TaqMan qPCR. Total RNA was extracted from DsRed+ T-cell clones and mock T cells using RNeasy Protect Cell Mini Kit (Qiagen) and treated with RNase-free DNase (Invitrogen). First-strand complementary DNA was synthesized from total RNA with SuperScript III reverse transcriptase (Invitrogen) and then treated with RNase H (Invitrogen). Human XpressRef Universal total RNA was purchased from SABiosciences (Frederick, MD) as reference. TaqMan qPCR was performed on ABI 7500 Real Time PCR System (Applied Biosystems, Carlsbad, CA) according to the manufacturer's instruction. TaqMan primers and probe sets for the different genes were purchased from Applied Biosystems. Each sample was analyzed in duplicate and repeated once. Human 18S ribosomal RNA was used for normalizing the sample loading. The reference total RNA was used as a calibrator. Three pooled mock T cells from UCB or PBL were used as negative control. A control with no template and reference total RNA were included in each plate. Gene expression level was presented as fold change 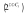 relative to the calibrator using the following formula: ΔΔCt = ΔCt of sample − ΔCt of calibrator; ΔCt = Ct of target gene − Ct of endogenous18S ribosomal RNA.
relative to the calibrator using the following formula: ΔΔCt = ΔCt of sample − ΔCt of calibrator; ΔCt = Ct of target gene − Ct of endogenous18S ribosomal RNA.
Bioinformatic analysis of integration sites. Integration sites of transposons were mapped to the human genome by Blat and Blast program (human genome freeze hg18; University of California at Santa Cruz Human Genome Project, http://genome.ucsc.edu/). Insertion sites were considered authentic only if they (i) contained sequence from the nested primer to the end of the 5′ terminal repeat sequence; (ii) matched a genomic location starting immediately after the end of the terminal repeat; (iii) showed ≥95% identity to the genomic sequence over the high-quality sequence region; and (iv) matched to no more than one genomic locus with ≥95% identity. The mapped integration sites were compared to 10,000 random computer-generated integration sites in the human genome excluding the gap regions. Customer Perl scripts were used to analyze the distributions of integration sites relative to genomic features, such as RefSeq genes, transcription start sites, CpG islands, etc., from the UCSC human genome hg18 annotation database (http://genome.ucsc.edu/). RefSeq genes were used as they were well annotated. Transcription start sites were derived from the hg18 RefSeq gene database. Statistical analysis was performed with the methods specified in the Results section.
Statistical analyses. A repeated measurement model was used to analyze the data in Figure 2. To make the outcome closer to a normal distribution, we chose the natural log transformation on the outcome. Kruskal–Wallis rank sum test was used to analyze the difference of median integrants and expression levels between groups in Figure 3b,c. Wilcoxon rank sum test was used to compare median difference between groups in Figure 4. Cochran–Mantel–Haenszel test was used to analyze the data set in Supplementary Table S1 to combine CD4+ and CD8+ T-cell subsets. All P values from pair-wise comparisons were adjusted using the Bonferroni method. Statistical analyses were conducted using SAS 9.1 software (Cary, NC). P values <0.05 were considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Stable transgene expression in human UCB-derived T cells by SB, Tol2 and PB transposons. Freshly isolated UCBMNCs (5 x 106) were co-nucelofected with 5 μg of transposon plasmids and different amounts of transposase-encoding plasmids (0, 2.5, 5, 10, 15, and 20 μg). pK/CMV-empty plasmid was used to adjust the total amount of DNA transfected in each sample. DsRed expression was analyzed on weeks 2, 3, 5, and 6. The data from 12 individual UCB units are shown. UCB1-7 and UCB8-12 nucleofected cells were exposed to anti-CD3/CD28 beads 2-4 h or overnight after transfection. Figure S2. Mapping integration sites in T-cell clones. (a) PB-PBL. * indicates that T-cell clones derived from PB-transfected PBL had similar PCR banding patterns. 24 out of 25 clones were subject to integration site analysis, and 20 integrant plasmids from each T-cell clone were sequenced (PCR bands were difficult to excise for sequencing). (b) PB-UCB. (c) Tol2 -PBL. (d) Tol2 -UCB. In PB-UCB, Tol2 -PBL, and Tol2 -UCB T-cell clones, 56-89% of PCR bands (68/∼76, 24/∼32 and 24/∼43, respectively) were recovered and sequenced for integration site analysis. Chr1 indicates chromosome 1. The number in the parenthesis indicates the start site of the integration. Human genome RefSeq (hg19) at UCSC human genome database was used to map integration sites. Figure S3. T-cell repertoire analysis of bulk T cells transfected with SB, Tol2 and PB. (a) Schematic representation of the SB, Tol2 and PB transposon constructs used for T-cell repertoire study. ΔNGFR, truncated nerve growth factor receptor; P, EM7 promoter; ZeoR, zeocin resistance gene; SD, splicing donor; SA, splicing acceptor. (b) Flow cytometric setting for T-cell repertoire analysis. A quadrant plot from UCB2 transfected with SB transposon and SB11 is representatively shown: PE+/FITC−, Vβ5.3; PE+/FICT+, Vβ7.1; PE−/FITC+, Vβ3. (c) % of Vβ usage in CD4+ and CD8+ subsets of transposon transfected PBL and UCB. Arrow indicates arbitrary 10-fold above mock. SB: black arrow. Tol2: green arrow. PB: purple arrow. Table S1. Summary of clonal expansion analyzed by TCR Vβ usage. Table S2. Linker and primer sequences used in linker-mediated PCR. Materials and Methods.
Acknowledgments
We thank Sonja Nodland (University of Minnesota Masonic Cancer Center) for major editing, R. Scott McIvor, David Largaespada, and Perry Hackett (University of Minnesota Center for Genome Engineering, Minneapolis, MN) for critical review of the manuscript. We also thank John E. Wagner for providing cord blood, Malcolm J. Fraser (University of Notre Dame, Notre Dame, IN) for providing piggyBac vectors, Andrew C. Wilber (Southern Illinois University, Springfield, IL) for providing Sleeping Beauty transposon recoverable vector, Kevin A.T. Silverstein (Bioinformatics Core Facility at Masonic Cancer Center, University of Minnesota) for the independent evaluation on the data, and Linan Ma (Biostatistics and Informatics Core at the Masonic Cancer Center, University of Minnesota) for initial statistical analysis. This work was supported by grants from the Children's Cancer Research Fund in Minneapolis, Alliance for Cancer Gene Therapy, the Gabrielle's Angel (formerly G&P) Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research Kimmel Scholar Program, the University of Minnesota Translational Research Grant, the University Minnesota Medical School Dean's Commitment (X.Z.), and Leukemia Research Fund. E.M. was a recipient of American Society of Hematology Research Trainee Award. P.B. was a recipient of the Undergraduate Research Opportunity Program Award and the Multicultural Summer Research Opportunities Award. This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E (X.W.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. Contribution statement: X.H. designed and performed the research, analyzed the data, and wrote the paper; H.G., S.T., E.M., and P.B. performed the research. Y.-C.J. performed DNA sequencing and analyzed the data; Q.C. performed the statistical analyses; Z.J.T. and Y.C.K. performed integration site analysis; S.C.E. provided miniTol2 construct and discussed the research; X.W. preformed integration bioinformatic analyses and wrote the paper; S.M.W. analyzed the data and wrote the paper; X.Z. oversaw the research and wrote the paper. The authors declare no competitive financial interests.
Supplementary Material
Stable transgene expression in human UCB-derived T cells by SB, Tol2 and PB transposons. Freshly isolated UCBMNCs (5 x 106) were co-nucelofected with 5 μg of transposon plasmids and different amounts of transposase-encoding plasmids (0, 2.5, 5, 10, 15, and 20 μg). pK/CMV-empty plasmid was used to adjust the total amount of DNA transfected in each sample. DsRed expression was analyzed on weeks 2, 3, 5, and 6. The data from 12 individual UCB units are shown. UCB1-7 and UCB8-12 nucleofected cells were exposed to anti-CD3/CD28 beads 2-4 h or overnight after transfection.
Mapping integration sites in T-cell clones. (a) PB-PBL. * indicates that T-cell clones derived from PB-transfected PBL had similar PCR banding patterns. 24 out of 25 clones were subject to integration site analysis, and 20 integrant plasmids from each T-cell clone were sequenced (PCR bands were difficult to excise for sequencing). (b) PB-UCB. (c) Tol2 -PBL. (d) Tol2 -UCB. In PB-UCB, Tol2 -PBL, and Tol2 -UCB T-cell clones, 56-89% of PCR bands (68/∼76, 24/∼32 and 24/∼43, respectively) were recovered and sequenced for integration site analysis. Chr1 indicates chromosome 1. The number in the parenthesis indicates the start site of the integration. Human genome RefSeq (hg19) at UCSC human genome database was used to map integration sites.
T-cell repertoire analysis of bulk T cells transfected with SB, Tol2 and PB. (a) Schematic representation of the SB, Tol2 and PB transposon constructs used for T-cell repertoire study. ΔNGFR, truncated nerve growth factor receptor; P, EM7 promoter; ZeoR, zeocin resistance gene; SD, splicing donor; SA, splicing acceptor. (b) Flow cytometric setting for T-cell repertoire analysis. A quadrant plot from UCB2 transfected with SB transposon and SB11 is representatively shown: PE+/FITC−, Vβ5.3; PE+/FICT+, Vβ7.1; PE−/FITC+, Vβ3. (c) % of Vβ usage in CD4+ and CD8+ subsets of transposon transfected PBL and UCB. Arrow indicates arbitrary 10-fold above mock. SB: black arrow. Tol2: green arrow. PB: purple arrow.
Summary of clonal expansion analyzed by TCR Vβ usage.
Linker and primer sequences used in linker-mediated PCR.
REFERENCES
- Ivics Z, Hackett PB, Plasterk RH., and, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Hackett PB, Ekker SC, Largaespada DA., and, McIvor RS. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- Ivics Z., and, Izsvák Z. Transposons for gene therapy! Curr Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- Huang X, Wilber AC, Bao L, Tuong D, Tolar J, Orchard PJ, et al. Stable gene transfer and expression in human primary T cells by the Sleeping Beauty transposon system. Blood. 2006;107:483–491. doi: 10.1182/blood-2005-05-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wilber A, McIvor RS., and, Zhou X. DNA transposons for modification of human primary T lymphocytes. Methods Mol Biol. 2009;506:115–126. doi: 10.1007/978-1-59745-409-4_9. [DOI] [PubMed] [Google Scholar]
- Huang X, Guo H, Kang J, Choi S, Zhou TC, Tammana S, et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett PB. Integrating DNA vectors for gene therapy. Mol Ther. 2007;15:10–12. doi: 10.1038/sj.mt.6300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E., and, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y., and, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, et al. piggyBac is a flexible and highly active transposon as compared to Sleeping Beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ., and, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P., and, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Inagaki H, Bessho Y., and, Hori H. Insertion of a novel transposable element in the tyrosinase gene is responsible for an albino mutation in the medaka fish, Oryzias latipes. Mol Gen Genet. 1995;249:400–405. doi: 10.1007/BF00287101. [DOI] [PubMed] [Google Scholar]
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi LR., and, Beverley SM. cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res. 2000;28:784–790. doi: 10.1093/nar/28.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Kaminski JM., and, George AL., Jr Functional zinc finger/Sleeping Beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S., and, Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- Yant SR, Huang Y, Akache B., and, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. The piggyBac transposon holds promise for human gene therapy. Proc Natl Acad Sci USA. 2006;103:14981–14982. doi: 10.1073/pnas.0607282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM., and, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, et al. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, International Human Genome Sequencing Consortium et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Zayed H, Izsvák Z, Khare D, Heinemann U., and, Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisko O, Izsvák Z, Szabó K, Kaufman CD, Herold S., and, Ivics Z. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc Natl Acad Sci USA. 2006;103:4062–4067. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JJ, Wang H, Rashid A, Tan TH, Hwang RF, Hamilton SR, et al. Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res. 2008;14:7043–7049. doi: 10.1158/1078-0432.CCR-08-0381. [DOI] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, et al. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JH, Wang X, Manning G, LaMere BJ, Le P, Zhu S, et al. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol. 2003;23:2068–2082. doi: 10.1128/MCB.23.6.2068-2082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman CE, Franovic A, Payette J., and, Lee S. ETS-1 oncogenic activity mediated by transforming growth factor alpha. Cancer Res. 2010;70:730–740. doi: 10.1158/0008-5472.CAN-09-2090. [DOI] [PubMed] [Google Scholar]
- Shibolet O, Giallourakis C, Rosenberg I, Mueller T, Xavier RJ., and, Podolsky DK. AKAP13, a RhoA GTPase-specific guanine exchange factor, is a novel regulator of TLR2 signaling. J Biol Chem. 2007;282:35308–35317. doi: 10.1074/jbc.M704426200. [DOI] [PubMed] [Google Scholar]
- Sterpetti P, Marucci L, Candelaresi C, Toksoz D, Alpini G, Ugili L, et al. Cell proliferation and drug resistance in hepatocellular carcinoma are modulated by Rho GTPase signals. Am J Physiol Gastrointest Liver Physiol. 2006;290:G624–G632. doi: 10.1152/ajpgi.00128.2005. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Xue X, Huang X, Nodland SE, Mátés L, Ma L, Izsvák Z, et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- Cadiñanos J., and, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Haley K, Wong M, Guo H, Lu C, Wilber A, et al. Hum Gene Ther. epub ahead of print; 2010. Unexpectedly high copy number of random integration but low frequency of persistent expression of the Sleeping Beauty transposase following trans delivery in primary T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lobo N, Bauser CA., and, Fraser MJ., Jr The minimum internal and external sequence requirements for transposition of the eukaryotic transformation vector piggyBac. Mol Genet Genomics. 2001;266:190–198. doi: 10.1007/s004380100525. [DOI] [PubMed] [Google Scholar]
- Li X, Harrell RA, Handler AM, Beam T, Hennessy K., and, Fraser MJ., Jr piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol. 2005;14:17–30. doi: 10.1111/j.1365-2583.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- Levine BL, Bernstein WB, Aronson NE, Schlienger K, Cotte J, Perfetto S, et al. Adoptive transfer of costimulated CD4+ T cells induces expansion of peripheral T cells and decreased CCR5 expression in HIV infection. Nat Med. 2002;8:47–53. doi: 10.1038/nm0102-47. [DOI] [PubMed] [Google Scholar]
- Riddell SR., and, Greenberg PD. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stable transgene expression in human UCB-derived T cells by SB, Tol2 and PB transposons. Freshly isolated UCBMNCs (5 x 106) were co-nucelofected with 5 μg of transposon plasmids and different amounts of transposase-encoding plasmids (0, 2.5, 5, 10, 15, and 20 μg). pK/CMV-empty plasmid was used to adjust the total amount of DNA transfected in each sample. DsRed expression was analyzed on weeks 2, 3, 5, and 6. The data from 12 individual UCB units are shown. UCB1-7 and UCB8-12 nucleofected cells were exposed to anti-CD3/CD28 beads 2-4 h or overnight after transfection.
Mapping integration sites in T-cell clones. (a) PB-PBL. * indicates that T-cell clones derived from PB-transfected PBL had similar PCR banding patterns. 24 out of 25 clones were subject to integration site analysis, and 20 integrant plasmids from each T-cell clone were sequenced (PCR bands were difficult to excise for sequencing). (b) PB-UCB. (c) Tol2 -PBL. (d) Tol2 -UCB. In PB-UCB, Tol2 -PBL, and Tol2 -UCB T-cell clones, 56-89% of PCR bands (68/∼76, 24/∼32 and 24/∼43, respectively) were recovered and sequenced for integration site analysis. Chr1 indicates chromosome 1. The number in the parenthesis indicates the start site of the integration. Human genome RefSeq (hg19) at UCSC human genome database was used to map integration sites.
T-cell repertoire analysis of bulk T cells transfected with SB, Tol2 and PB. (a) Schematic representation of the SB, Tol2 and PB transposon constructs used for T-cell repertoire study. ΔNGFR, truncated nerve growth factor receptor; P, EM7 promoter; ZeoR, zeocin resistance gene; SD, splicing donor; SA, splicing acceptor. (b) Flow cytometric setting for T-cell repertoire analysis. A quadrant plot from UCB2 transfected with SB transposon and SB11 is representatively shown: PE+/FITC−, Vβ5.3; PE+/FICT+, Vβ7.1; PE−/FITC+, Vβ3. (c) % of Vβ usage in CD4+ and CD8+ subsets of transposon transfected PBL and UCB. Arrow indicates arbitrary 10-fold above mock. SB: black arrow. Tol2: green arrow. PB: purple arrow.
Summary of clonal expansion analyzed by TCR Vβ usage.
Linker and primer sequences used in linker-mediated PCR.