Abstract
The transfection efficiency (TE) of chitosan–plasmid DNA (pDNA) polyplexes can be critically modulated by the polymer degree of deacetylation (DDA) and molecular weight (MW). This study was performed to test the hypothesis that the TE dependence on chitosan MW and DDA is related to the polyplex stability, hence their intracellular decondensation/unpacking kinetics. Major barriers to nonviral gene transfer were studied by image-based quantification. Although uptake increased with increased DDA, it did not appear to be a structure-dependent process affecting TE, nor was nuclear entry. Colocalization analysis showed that all chitosans trafficked through lysosomes with similar kinetics. Fluorescent resonant energy transfer (FRET) analysis revealed a distinct relationship between TE and polyplex dissociation rate. The most efficient chitosans showed an intermediate stability and a kinetics of dissociation, which occurred in synchrony with lysosomal escape. In contrast, a rapid dissociation before lysosomal escape was found for the inefficient low DDA chitosan whereas the highly stable and inefficient complex formed by a high MW and high DDA chitosan did not dissociate even after 24 hours. This study identified that the kinetics of decondensation in relation to lysosomal escape was a most critical structure-dependent process affecting the TE of chitosan polyplexes.
Introduction
The identification of an efficient and safe DNA delivery vector remains a critical impediment to successful clinical translation of gene therapy. Polyplexes formed by combining DNA with natural polymers are regarded as promising alternatives to viral vectors, considering their reduced immunogenicity, ability to carry large DNA loads, and inherent formulation flexibility. However, polyplexes suffer from significantly lower transfection efficiency (TE) than their viral counterparts. To improve and optimize their transfection ability, it is critical to improve the understanding of intracellular trafficking of these gene vectors to identify specific rate-limiting steps to gene expression, thus allowing the development of strategies to overcome these barriers and to ultimately design improved DNA delivery vectors.1,2
Chitosan, a natural polycation with high positive charge density, has been reported as an effective polynucleotide delivery vector3,4,5,6 with the ability to bind and condense plasmid DNA (pDNA)7 into a system protective against nuclease degradation.8 Chitosan is produced by deacetylation of chitin resulting in a polymer with glucosamine and N-acetylated glucosamine monomers, the proportion of which is described by the degree of deacetylation (DDA). Chitosan physicochemical properties depend on molecular weight (MW) and DDA and these two factors can influence TE to a significant extent.3,6,9,10
Timely intracellular DNA unpacking and release is generally regarded as one important rate-limiting step for nonviral condensing polycations.11 It has been suggested that the most efficient chitosan polyplexes have a subtle balance between polyplex stability (maximized at high MW and high DDA) to protect DNA against nucleases, and complex instability (occurring at low MW and/or DDA), to permit DNA dissociation from chitosan and access to transcriptional machinery.3,7,12,13 A positive correlation between an increase in either DDA and/or MW and the binding affinity of DNA to chitosans was suggested by Strand et al.12 and was recently quantified by isothermal calorimetry.14 Despite the potential of chitosan as a gene delivery vehicle and the ability to tailor its functional properties through control of MW and DDA, very little is known about how these structural parameters of chitosan influence intracellular trafficking of chitosan–DNA polyplexes and impact TE.
Prior assessment of the role of DDA and MW on the transfection process has largely been based on end point reporter gene expression levels, providing little or no information concerning intracellular events.6,7,15 Although intracellular trafficking studies of nonviral vectors are numerous,1,16,17,18,19 few have examined chitosan, and most are comparative to other polycations2,20,21 examining only one chitosan type, without accounting for the strong influence of DDA and MW on the transfection process.6 The few studies that have examined changes in DDA and MW simultaneously3,9,10 have not assessed their influence on intracellular events in terms of polyplex stability. Hence, presently, no consensus exists as to what major barriers, including internalization, endolysosomal escape, unpacking, and nuclear entry, limit the TE of chitosan–DNA polyplexes.
In this study, we have investigated the intracellular trafficking of specific chitosan polyplexes of varying DDA and MW with the intent of identifying both formulation-dependent and formulation-independent rate-limiting barriers. Our hypotheses were that the variation in TE for different chitosans is mainly due to chitosan–DNA binding stability and that barriers to chitosan–DNA systems generally lie upstream of polyplex decondensation. Polyplex cellular binding, uptake, and endolysosomal trafficking were assessed by both fluorescence-activated cell sorting (FACS) and live-cell confocal imaging. Polyplex stability was examined using fluorescent resonant energy transfer (FRET).2,18,20,22 The spatiotemporal monitoring of specific chitosan polyplexes revealed a similar pattern of cellular internalization and sequestration in lysosomes for all chitosans where only their decondensation kinetics varied with chitosan MW and DDA. These results suggest that the timing of intracellular dissociation of chitosan and pDNA in relationship to lysosomal transit and escape is a determining factor of efficient gene expression and that polyplexes of intermediate stability combining specific MW and DDA offer the greatest potential for high levels of gene expression as they decondense in synchrony with lysosomal escape.
Results
Kinetics of gene expression
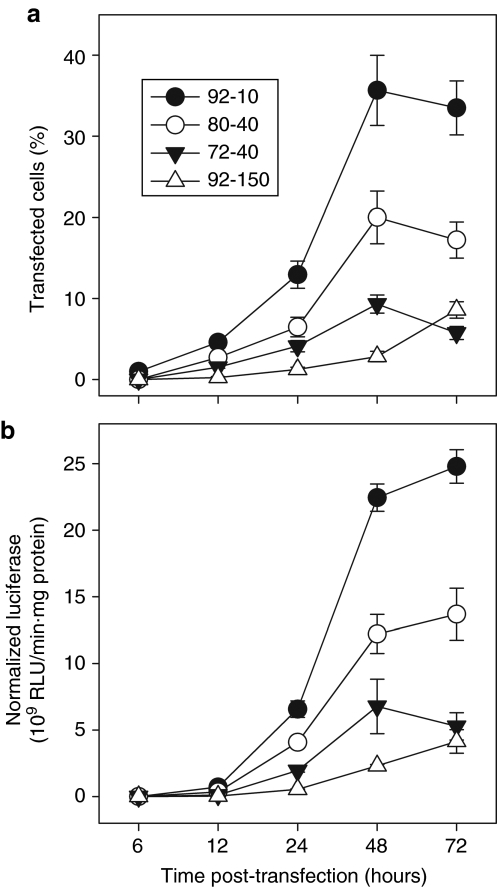
The kinetics of transgenic gene expression of polyplexes formed with the four selected chitosans were characterized to establish a time frame for intracellular events leading to gene expression. The results confirmed that TE is formulation-dependent, with chitosan 92-10 as the most efficient, followed by 80-40, 72-10, and finally 92-150 with very slow kinetics and low transgene expression (Figure 1a,b).6 In this study, chitosan 92-10 served as positive control as it has previously been shown by us6 to be more efficient than lipofectamine (Sigma, St Louis, MO) in HEK293. Slight protocol modifications have been introduced since these previous studies6 including a change of media to pH 7.4 at 24 hours, which results in increased TE, from around 28% to nearly 40% positive cells, with corresponding increases in luciferase reporter activity.23 The time to gene expression is comparable for all chitosans tested here, starting at 24 hours and peaking at 48 hours post-transfection (Figure 1a,b), suggesting that they follow similar intracellular pathways, although the slowest vector, chitosan 92-150, still shows increasing expression at this time. Some marginal gene expression was seen at 12 hours for 92-10, which could imply that the onset of significant gene expression for optimal chitosans is between 12 and 24 hours, as reported with other chitosans in HEK293.3
Figure 1.
Transfection kinetics of polyplexes prepared with chitosans of different degree of deacetylation (DDA) and molecular weight (MW). (a,b) Analysis of the kinetics of gene expression for HEK 293 cells transfected with chitosan polyplexes using chitosans with different DDA and MW. Flow cytometry provided (a) % cells expressing green fluorescent protein and (b) luminometry the level of luciferase expression. Chitosan 92-10 was the most efficient of the four formulations tested where gene expression occurs at 24 hours and peaks at 48 hours. Results are the average of three (N = 3) independent experiments ± SD where each experiment included two replicates. RLU, relative light units.
Cell binding and uptake
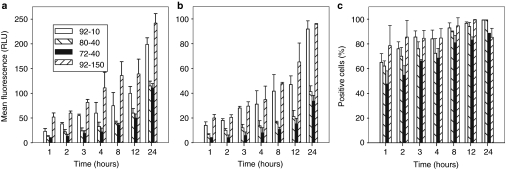
FACS analysis on transfected cells was performed to examine whether binding or uptake of polyplexes is responsible for the measured differences in TE between chitosan types (Figure 1). Polyplexes were prepared with either the rhodamine B isothiocyanate (RITC)-labeled chitosans and unlabeled pDNA or with unlabeled chitosan complexed with labeled Cy5-pDNA. Figure 2 shows cell binding and uptake data obtained with polyplexes prepared with labeled chitosans. Similar kinetics and trends were seen with polyplexes prepared with labeled DNA (Supplementary Figure S1a), revealing that both methods provide similar measures of polyplex uptake. This can be partly explained by the fact that when chitosan–DNA polyplexes are prepared at N:P = 5, free unbound chitosan is at most 50% of total chitosan24 and that no free DNA exists at early time points, as demonstrated below by the FRET data using labeled DNA. All four polyplex types had detectable binding to cells at 1 hour, with a subsequent strong increase in binding over time. Higher DDA polyplexes bind to a greater level to cells for all time points. There was a strong correlation between cell binding (Figure 2a) and cell uptake (Figure 2b), with a notable increase in uptake for the two chitosans with high DDA (92%) compared to lower DDA chitosans. The correlation between binding and uptake underscores the necessity of establishing cell contact with polyplexes. The number of cells positive for uptake (Figure 2c) peaked at around 8 hours when nearly 100% of the cells contained internalized polyplexes. In contrast, the total amount taken up per cell (Figure 2b) increased continuously throughout time until the media was changed, indicating that no saturation of internalization occurs at any time point and also revealing, by simple morphological assessment, a lack of toxicity of chitosan-based vectors with increasing dose as reported previously.3 These results were confirmed by confocal microscopy using polyplexes prepared with both labeled DNA and labeled chitosan (Supplementary Figure S1b). Although some differences in TE between lower DDA chitosan (80-40 and 72-40) and chitosan 92-10 can be accounted for by lower uptake, especially in terms of levels, uptake cannot account for the poor TE of 92-150, as uptake was similar to 92-10 for both the total amount per cell (Figure 2b) and the number of positive cells (Figure 2c). These results suggest that the formulation-dependent process resulting in low TE for chitosans 72-40 and 92-150 lies downstream of cell uptake.
Figure 2.
Kinetics of cellular binding and uptake of polyplexes prepared with chitosans of different degree of deacetylation (DDA) and molecular weight (MW). HEK293 cells were incubated with fluorescent chitosan polyplexes for the indicated periods of time and analyzed by flow cytometry. Flow cytometry quantitative analysis of mean fluorescence per cell for polyplex (a) binding and (b) uptake and (c) of % cells with internalized polyplexes was performed following trypsinization and extensive washes, except for (a) cell binding, where cells were detached by enzyme-free cell dissociation buffer and analyzed directly. (b) Mean uptake levels per cell and (c) % positive cells were obtained from the same set of flow cytometry data. Graphs show that binding and uptake are time and DDA-dependent, with both 92% DDA chitosans binding more effectively than the lower DDA chitosans, resulting in increased uptake. Results are the average of three (N = 3) independent experiments ± SD where each experiment included two replicates. RLU, relative light units.
Colocalization of polyplexes with lysosomes
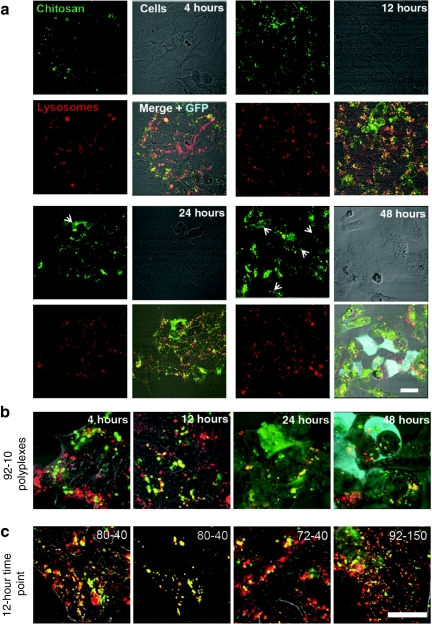
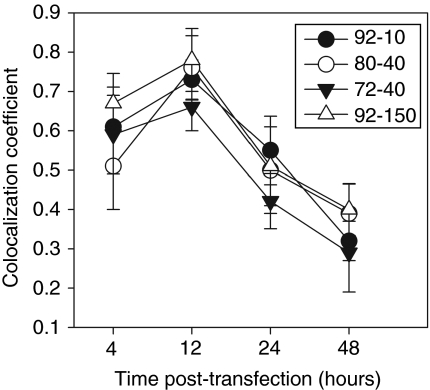
Because cationic polyplex systems may traffic through lysosomes,3,16 we assessed the colocalization between polyplexes and lysosomes. Lysosomal labeling by specific accumulation of fluorescent-dextran following endocytosis has been demonstrated by colocalization with established lysosomal markers such as lysosome-associated membrane protein-2 in many cell types including HEK293.25,26,27 After 4 hours (Figure 3a,b) of incubation, the intracellular transit of 92-10 polyplexes show a mixture of polyplexes in lysosomes (yellow) and a separate population that is not in lysosomes (red). Subsequently at 12 hours, a stronger colocalization in lysosomes was seen which then diminished at 24–48 hours indicating a gradual loss of colocalization in parallel with a gradual increase of diffuse RITC-chitosan signal in the cell cytoplasm as shown by arrows (Figure 3a). These observations suggest that at 4 hours polyplexes are still in endosomes and in transit to lysosomes, respecting the kinetics of about 4 hours for endosomal fusion with lysosomes.18 Nearly all polyplexes reach lysosomes at 12 hours, with little or no escape. Chitosan 92-10 polyplexes then escape lysosomes relatively slowly from 12 to 48 hours, as indicated by increasing diffuse staining seen in many cells at 48 hours. Importantly, this same kinetic of lysosomal transit applies to all four chitosans where images at 12 hours (Figure 3c) show strong colocalization with lysosomes in all cases. Similar results were obtained when DNA was labeled (data not shown). The second image from the left in Figure 3c is a representation of the mask function in the LSM software that exclusively extracts colocalized pixels, from which the calculated colocalization coefficient in this case was 0.88. The mask image serves here as an example of what was done for all conditions and time points to quantify the data in Figure 4. The monitoring of the four types of chitosan-based polyplexes revealed a time-dependent accumulation of all polyplex systems in lysosomes with a steady increase in colocalization to ~80% within the first 12 hours, followed by steady decrease to ~40% at 48 hours (Figure 4).
Figure 3.
Intracellular lysosomal transit of chitosan-based polyplexes. (a,b) Live-cell confocal imaging of the intracellular lysosomal transit of chitosan-based polyplexes for chitosan 92-10 and (c) for the three additional chitosans used in this study at 12 h. (a) Lysosomes of HEK293 were labeled by dextran pulse-chase (red) before transfecting cells with labeled chitosan–plasmid DNA complexes (green) for 8 hours. Colocalization of chitosan and lysosomes is indicated as yellow overlap of green and red pixels in the merged channel. A generalized punctate pattern for both 92-10 polyplexes and lysosomes with strong colocalization peaking at 12 hours followed by a subsequent gradual increase in a diffuse pattern of chitosan and loss of colocalization to lysosomes from 24 hours (arrows heads) to 48 hours. Most of the cells transfected using chitosan 92-10 show this typical diffuse staining at 48 hours where GFP expression can also be seen (blue). (b,c) Zoomed and merged images of polyplexes prepared (b) with chitosan 92-10 at various time points or (c) with additional chitosans imaged at 12 hours showing strong colocalization of polyplexes with lysosomes (mostly yellow pixels). The second image from the left in c shows an example of exclusively colocalized pixels from the first image on the left (80-40 chitosan), where the colocalization coefficient between polyplexes and lysosomes was calculated to be 0.88. Bar = 10 µm. GFP, green fluorescent protein.
Figure 4.
Kinetics of colocalization of chitosan–plasmid DNA polyplexes and lysosomes following transfection. Colocalization coefficients between labeled polyplexes and fluorescent dextran-labeled lysosomes were calculated from confocal live-cell images of multiple cells taken at specific time points following transfection using the LSM 510 colocalization analysis module. The analysis reveals an initial accumulation of the polyplex in the lysosomes followed by a time-dependent lysosomal escape from 12 to 48 hours that was similar for all chitosans. Results shown are the average of three (N = 3) independent experiments ± SD where each experiment quantified a minimum of 15 cells with three optical sections each.
Intracellular decondensation kinetics of chitosan-based polyplexes
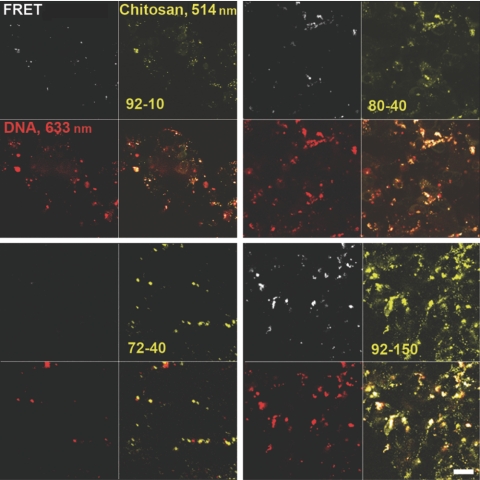
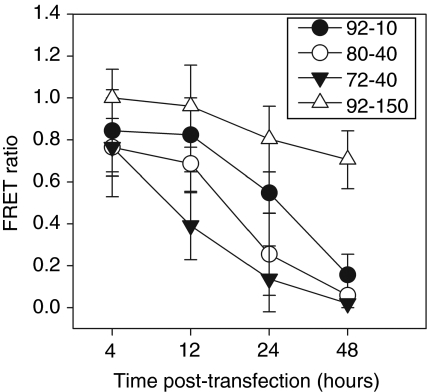
The dissociation of pDNA from the polyplex depends on the physicochemical characteristics of the polyplex that determines its stability. We monitored decondensation using double-labeled polyplexes characterized under various conditions for sensitized FRET analysis (Supplementary Figure S2), an approach that accounts for acceptor and donor channel crosstalk to provide the fraction of unpacked/decondensed versus complexed/condensed pDNA.28,29 Representative images for all chitosans at 24 hours (Figure 5) show channels for chitosan, DNA, and FRET, the latter indicating tight binding in condensed polyplexes. Decondensation results in loss of FRET. Comparison of the FRET channel to the DNA channel for all polyplexes reveals that polyplexes prepared with both 92-10 and 80-40 chitosans have a mixture of condensed FRET-generating (white dots) polyplexes and decondensed unpackaged DNA (red dots) at 24 hours. In contrast, chitosan 92-150 polyplexes were nearly all condensed at 24 hours, generating strong FRET signals, whereas 72-40 polyplexes were nearly all decondensed with no FRET. The change in FRET ratio [Eq. (1) in Methods] over time as shown in Figure 6 provides a quantitative assessment of the decondensation state of the polyplexes based on these images. At early times (4 hours), 92-150 generates the strongest FRET signal indicating a tighter pDNA condensation state than with the other lower DDA or lower MW chitosans (Figure 6). We observed a strong time-dependent decrease in condensation for all chitosans except for 92-150, which remained 80% condensed even after 48 hours. The decondensation rate was fastest for chitosan 72-40, with rapid decondensation within the first 12 hours, before lysosomal escape at 12 hours (Figure 4), and losing nearly all FRET signal within 24 hours. In contrast, the two most efficient chitosans displayed intermediate stability profiles with a relatively minor decondensation at 12 hours followed by a rapid loss of FRET and therefore a loss of the condensed state in the 12- to 24-hour period, simultaneous with lysosomal escape (Figure 4).
Figure 5.
FRET observation of the decondensation of plasmid DNA (pDNA)–chitosan based polyplexes of different degree of deacetylation (DDA) and molecular weight (MW) inside cells. HEK293 cells were transfected for 8 hours with rhodamine B isothiocyanate (RITC)-labeled chitosan vectors and Cy5-labeled pDNA and observed by confocal microscopy at 24 hours. For FRET analysis, the donor fluorophore (RITC) was excited at 514 nm and the FRET signal corresponding to acceptor (Cy5) emission was detected. Signal from RITC-chitosan (514 nm excitation) and of total Cy5-pDNA (633 nm excitation) were detected as well. Chitosan formulation is indicated in the quadrant corresponding to the chitosan channel. Comparison between the FRET channel and the DNA channel for all polyplexes reveals that polyplexes prepared with both 92-10 and 80-40 chitosans have a mixture of condensed FRET-generating (white in the lower right panel) and decondensed (red in the lower right panel) polyplexes at 24 hours post-transfection. In contrast, 92-150 polyplexes are still nearly all condensed (white in the lower right panel), whereas 72-40 polyplexes are nearly all unpacked (red in lower right panel). Quantitative analysis of decondensation based on these FRET images is found in Figure 6. Bar = 10 µm. FRET, fluorescent resonant energy transfer.
Figure 6.
Intracellular decondensation/dissociation kinetics of plasmid DNA (pDNA)–chitosan polyplexes of different degree of deacetylation (DDA) and molecular weight (MW). FRET ratio values for each time point were determined for polyplexes prepared with four different chitosans. FRET ratio profiles illustrate that unpacking increases over time with kinetics depending on the formulation. Chitosan 92-150 unpacking is slow with most particles remaining condensed (FRET ratio close to 1) after 48 hours, whereas 72-40 unpacking was fast and largely unpacked at 12 hours. The intermediate profiles of 80-40 and 92-10 correspond to the most efficient chitosans for transfection (Figure 1), suggesting the importance of achieving a balance between protective stability and dissociation of the polyplexes for maximal transfection efficiency. Particle dissociation, as indicated by a large drop in FRET between 12 and 24 hours for the most efficient chitosans, was seen to occur at the same time as lysosomal escape in Figure 4, suggesting synchronization of these two events to be important. Results are the average of three (N = 3) independent experiments ± SD where each experiment also involved two replicates. FRET, fluorescent resonant energy transfer.
Assessment of chitosan localization to the nucleus
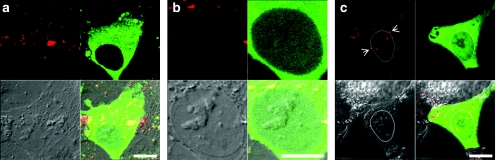
We imaged transfected cells to determine the presence of chitosan in nuclei. Imaging started at 18 hours, to examine positively expressing cells and reduce the chance that the transgene-expressing cell had undergone division, which could passively allow chitosan entry. Chitosan could not be visualized within nuclei in any cells imaged between 18 and 26 hours (Figure 7a,b). When positively expressing cells were imaged at 48 hours (Figure 7c), occasional observations (about 2–5%) of chitosan inside the nucleus occurred. Therefore, we may assume that on the rare occasions where chitosan is seen in the nuclei of transfected cells, its presence is possibly due to nuclear division in mitotic cells. Most of the polyplexes after 24 hours seemed to collect in a perinuclear distribution, as depicted by arrows in a representative zoomed image (Figure 7c).
Figure 7.
Analysis of the presence of chitosan in the nucleus of cells positive for transgene expression of green fluorescent protein (GFP). (a,b) At early time points (between 18 and 26 hours) of transfection, GFP-positive (green) cells were analyzed for the presence of rhodamine B isothiocyanate-chitosan (red) in the nucleus, where the nucleus was distinguished by a reduced intensity of GFP and by disseminated intravascular coagulation (bottom left quadrant). (a) A cell representative of the vast majority of GFP-expressing cells with no intranuclear chitosan. (b) The zoomed image of the nucleus of the cell in a showing an absence of chitosan. (c) At 48 hours, a small fraction of cells (~2–5%) show some chitosan inside the nucleus, delineated here by a white border to increase contrast. Arrowheads indicate polyplexes on the edge of the nucleus, as seen in most transfected cells.
Discussion
The intracellular trafficking of chitosan-based polyplexes was investigated to identify processing events that are sensitive to MW and DDA and thus responsible for the dramatic dependence of TE on chitosan MW and DDA. We have compared the ability of different formulation to traverse the major barriers to nonviral gene transfer. In agreement with our hypothesis, we have shown by real-time FRET monitoring, that the kinetics of polyplex decondensation was the most critical formulation-dependent intracellular process that could account for the intriguing and important relationship between TE and chitosan structure under these experimental conditions. According to our results, the tailoring of chitosan MW and DDA can produce polyplexes with an optimal stability needed to achieve the correct kinetics of decondensation that are in tune with the overall intracellular trafficking of the polyplexes, specifically the long transit time in lysosomes (above 12 hours). Our findings further validate the generally accepted hypothesis, based mostly on physicochemical characterization and end-point TE, that the most efficient chitosan polyplexes have physicochemical properties that endow them with a subtle balance between polyplex stability for pDNA protection but with the ability to dissociate in coordination with lysosomal escape.3,6,7,12,13 A recent elegant study comparing the unpacking rate of chitosan (380 kd–83% DDA) to other polycations found higher rates of unpacking for chitosan than our study with about 60% unpacking at 2 hours and nearly 80% at 24 hours.2 In terms of DDA, the chitosan used was similar to the 80-40 chitosan tested here, but because of the higher MW, the stability of this chitosan should have been increased, rather than decreased as suggested by the observed fast decondensation rate. This discrepancy may be due to the higher pH (pH 7.4 versus 6.5) of transfection in Chen's study, a pH where Ma et al.14 and Strand et al.12 found a significantly reduced binding affinity as compared to pH 6.5 in our study. Notably TE is also significantly reduced at this higher pH.
The uptake and lysosomal colocalization data from this study provided important data on the mechanism of intracellular trafficking of chitosan-based polyplexes in general. We found first that uptake was not a limiting step in cell trafficking of polyplexes for conditions tested here and, second, that all polyplexes traffic intracellularly via lysosomes. When compared to other polycations such as polyethyleneimine, with vesicular escape observed within the first 4 hours after transfection30 and with significant gene expression detected as early as 5–6 hours post-transfection18,31 and peaking at 24 hours,3 the sequence of lysosomal escape at 12–24 hours (Figure 4) and maximal gene expression at 48 hours (Figure 1) seen here with chitosan–pDNA systems suggests that the initial lysosomal sequestration accounts for the late onset of gene expression, as noted elsewhere.3 It appears that lysosomal sequestration constitutes a rate-limiting step for chitosan vectors that is independent of chitosan MW and DDA, limiting the speed of gene expression to those currently observed and possibly the attained number of transfected cells to 40% of all cells.
Although vector transit through lysosomes appears to be formulation independent, the kinetics of polyplex decondensation in relation to lysosomal sequestration and escape was found to be critically dependent on chitosan structure. There may be substantial and accelerated pDNA degradation in lysosomes for chitosans with weak stability (72-40), and inhibited enzymatic digestion of polyplexes exhibiting high TE (80-40 and 92-10), which necessarily have the ability to protect pDNA up until lysosomal escape to the cytosol. This interpretation of our data highlights the importance of DNA protection against lysosomal enzymes, and hence of selecting chitosans that promote relatively stable polyplexes, but not overly stable, with the latter exemplified by chitosan 92-150, where polyplexes are still condensed after 48 hours and are inefficient gene expressors. Co-incubation with chitosanase published recently32 may potentiate lysosomal degradation and increase the TE of highly condensed and stable polyplexes. These mechanisms involving lysosomal transit and degradation are supported by earlier studies hypothesizing that the escape of chitosan from lysosomes is driven by intravesicular osmotic swelling induced by enzymatic degradation.9 Prior studies have also concluded that the enzymatic cleavage site on chitosan consists of triplets of acetylated glucosamine that would give high DDA chitosan an increased resistance to lysosomal degradation.33 Reduced enzymatic susceptibility may also play a role in the increased resistance of 92-150 to lysosomal transit and the apparent substantial degradation of 72-40. For chitosans 92-10 and 80-40, the lysosomal transit may be beneficial, which is in opposition to the generalized notion that lysosomal colocalization is to be avoided. Instead, taken together, a dual-phase mechanism of the dissociation of pDNA from chitosan can be hypothesized for efficient formulations, with a partial lysosomal degradation of the polyplex, necessary not only for escape but also to weaken the polyplex, facilitating the second phase of complete cytosolic dissociation of pDNA, thought to be mediated by competition with intracytoplasmic polyanions.34 This model is consistent with the lack of significant increase of TE of chitosan polyplexes observed in studies using agents to accelerate endosomal escape.5,7
Nonetheless, our findings involving lysosomal sequestration are in contradiction with other studies suggesting chitosan polyplexes do not transit through lysosomes at all2 or only for short periods (within 4 hours of transaction).10,35 The lack of consensus between our results and these studies could stem in part from the use of different cell types, different chitosans, and by the use of different methodologies for transfection and lysosomal staining, including different transfection pH or of the use of an acidophilic Lysotracker dye. In this context, it is important to note that inhibition of lysosomal acidification can abolish Lysotracker lysosomal staining.36 Therefore, this agent may not be a precise marker of lysosomes when loaded with polycations that provide proton buffering capacity such as polyethyleneimine16 and chitosan,37 as their accumulation may inhibit lysosomal acidification.
As stated above, it appears that binding and uptake are not limiting steps in the transfection process for HEK293 cells tested here. Thus, despite the increase in binding and uptake with increasing DDA seen here, no relationship between uptake and TE was observed, as is the case elsewhere.10,13 The use of only HEK293, known for a high endocytic activity, in the study may be viewed as a limitation, although the lack of impediment at the uptake level, possibly even upregulated by the fact that uptake and binding are aided by the two-dimensional morphology of in vitro tests,38 provides a useful model to study downstream processes. The positive correlation between DDA, binding and uptake, and the fact that no receptor specific to chitosan has been reported to date strongly suggests that the nature of the cell binding of chitosan is a nonspecific electrostatic interaction with the negatively charged cell membrane.
The last important barrier in the intracellular traffic examined here was the nuclear translocation of pDNA, and whether chitosan facilitated pDNA nuclear entry. Our result suggests nuclear uptake is not facilitated by chitosan but that nuclear uptake is rather more simply a function of the availability of cytosolic pDNA, resulting from formulation-dependent processes upstream of nuclear localization. Once the pDNA is released to the cytosol, the SV40 enhancer sequence of the plasmid, known to increase nuclear translocation of plasmids in the absence of cell division, would allow nuclear entry of the pDNA independent of vector design.39 The perinuclear distribution of chitosan observed here is similar to previous observations with other polycations and supports lysosomal transit,22 as passive diffusion of pDNA or other macromolecules toward the nucleus40 does not occur and significant nuclease degradation is observed within 1 hour postmicroinjection.41 Rather, perinuclear distribution of gene vectors has been attributed to a dynein-driven movement of endolysosomal vesicles along microtubules from the cell periphery to the microtubule-orienting center near the nucleus.42
The study presented here complements existing methodologies for the study of the intracellular pharmacokinetics of polyplexes, including the use of confocal-based analytical modeling,1,2 electron microscopy,3 and particle tracking.43 The imaging tools allowed us to identify both chitosan structure-dependent and chitosan structure-independent rate-limiting barriers, adding to the knowledge of the complex transfection mechanisms of chitosan-based polyplexes. The importance of understanding the mechanisms of intracellular trafficking was highlighted as they lead to detailed structure–activity relationships and the rational design of optimal gene delivery vectors.
Materials and Methods
Preparation of chitosan/pDNA polyplexes. Specific chitosans were chosen based on previously described DNA-binding properties14 and TE.6 These considerations led to the choice of two chitosans (80-40 and 92-10) with high TE and with an expected intermediate stability (not too high, not too low) and two other chitosans with low TE and expected stability properties that were either too high (high DDA and MW = 92-150) or too low (low DDA and MW = 72-40) to be effective. These four chitosans with specific DDA–MW were prepared from chitosans (Biosyntech, Laval, Quebec, Canada) with specific starting DDA characterized by 1H NMR (ref. 44) and subsequent nitrous acid depolymerization and characterization by gel permeation chromatography.6 Chitosans were labeled with fluorescent RITC (Sigma) to 0.9–1% degree of substitution as described in Ma et al.45 The plasmid eGFPLuc (Clonetech Labs, Mountain View, CA) of 6.4 kb encodes for a fusion of enhanced green fluorescent protein and luciferase driven by a human cytomegalovirus promoter. When indicated, pDNA was either labeled with Cy 5 or fluorescein LabelIt (Mirus, Madison, WI) according to Mirus's protocol at a volume/mass ratio of 0.25/1 for a final labeling concentration of 1 fluorophore unit for every 220 or 160 base pairs for Cy 5 and fluorescein, respectively.
Polyplexes were prepared according to Lavertu et al.6 Briefly, depolymerized chitosans (92-10, 80-40, 72-40, and 92-150) were dissolved at 0.5% (wt/vol) in hydrochloric acid and then diluted to reach the amine (deacetylated groups) to phosphate (on DNA) ratio of 5 (N:P ratio of 5). Chitosan solutions were then added to an equal volume of 330 µg/ml solution of pDNA and allowed to form polyplexes for 30 minutes. Size and zeta potentials of polyplexes prepared with all four chitosans (92-10, 80-40, 72-40, and 92-150) used in this study were previously characterized.6 We repeated these measurements (Zeta Sizer Nano; Malvern Instruments, Malvern, UK) with either labeled DNA or labeled chitosan or with unlabeled polyplexes and observed no significant differences in size or zeta potential compared to the those obtained in Lavertu et al.6 and between labeled and unlabeled polyplexes (data not shown).
In vitro transfection of HEK293. HEK293 cells (ATCC, Manassas, VA) were cultured in complete media (Dulbecco's modified Eagle medium high glucose supplemented with 10% fetal bovine serum; Invitrogen, Carlsbad, CA) in a 5% CO2 37 °C incubator. For transfection, cells were plated in 24-well culture plates at a density of 50,000 cells/well in complete medium (Dulbecco's modified Eagle medium high glucose, 10% fetal bovine serum, pH 7.4) for 24 hours before transfection in modified transfection media [Dulbecco's modified Eagle medium high glucose, 2-(N-morpholino) ethanesulfonic acid (Sigma), 10% serum, pH 6.5]. For all imaging performed in this study, cells were seeded 24 hours before transfection in 35-mm glass bottom culture dishes (MatTek, Ashland, MA) using 500 µl of complete medium at 50,000 cells/dish. Chitosan–pDNA polyplexes were incubated with cells at a concentration of 2.5 µg pDNA/wells in media containing 10% serum at pH 6.5 for 24 hours, or until analysis for earlier time points, depending on the experiment. For colocalization with lysosomes and in FRET studies, cells were incubated with polyplexes for 8 hours and media was replaced with polyplex-free transfection media. For time points longer than 24 hours, transfection media was replaced at 24 hours with chitosan-free standard cell culture media containing 10% serum at pH 7.4 until analysis, usually at 48 hours unless otherwise noted. Transfection efficiencies (% cells) and transgene expression levels (luciferase normalized to protein content determined by bicinchoninic acid) were quantitatively assessed by flow cytometry and luciferase assay, respectively, as described previously.6 All experiments were done in duplicates, and with a minimum of three independent experiments that were averaged to represent final results.
Cellular binding and uptake of chitosan-based polyplexes. Cells were transfected as described above with polyplexes prepared with one of four different RITC-labeled chitosans and uptake/binding was analyzed by FACS (MOFLO BTS, Cytomation; Beckman Coulter, Fullerton, CA). Additional experiments were performed with polyplexes prepared with unlabeled chitosans but with Cy5-labeled pDNA (Supplementary Figure S1a). After predetermined incubation times at 37 °C in the presence of fluorescent polyplexes, cells were washed twice with phosphate-buffered saline, treated with trypsin–EDTA (Invitrogen) for 10 minutes to detach cells and to remove membrane-bound polyplexes and then washed two times by centrifugation and resuspension in phosphate-buffered saline. Effective removal of cell-bound polyplexes for uptake analysis was validated by confocal microscopy (data not shown) and by FACS showing that further treatment with heparin, chitosanase, or pronase had no effect on the fluorescence of RITC-chitosan treated cells (data not shown). The level of membrane-bound polyplexes at each time point was also determined by subtracting the mean fluorescence of internalized polyplexes, as determined with trypsinized cells, from the total fluorescence obtained from cells detached with nonenzymatic cell dissociation buffer (Invitrogen), which contains both internalized and cell-bound polyplexes. Cells were directly analyzed by FACS by double gating against noncell events (residues) and on control nontransfected cells for autofluorescence with a minimum of 20,000 detected events per sample. Data are presented as % cells with internalized (or associated) polyplexes, whereas the level of uptake is presented as mean fluorescence per cell. Samples were prepared in duplicate and all experiments were performed three times. Qualitative assessment of the cellular uptake was made with live-cell confocal microscopy as described in Supplementary Materials and Methods.
Imaging and analysis of the intracellular trafficking of polyplexes. Lysosomes were labeled by a pulse-chase method similar to that previously published.25 Cells were pulse-loaded with fluorescently labeled dextran (10,000 d MW, 5 µg/ml, Rhodamine green or Alexa 646; Invitrogen) in complete media for 18 hours, washed twice with phosphate-buffered saline, chased with dextran-free complete media for 4–6 hours, and then transfected with labeled polyplexes. The live-cell configuration presented throughout this study was chosen because fixation has been shown to cause artifactual internalization to both the cytoplasm and the nucleus of cell-binding cationic molecules.46,47
Colocalization was first assessed qualitatively by the occurrence of yellow pixels48 resulting from the spatial overlap of green (polyplex pseudocolor) and red pixels (lysosomes pseudocolor) from two separate channels. Colocalization between all four polyplexes and lysosomes in selected regions of interest corresponding to whole cells from images taken at 4, 12, 24, and 48 hours was then quantified in terms of the colocalization coefficient. The colocalization coefficient, defined as the sum of intensities of colocalizing pixels from the “RITC-chitosan” and “fluorescent dextran-lysosomes” channels compared with the overall sum of pixel intensities (above threshold after background substraction), was calculated using the LSM 3.2 Mander's colocalization coefficient macro (Zeiss, Jena, Germany) and plotted over time. A minimum of 15 cells with at least three optical sections per chitosan for each time point were analyzed in three independent experiments. Details of the imaging methodology are described in Supplementary Materials and Methods.
Imaging and analysis of polyplex decondensation kinetics using FRET. Intracellular polyplex stability was assessed by monitoring the unpacking or decondensation kinetics by FRET using the probes described in Supplementary Materials and Methods. Because the energy transfer process only occurs over distances ranging from 10 to 100 Å, FRET provides an accurate measure of the polyplex condensation state. FRET was demonstrated by sensitized emission, a method that selectively images the FRET-induced emission of the acceptor upon excitation of the donor. Sensitized emission FRET is nondestructive, lends well to time lapse imaging of live cells, and is ideal for intermolecular FRET.28,49 The imaging system for FRET was as described above except for FRET-specific settings in the channel configurations. For each time point, images were captured using three channels: (i) FRET channel (excitation 514 nm, dichroic 660 longpass, emission 680 longpass META), (ii) donor RITC channel (excitation 514 nm, dichroic 565 nm longpass, acceptor excitation/emission), and (iii) acceptor Cy5 channel (excitation 633 nm, dichroic 660 longpass, emission 680 longpass META) for the detection of FRET emission (FRETem), RITC-Chitosan emission (RITC-Chitoem), and Cy5 DNA emission (Cy5 DNAem), respectively. We have simultaneously quantified the FRET intensity of polyplexes and the ratio of complexed DNA to uncomplexed DNA over time with FRET measurements performed in background-subtracted regions of interest corresponding to individual cells. FRET pixel intensities (FRETem) in the regions of interest from channel 1 (FRET channel: RITC excitation and Cy5 emission) were normalized to the acceptor emission (Cy5-DNAem) from channel 3 (DNA-Cy5 channel: DNA Cy5 excitation, Cy5 emission), measured after specific excitation of the acceptor according to Eq. (1).28,29,50 This FRET ratio defines the apparent energy transfer efficiency of the polyplexes in each regions of interest corresponding to a cell and is also proportional to the fraction of pDNA molecules bound to chitosan.
Correction factors α and β are used in sensitized FRET to account for crosstalk of fluorophores into the FRET channel determined by incubating cells with polyplexes labeled with only Cy5 or only RITC and measuring the contribution of both fluorophores to the FRET measurement (excitation 514 nm, dichroic LP 660, emission 680 LP META). They were determined to be α = 6.9% (for RITC bleedthrough into acceptor channel) and β = 2.9% (for Cy5 crosstalk by donor excitation). FRET ratios were measured for each polyplex at each time point and normalized to the FRET ratio obtained with chitosan 92-150 at 4 hours, which represented the highest FRET signal intensity corresponding to the most condensed state and essentially 100% complexation of DNA to chitosan. Although less condensed, all formulations showed near 100% complexation at 4 hours. The FRET signal decrease in intensity relative to this reference point was then calculated and plotted versus time.
Characterization of nuclear localization of chitosan in GFP-positive cells. The presence or absence of nuclear translocation of chitosan in GFP-positive cells was determined by imaging GFP-expressing cells for each chitosan at early time points in the transfection kinetics (between 18 and 26 hours) for cells that have likely not divided and then at 48 hours to image cells that may have possibly divided. Nuclei peripheries were visualized in differential interference contrast to determine whether chitosans entered the nucleus along with plasmids.
SUPPLEMENTARY MATERIAL Figure S1. Comparison of uptake of polyplexes prepared with labeled DNA and labeled chitosan. (a) Cellular uptake by FACS with pDNA-chitosan (92-10) polyplexes prepared with either labeled DNA or labeled chitosan were similar. (b) Uptake was also qualitatively assessed at 8 hrs by confocal microscopy on live cells transfected with polyplexes prepared with fluorescein-DNA (green) and RITC-chitosan 92-10 (red). Images are for focal planes passing through cell bodies where cell membranes were stained prior to imaging with cell mask (blue) to clearly differentiate internalized polyplexes from surface bound polyplexes. Each image also shows a DIC image in grey and an image merging all 4 channels (bottom right), where yellow indicates overlap between DNA and chitosan. After 8hrs incubation time, there is a final homogenous distribution for both DNA and chitosan consistent with FACS results, where all cells have taken up polyplexes. All DNA appear complexed (yellow overlap) with chitosan although only FRET can determine molecular interaction. Scale bar = 10 um. Figure S2. FRET probe characterization. (a)Fluorescent intensities of chitosan (80-40 in this case) labeled with rhodamine (donor) complexed to Cy5-labeled (acceptor) pDNA were measured in buffers of different pH. No significant variations were observed for either probe in the range of pH pertinent to intracellular studies. (b-d) The FRET signal generated by excitation at 514nm upon addition of heparin was also assessed by spectrophotometry (b) and by microscopy (c,d). (b) The addition of heparin caused an unpacking of the complexes and a decrease in FRET signal intensity, as expected. (c-d) Complexes were formed using labeled RITC-chitosan and Cy5-pDNA and imaged before (c) and 1 hour after the addition of heparin (d). Images in c) show polyplexes with FRET signal and no free pDNA since 633 excitation light (DNA 633 in c) illuminates only FRET generating particles (FRET in c) compared to polyplexes in the presence of heparin (DNA 633 in d) with no FRET but rather free DNA dissociated from chitosan, thus confirming heparin-induced polyplexe dissociation. (a,b) Results are the average of two independent experiments ± standard deviation where each experiment included 2 replicates. Materials and Methods.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research, the Natural Sciences and Engineering Research Council, le Fond de la recherche en santé du Québec, and the Canada Foundation for Innovation.
Supplementary Material
Comparison of uptake of polyplexes prepared with labeled DNA and labeled chitosan. (a) Cellular uptake by FACS with pDNA-chitosan (92-10) polyplexes prepared with either labeled DNA or labeled chitosan were similar. (b) Uptake was also qualitatively assessed at 8 hrs by confocal microscopy on live cells transfected with polyplexes prepared with fluorescein-DNA (green) and RITC-chitosan 92-10 (red). Images are for focal planes passing through cell bodies where cell membranes were stained prior to imaging with cell mask (blue) to clearly differentiate internalized polyplexes from surface bound polyplexes. Each image also shows a DIC image in grey and an image merging all 4 channels (bottom right), where yellow indicates overlap between DNA and chitosan. After 8hrs incubation time, there is a final homogenous distribution for both DNA and chitosan consistent with FACS results, where all cells have taken up polyplexes. All DNA appear complexed (yellow overlap) with chitosan although only FRET can determine molecular interaction. Scale bar = 10 um.
FRET probe characterization. (a)Fluorescent intensities of chitosan (80-40 in this case) labeled with rhodamine (donor) complexed to Cy5-labeled (acceptor) pDNA were measured in buffers of different pH. No significant variations were observed for either probe in the range of pH pertinent to intracellular studies. (b-d) The FRET signal generated by excitation at 514nm upon addition of heparin was also assessed by spectrophotometry (b) and by microscopy (c,d). (b) The addition of heparin caused an unpacking of the complexes and a decrease in FRET signal intensity, as expected. (c-d) Complexes were formed using labeled RITC-chitosan and Cy5-pDNA and imaged before (c) and 1 hour after the addition of heparin (d). Images in c) show polyplexes with FRET signal and no free pDNA since 633 excitation light (DNA 633 in c) illuminates only FRET generating particles (FRET in c) compared to polyplexes in the presence of heparin (DNA 633 in d) with no FRET but rather free DNA dissociated from chitosan, thus confirming heparin-induced polyplexe dissociation. (a,b) Results are the average of two independent experiments ± standard deviation where each experiment included 2 replicates.
REFERENCES
- Akita H, Ito R, Khalil IA, Futaki S., and, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH., and, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köping-Höggård M, Tubulekas I, Guan H, Edwards K, Nilsson M, Vårum KM, et al. Chitosan as a nonviral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo. Gene Ther. 2001;8:1108–1121. doi: 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- Roy K. Chitosan-DNA Nanoparticles: Synthesis, Characterization, Subcellular Transport and Oral Delivery of Genetic Vaccines. Johns Hopkins University: Baltimore, MD; 1999. [Google Scholar]
- Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, et al. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- Lavertu M, Méthot S, Tran-Khanh N., and, Buschmann MD. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials. 2006;27:4815–4824. doi: 10.1016/j.biomaterials.2006.04.029. [DOI] [PubMed] [Google Scholar]
- MacLaughlin FC, Mumper RJ, Wang J, Tagliaferri JM, Gill I, Hinchcliffe M, et al. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release. 1998;56:259–272. doi: 10.1016/s0168-3659(98)00097-2. [DOI] [PubMed] [Google Scholar]
- Richardson SC, Kolbe HV., and, Duncan R. Potential of low molecular mass chitosan as a DNA delivery system: biocompatibility, body distribution and ability to complex and protect DNA. Int J Pharm. 1999;178:231–243. doi: 10.1016/s0378-5173(98)00378-0. [DOI] [PubMed] [Google Scholar]
- Köping-Höggård M, Vårum KM, Issa M, Danielsen S, Christensen BE, Stokke BT, et al. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11:1441–1452. doi: 10.1038/sj.gt.3302312. [DOI] [PubMed] [Google Scholar]
- Huang M, Fong CW, Khor E., and, Lim LY. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J Control Release. 2005;106:391–406. doi: 10.1016/j.jconrel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Schaffer DV, Fidelman NA, Dan N., and, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Strand SP, Danielsen S, Christensen BE., and, Vårum KM. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules. 2005;6:3357–3366. doi: 10.1021/bm0503726. [DOI] [PubMed] [Google Scholar]
- Strand SP, Lelu S, Reitan NK, de Lange Davies C, Artursson P., and, Vårum KM. Molecular design of chitosan gene delivery systems with an optimized balance between polyplex stability and polyplex unpacking. Biomaterials. 2010;31:975–987. doi: 10.1016/j.biomaterials.2009.09.102. [DOI] [PubMed] [Google Scholar]
- Ma PL, Lavertu M, Winnik FM., and, Buschmann MD. New insights into chitosan-DNA interactions using isothermal titration microcalorimetry. Biomacromolecules. 2009;10:1490–1499. doi: 10.1021/bm900097s. [DOI] [PubMed] [Google Scholar]
- Köping-Höggård M, Mel'nikova YS, Vårum KM, Lindman B., and, Artursson P. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in vitro and in vivo. J Gene Med. 2003;5:130–141. doi: 10.1002/jgm.327. [DOI] [PubMed] [Google Scholar]
- Akinc A, Thomas M, Klibanov AM., and, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- Kulkarni RP, Mishra S, Fraser SE., and, Davis ME. Single cell kinetics of intracellular, nonviral, nucleic acid delivery vehicle acidification and trafficking. Bioconjug Chem. 2005;16:986–994. doi: 10.1021/bc050081u. [DOI] [PubMed] [Google Scholar]
- Itaka K, Harada A, Yamasaki Y, Nakamura K, Kawaguchi H., and, Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J Gene Med. 2004;6:76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- Hama S, Akita H, Iida S, Mizuguchi H., and, Harashima H. Quantitative and mechanism-based investigation of post-nuclear delivery events between adenovirus and lipoplex. Nucleic Acids Res. 2007;35:1533–1543. doi: 10.1093/nar/gkl1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YP, Chen HH, Leong KW., and, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J Control Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH., and, Leong KW. Simultaneous non-invasive analysis of DNA condensation and stability by two-step QD-FRET. Nano Today. 2009;4:125–134. doi: 10.1016/j.nantod.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielson NP., and, Pack DW. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules. 2006;7:2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- Nimesh S, Thibault MM, Lavertu M., and, Buschmann MD. Mol Biotechnol. epub ahead of print; 2010. Enhanced gene delivery mediated by low molecular weight chitosan/DNA complexes: effect of pH and serum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PL, Winnik FM., and, Buschmann MD.Simultaneous determination of unbound polycation and particle size of DNA/chitosan complexes by asymmetrical flow field-flow fractionation Biomacromoleculesin press.
- Bright NA, Gratian MJ., and, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Harada M, Kawaguchi T, Kumemura H, Terada K, Ninomiya H, Taniguchi E, et al. The Wilson disease protein ATP7B resides in the late endosomes with Rab7 and the Niemann-Pick C1 protein. Am J Pathol. 2005;166:499–510. doi: 10.1016/S0002-9440(10)62272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves DC, Liebelt DA, Lakshmanan V, Roepe PD, Fidock DA., and, Akabas MH. Chloroquine-resistant isoforms of the Plasmodium falciparum chloroquine resistance transporter acidify lysosomal pH in HEK293 cells more than chloroquine-sensitive isoforms. Mol Biochem Parasitol. 2006;150:288–299. doi: 10.1016/j.molbiopara.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe A, Christensen K., and, Swanson JA. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys J. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., and, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichler A, Leborgne C, Coeytaux E., and, Danos O. Polyethylenimine-mediated gene delivery: a mechanistic study. J Gene Med. 2001;3:135–144. doi: 10.1002/jgm.173. [DOI] [PubMed] [Google Scholar]
- Zanta MA, Belguise-Valladier P., and, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci USA. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DC, Liu WG, Zuo AJ, Sun SJ, Cheng N, Guo G, et al. Pre-deliver chitosanase to cells: a novel strategy to improve gene expression by endocellular degradation-induced vector unpacking. Int J Pharm. 2006;314:63–71. doi: 10.1016/j.ijpharm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Nordtveit RJ, Varum KM., and, Smidsrod O. Degradation of partially N-acetylated chitosans with hen egg white and human lysozyme. Carbohydr Polym. 1996;29:163–167. [Google Scholar]
- Labat-Moleur F, Steffan AM, Brisson C, Perron H, Feugeas O, Furstenberger P, et al. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- Kiang T, Bright C, Cheung CY, Stayton PS, Hoffman AS., and, Leong KW. Formulation of chitosan-DNA nanoparticles with poly(propyl acrylic acid) enhances gene expression. J Biomater Sci Polym Ed. 2004;15:1405–1421. doi: 10.1163/1568562042368112. [DOI] [PubMed] [Google Scholar]
- Steen M, Kirchberger T., and, Guse AH. NAADP mobilizes calcium from the endoplasmic reticular Ca(2+) store in T-lymphocytes. J Biol Chem. 2007;282:18864–18871. doi: 10.1074/jbc.M610925200. [DOI] [PubMed] [Google Scholar]
- Ishii T, Okahata Y., and, Sato T. Mechanism of cell transfection with plasmid/chitosan complexes. Biochim Biophys Acta. 2001;1514:51–64. doi: 10.1016/s0005-2736(01)00362-5. [DOI] [PubMed] [Google Scholar]
- Schaffert D., and, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- Dowty ME, Williams P, Zhang G, Hagstrom JE., and, Wolff JA. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc Natl Acad Sci USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, et al. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- Suh J, Wirtz D., and, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci USA. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavertu M, Xia Z, Serreqi AN, Berrada M, Rodrigues A, Wang D, et al. A validated 1H NMR method for the determination of the degree of deacetylation of chitosan. J Pharm Biomed Anal. 2003;32:1149–1158. doi: 10.1016/s0731-7085(03)00155-9. [DOI] [PubMed] [Google Scholar]
- Ma O, Lavertu M, Sun J, Nguyen S, Buschmann MD, Winnik FM, et al. Precise derivatization of structurally distinct chitosans with rhodamine B isothiocyanate. Carbohydr Polymers. 2008;72:616–624. [Google Scholar]
- Lundberg M., and, Johansson M. Is VP22 nuclear homing an artifact. Nat Biotechnol. 2001;19:713–714. doi: 10.1038/90741. [DOI] [PubMed] [Google Scholar]
- Kaplan IM, Wadia JS., and, Dowdy SF. Cationic TAT peptide transduction domain enters cells by macropinocytosis. J Control Release. 2005;102:247–253. doi: 10.1016/j.jconrel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Bolte S., and, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Elder AD, Domin A, Schierle GSK, Lindon C, Pines J, Esposito A, et al. A quantitative protocol for dynamic measurements of protein interactions by Forster resonance energy transfer-sensitized fluorescence emission. J Royal Soc Interface. 2009;6:S59–S81. [Google Scholar]
- Gordon GW, Berry G, Liang XH, Levine B., and, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of uptake of polyplexes prepared with labeled DNA and labeled chitosan. (a) Cellular uptake by FACS with pDNA-chitosan (92-10) polyplexes prepared with either labeled DNA or labeled chitosan were similar. (b) Uptake was also qualitatively assessed at 8 hrs by confocal microscopy on live cells transfected with polyplexes prepared with fluorescein-DNA (green) and RITC-chitosan 92-10 (red). Images are for focal planes passing through cell bodies where cell membranes were stained prior to imaging with cell mask (blue) to clearly differentiate internalized polyplexes from surface bound polyplexes. Each image also shows a DIC image in grey and an image merging all 4 channels (bottom right), where yellow indicates overlap between DNA and chitosan. After 8hrs incubation time, there is a final homogenous distribution for both DNA and chitosan consistent with FACS results, where all cells have taken up polyplexes. All DNA appear complexed (yellow overlap) with chitosan although only FRET can determine molecular interaction. Scale bar = 10 um.
FRET probe characterization. (a)Fluorescent intensities of chitosan (80-40 in this case) labeled with rhodamine (donor) complexed to Cy5-labeled (acceptor) pDNA were measured in buffers of different pH. No significant variations were observed for either probe in the range of pH pertinent to intracellular studies. (b-d) The FRET signal generated by excitation at 514nm upon addition of heparin was also assessed by spectrophotometry (b) and by microscopy (c,d). (b) The addition of heparin caused an unpacking of the complexes and a decrease in FRET signal intensity, as expected. (c-d) Complexes were formed using labeled RITC-chitosan and Cy5-pDNA and imaged before (c) and 1 hour after the addition of heparin (d). Images in c) show polyplexes with FRET signal and no free pDNA since 633 excitation light (DNA 633 in c) illuminates only FRET generating particles (FRET in c) compared to polyplexes in the presence of heparin (DNA 633 in d) with no FRET but rather free DNA dissociated from chitosan, thus confirming heparin-induced polyplexe dissociation. (a,b) Results are the average of two independent experiments ± standard deviation where each experiment included 2 replicates.