Abstract
Retroviral vectors have been used to treat patients with the X-linked severe combined immunodeficiency disease and chronic granulomatous disease. In both cases, success has been undermined by clonal expansion of transduced cells in some patients due to insertional mutagenesis induced by random vector integration. This outcome underscores the importance of designing vectors for site-specific gene insertion to avoid unanticipated gene disruption or gene activation. In the present study, we incorporated the sequence-specific Cre protein into lentiviral virions. We demonstrated that the virion-associated Cre protein remained enzymatically active and was capable of directing site-specific insertion of a gene in the vector into a defined loxP site in the host genome. As there are loxP-like sequences throughout human genome that can be recognized by either wild-type Cre or Cre variants, our study demonstrates a new strategy of designing lentiviral-based vector for gene targeting.
Introduction
Retroviral vectors have been used to treat patients with the X-linked severe combined immunodeficiency disease and chronic granulomatous disease. In both cases, success has been undermined by clonal expansion of transduced cells in some patients,1,2,3 and activation of cellular proto-oncogenes by random vector integration most likely contributed to the clonal expansion. The bias of retrovirus to integrate near transcription start sites of active genes and their preferential integration near a subset of cellular genes regulating cell cycle and apoptosis may account for the high incidence of insertional mutagenesis.4,5 In contrast, lentivirus has reduced capacity to induce genotoxicity in transduced cells. Like retrovirus, lentivirus also integrates preferentially near active genes.4,6 However, lentivirus integration does not exhibit bias toward transcription start sites or preferential integration into a subset of cellular genes. The cis-regulatory elements in lentiviral long-terminal repeats (LTRs) were also removed to generate the so-called self-inactivating vector, further reducing their risk to generate genotoxicity.7 However, in a recent β-thalassemia gene therapy trial, clonal expansion was detected in a patient infused with hematopoietic stem cells transduced with a lentiviral vector (http://www.pei.de/cln_109/nn_154420/EN/infos-en). The dominant clone demonstrated elevated expression of high-mobility group A2 proteins which might result from lentiviral integration nearby.8 High-mobility group A2 proteins expression is altered in some malignant tumors,9,10,11 and rearrangement of this gene has been reported in hematological malignancies and myelodysplastic syndromes.12 Thus, the risk of insertional mutagenesis cannot be completely avoided even with lentiviral vectors. These studies underscore the importance to develop alternative strategies to minimize the risk of vector-induced genotoxicity in gene therapy.
Insertion of a gene into a predetermined “safe” genomic locus avoids the problem of genotoxicity. Site-specific gene insertion by homologous recombination is a very useful but typically inefficient technique: gene insertion into a desired locus typically occurs in about 1 out of every 106 cells treated. Retrovirus can deliver genes at extremely high efficiency but its integration is not site-specific. Modulating retroviral integration, mainly through the fusion of the retroviral integrase with sequence-specific DNA-binding proteins, represents an attractive strategy for site-specific gene insertion.13,14,15,16 However, such a strategy is limited by reduced DNA-binding specificity of the fusion protein and the difficulty to incorporate the fusion protein into infectious virions. The use of site-specific recombinases such as Cre has been shown to significantly enhance the efficiency of gene targeting in mammalian cells.17,18 Cre, a 38-kd recombinase from bacteriophage P1, utilizes its endonuclease activity to catalyze recombination between two identical loxP sites. The enzyme requires no accessory proteins or cofactors for its reaction and functions efficiently in vitro and under a wide variety of cellular conditions. Furthermore, natural occurring loxP sites pre-exist in human genome and can potentially serve as targets for site-specific gene insertion.19 Although some of these so-called “pseudo”-loxP sites diverge significantly in sequences from the native loxP site, Cre nevertheless binds to these sequences and mediates recombination. In the present study, we evaluate the feasibility of combining the highly efficient Cre-loxP system for gene targeting and the extremely versatile gene delivery system of lentivirus for site-specific gene insertion in human cells. We generated three components to evaluate this strategy: (i) a cell line containing a single loxP site in the genome to serve as the target for gene insertion; (ii) a targeting lentiviral vector containing the gene of interest adjacent to a loxP site; (iii) a modified Cre protein that was incorporated into the lentivirial virion through fusion with the viral protein R (Vpr) protein of human immunodeficiency virus-1 (HIV-1). Vpr is a 96-amino acid virion-associated protein that regulates nuclear import of the HIV-1 preintegration complex.20,21 We hypothesized that close association of the Cre protein with the targeting vector genome containing the gene of interest in the virion improved the probability for site-specific gene insertion. Our result shows that virion-associated Cre proteins mediated the insertion of the gene of interest from the vector into the loxP site present in the host genome. Our work provides supports for using lentiviral vectors to deliver a therapeutic gene into potentially “safe” loci in the host genome for human gene therapy.
Results
Establishment of a cell line containing the target loxP site for gene insertion
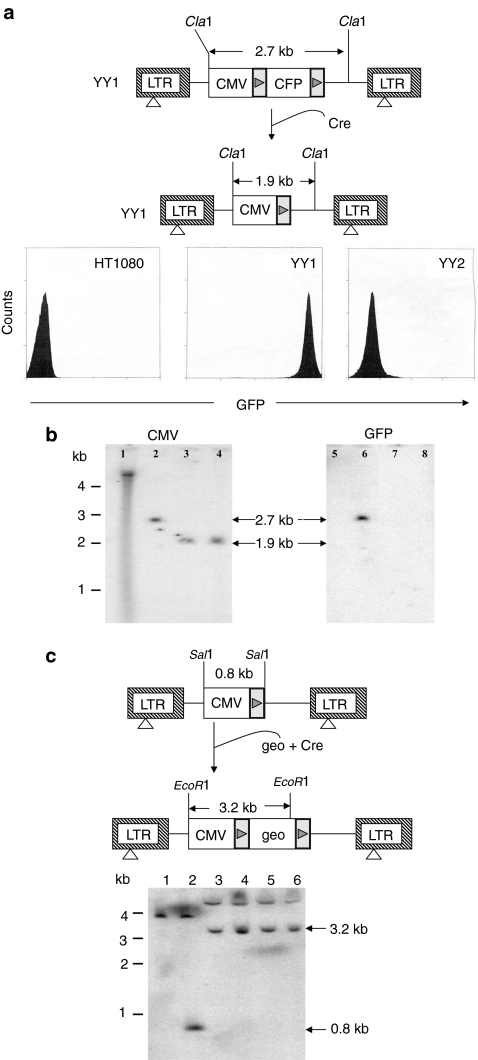
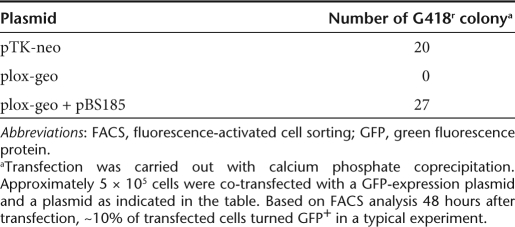
To evaluate the capacity of lentiviral vectors for site-specific gene insertion, we first established a cell line containing the target loxP site. Human fibrosarcoma HT1080 cells were transduced with CCGLΔEn, a retroviral vector containing the gene encoding green fluorescence protein (GFP) controlled by the promoter of cytomegalovirus (CMV) immediate early (IE) gene (Figure 1a). The GFP gene is flanked by loxP sites and can readily be removed by Cre, resulting in the generation of a single loxP site immediately adjacent to the CMV promoter. A stable clone, YY1, was isolated for high GFP expression by fluorescence-activated cell sorting (FACS) (Figure 1a). Southern blot analysis of the YY1 genomic DNA showed single-copy integration of CCGLΔEn (data not shown). To create the target loxP site for gene insertion, YY1 cells were transiently transfected with cre-expression plasmid pBS18518 that mediated recombination between the two loxP sites in YY1 and removed the GFP gene. GFP− clones were isolated and Southern blot analysis showed the expected digestion patterns upon the removal of the GFP gene in two of such clones (Figure 1b). One of the clones, named YY2, which showed background GFP expression was picked for subsequent studies (Figure 1a). To determine whether the single loxP site in YY2 cells is suitable for site-specific gene insertion, we constructed plox-geo containing the geo gene encoding a neomycin phosphotransferase-β-galactosidase (β-gal) fusion protein flanked by two loxP sites. This plasmid contains neither a promoter nor a polyadenylation signal, reducing the likelihood of generating G418-resistant colonies from random insertion into the host genome. YY2 cells were transiently transfected with plox-geo with or without pBS185 and G418-resistant colonies were scored (Table 1). Transfection with plox-geo alone failed to generate any colony whereas co-transfection with pBS185 generated G418-resistant colonies at similar levels as the positive control, pTK-neo, containing the gene encoding neomycin phosphotransferase (neo) controlled by the herpes thymidine kinase (TK) promoter. To confirm site-specific insertion of the geo gene, Southern blot analysis of the genomic DNA from four randomly picked G418-resistant colonies was carried out. As shown in Figure 1c, EcoR1 digestion exhibited the expected 3.2-kb fragment containing the CMV promoter and the geo gene. These results suggested that the single loxP site in YY2 cells could serve as a target for Cre-mediated gene insertion.
Figure 1.
Establishment of the YY2 cell line for site-specific gene insertion. (a) To establish the target cell line for site-specific gene insertion, HT1080 cells were transduced with the retroviral vector, CCGLΔEn, containing a loxP-flanked GFP gene controlled by the CMV IE promoter. A cell line, YY1, with high levels of GFP expression was established. To create a single loxP site as the insertion target, YY1 cells were transiently transfected with cre-expression plasmid pBS185. A cell line, YY2, that lost GFP expression was established. The expected fragment size of the proviral DNA in YY1 and YY2 cells digested with Cla1 was also shown. FACS of GFP expression in both cell lines was shown on the bottom. (b) Southern blot analysis of Cla1-digested genomic DNA from YY1 and two GFP− clones (YY2 and YY3) derived from pBS185-transfected YY1 cells. The probe used was shown on top of each gel. Lanes 1 and 5: HT1080; lanes 2 and 6: YY1; lanes 3 and 7: YY2; lanes 4 and 8: YY3. (c) Southern blot analysis of the genomic DNA of YY2 cells containing the gene insertion event. The restriction enzyme used and expected fragment size before and after geo gene insertion into YY2 cells were shown. The blot was hybridized with a CMV IE promoter-specific probe. Lane 1: HT1080 digested with Sal1; lane 2: YY2 digested with Sal1; lane 3–6: YY2-derived clones containing site-specific insertion of the geo gene digested with EcoR1. CMV, cytomegalovirus; FACS, fluorescence-activated cell sorting; GFP, green fluorescence protein; IE, immediate early; LTR, long-terminal repeat.
Table 1. Site-specific insertion of the geo gene into the YY2 genome.
Incorporation of Cre into lentiviral virions
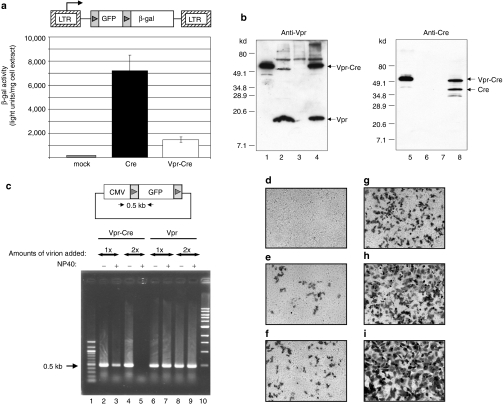
We next determined whether the Cre protein could get incorporated into lentiviral virions by constructing a vpr-cre gene encoding the entire Cre protein fused to the C-terminus of Vpr. To determine whether such a fusion disrupts the Cre recombinase activity, we co-transfected 293T cells with pCMV-vpr-cre, an expression plasmid for Vpr-Cre, and a retroviral reporter plasmid, pCG-β-gal containing the β-gal gene preceded by a loxP-flanked GFP gene (Figure 2a). Removing the GFP gene by Cre would activate β-gal expression. As shown in Figure 2a, Vpr-Cre was able to activate β-gal expression, albeit at a reduced level than wild-type Cre. Thus, Vpr-Cre fusion somewhat compromised the Cre recombinase activity although a fraction of the activity was still retained. To determine whether Vpr-Cre could be incorporated into virions, lentiviral vectors were generated from 293T cells co-transfected with pCMV-vpr-cre and pCMV-HIV-1Δvpr, the packaging plasmid for lentiviral vectors. The vpr gene in pCMV-HIV-1Δvpr was inactivated to avoid competition between wild-type Vpr and Vpr-Cre for packaging. Harvested cell extracts or virions were analyzed by western blotting. When a Vpr-specific antibody was used, the cell lysate from the co-transfection exhibited a 50-kd band which was the expected size for Vpr-Cre (Figure 2b, lane 1). This band was absent from virions derived from pCMV-HIV-1Δvpr alone (lane 3). Virions derived from pCMV-HIV-1,22 the packaging plasmid with a functional vpr gene, exhibited the expected wild-type Vpr band (lane 2). Interestingly, virions derived from co-transfection with pCMV-vpr-cre and pCMV-HIV-1Δvpr showed two reacting bands (lane 4). The upper band corresponded in size to Vpr-Cre whereas the lower band was consistent with Vpr. Because the lower band, presumably Vpr, was absent from the total cell lysate (compare lanes 1 and 4), it was possible that Vpr observed in the harvested virions was actually derived from cleavage of Vpr-Cre during or after encapsidation of the fusion protein into virions. The protease that mediated this cleavage remained unclear. Consistent with this hypothesis, a 38-kd protein corresponding to the size of cleaved Cre was detected when the same blot was reprobed with a Cre-specific antibody (lane 8). These results suggested that Vpr-Cre could be efficiently incorporated into virions although partial cleavage of the virion-associated Vpr-Cre occurred after packaging.
Figure 2.
The recombinase activity of Vpr-Cre and its incorporation into HIV virions. (a) The recombinase activity of Vpr-Cre. 293T cells were co-transfected with pCMV-cre or pCMV-vpr-cre as indicated with pCG-β-gal, a retroviral vector containing a loxP-flanked GFP gene preceding the β-gal gene. The arrow indicates the direction of transcription from the 5′ LTR and the arrowheads represent the loxP sites. Cell lysates were prepared 40 hours after transfection, and the β-gal activity was determined and normalized to the total protein used in each reaction. (b) Western blot analysis of Vpr-Cre incorporation into HIV virions. 293T cells were co-transfected by pCMV-vpr-cre with either pCMV-HIV-1 or pCMV-HIV-1Δvpr. Virions were harvested 40 hours later and purified through a 20% sucrose cushion. The viral proteins were revealed by western blot analysis with a monoclonal antibody specific for Vpr (lanes 1–4) or Cre (lanes 5–8). Lanes 1 and 5: cell lysates derived from pCMV-vpr-cre and pCMV-HIV-1Δvpr co-transfection; lanes 2 and 6: virions derived from pCMV-HIV-1 transfection; lanes 3 and 7: virions derived from pCMV-HIV-1Δvpr transfection; lanes 4 and 8: virions derived from pCMV-vpr-cre and pCMV-HIV-1Δvpr co-transfection. (c) The in vitro assay to detect the recombinase activity of virion-associated Vpr-Cre. The assay was performed with purified virions containing either Vpr-Cre (lanes 2–5) or Vpr (lanes 6–9). Each reaction contained the GFP plasmid shown on top and the virions were either mock treated or treated with 0.25% NP40 as indicated. The reaction was allowed to proceed for 30 minutes at 30 °C and recombination was detected by PCR using a pair of primer specific for the CMV IE promoter and the GFP gene, respectively. The arrowheads in the GFP plasmid represent the loxP sites. (d–i) The in vivo assay to detect the recombinase activity of virion-associated Vpr-Cre. The HIV7/CMV-GFP vector containing either Vpr or Vpr-Cre was prepared from 293T cells. HT1080/Gβ cells were transduced with either the (d) Vpr-containing vector or increasing amounts of the (e–i) Vpr-Cre-containing vector. (e) 10 µl, (f) 20 µl, (g) 50 µl, (h) 100 µl, and (i) 200 µl. The titer of each vector was shown in Table 2. The β-gal expression in transduced cells was detected 72 hours after transduction. CMV, cytomegalovirus; GFP, green fluorescence protein; HIV, human immunodeficiency virus; IE, immediate early; LTR, long-terminal repeat; Vpr, viral protein R.
To determine whether incorporated Vpr-Cre retained its recombinase activity, we subjected the harvested virions to an in vitro recombination assay. Virions were treated with NP40 to allow the release of incorporated Vpr-Cre, and its activity was monitored by mixing the treated virions with a plasmid containing the GFP gene flanked by two loxP sites (Figure 2c). Without recombination, PCR was expected to amplify a 0.5-kb fragment. The fragment would be absent if the GFP gene was removed by Cre. As shown in Figure 2c, NP40 treatment of virions-containing Vpr-Cre resulted in a decrease or complete absence of the 0.5-kb fragment (lanes 3 and 5, respectively). In contrast, the 0.5-kb fragment continued to be present if the same vector preparation was not subjected to NP40 treatment (lanes 2 and 4). Vectors-containing Vpr exhibited the 0.5-kb PCR product irrespective of the NP40 treatment (lanes 6–9). These results suggested that Vpr-Cre was incorporated into virions and was released upon detergent treatment to mediate the recombination reaction in vitro.
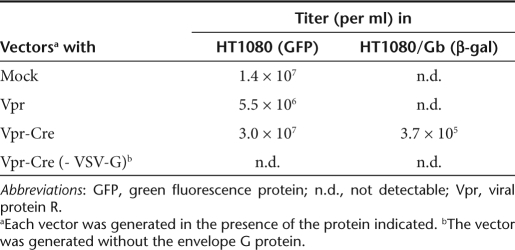
To determine whether virion-associated Vpr-Cre can function in cells, we established a stable cell line, HT1080/Gβ, derived from the transduction of HT1080 cells with the CG-β-gal retroviral vector (Figure 2a). HIV7/CMV-GFP, a GFP gene-containing lentiviral vector,23 was prepared in the presence of Vpr-Cre. The vector titer was determined in HT1080 cells by GFP expression and the same amount of the vector was used to transduce HT1080/Gβ cells and scored for β-gal expression. Increasing input of HIV7/CMV-GFP containing Vpr-Cre resulted in the emergence of β-gal+ cells (Figure 2e–i and Table 2) whereas vectors with Vpr only failed to activate β-gal expression (Figure 2d). This result demonstrated that only the vector-containing Vpr-Cre was able to activate β-gal expression in HT1080/Gβ cells. β-Gal activation relied on vector cell entry as HIV7/CMV-GFP without the VSV-G envelope protein failed to generate any β-gal+ cell (Table 2). These studies confirmed that the virion-associated Vpr-Cre remained enzymatically active. The almost 100-fold difference in titer between HT1080 and HT1080/Gβ cells of the Vpr-Cre-containing vector (Table 2) could be explained by three possibilities: (i) susceptibility of the two cell lines to vector transduction was different; (ii) a difference in the assay used for titer determination (GFP versus β-gal); (iii) only a fraction of harvested virions contained sufficient Vpr-Cre to mediate recombination and activate β-gal expression in HT1080/Gβ cells.
Table 2. β-Gal activation by a Vpr-Cre-containing lentiviral vector in HT1080/Gβ cells.
Lentiviral vector-mediated site-specific gene insertion
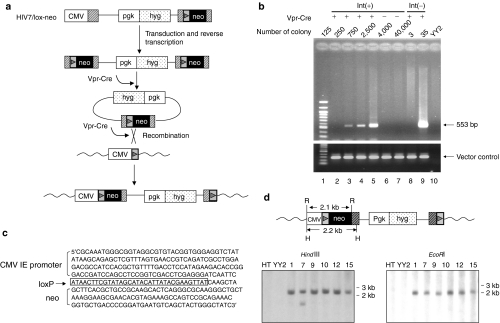
To test for site-specific gene insertion, a lentiviral-based targeting vector, pHIV7/lox-neo, was constructed. The hygromycin-resistant (hyg) gene controlled by the 3-phosphoglycerate kinase (pgk) promoter was inserted into the backbone of the vector for titer determination (Figure 3a). A cassette containing the loxP site followed by the neo gene was inserted into the U3 region of the 3′ LTR. Upon replication, the loxP-neo cassette would appear in both LTRs, thereby providing a substrate for recombination that could be catalyzed by virion-associated Vpr-Cre. We hypothesized that the circular recombination product containing the loxP-neo cassette was able to be inserted into the loxP site in the YY2 genome, again catalyzed by virion-associated Vpr-Cre (Figure 3a). This insertion directly places the neo gene under the control of the CMV promoter, permitting G418 selection. To test this hypothesis, YY2 cells were transduced with increasing amounts of HIV7/lox-neo encapsidated with Vpr-Cre and G418-resistant colonies were pooled. The genomic DNA from the pool was subjected to PCR amplification using a pair of CMV- and neo-specific primers. Site-specific gene insertion was expected to generate a 553-bp PCR fragment. As shown in Figure 3b, the expected PCR product was observed from the genomic DNA of ~250, 750, and 2,500 pooled colonies (lanes 3–5). A pool of 125 colonies failed to exhibit this band. Other two primer pairs spanning the CMV-neo fragment also confirmed this observation (data not shown). As a control, we pooled G418-resistant colonies from cells transduced with the same vector without Vpr-Cre. We failed to detect the expected fragment with the genomic DNA isolated from up to 40,000 pooled colonies (lanes 6 and 7), indicating that virion-associated Vpr-Cre was required for the gene targeting event. The 553-bp fragment was cloned and sequenced, and the sequencing data confirmed that the neo gene was positioned downstream from the CMV IE promoter with a loxP site present in between (Figure 3c). To assess the targeting efficiency, the titer of the targeting vector was determined by selecting the transduced YY2 cells in hygromycin. Based on the hygromycin-resistant titer and the neo-resistant colony required to detect the PCR fragment, the efficiency for site-specific gene insertion was determined to be in the range between 10−3 and 0.5 × 10−3.
Figure 3.
Site-specific gene insertion with a Vpr-Cre containing lentiviral vector. (a) Steps involved in site-specific gene insertion with the targeting vector, HIV7/lox-neo. The vector backbone of HIV7/lox-neo contains the hyg gene controlled by the pgk promoter. The loxP-neo cassette was inserted into the 3′ LTR. Upon transduction and reverse transcription in transduced YY2 cells, this cassette would appear in both LTRs, thereby providing a substrate for recombination mediated by virion-associated Vpr-Cre. The resulting circular product containing a single loxP site can recombine with the target loxP site in the YY2 genome and insert the neo gene downstream from the CMV IE promoter. The insertion reaction presumably is catalyzed again by virion-associated Vpr-Cre. Wavy lines indicate the YY2 genomic DNA. (b) PCR detection of site-specific gene insertion events. YY2 cells were transduced with increasing amounts of HIV7/lox-neo with or without Vpr-Cre and selected in G418-containing medium. Upon selection for 2 weeks, various numbers of G418-resistant colonies as indicated were pooled and the genomic DNA was subjected to PCR amplification using a pair of primer specific for the CMV IE promoter and the neo gene. This pair of primer was expected to amplify a 533-bp fragment if gene targeting occurred (top). As a control, a second pair of primers that specifically amplified a 5′ HIV gag region present in the vector was also used in the PCR to demonstrate vector integration irrespective of the presence or absence of Vpr-Cre (bottom). Int(+), vectors containing a functional integrase; Int(−), vectors containing a defective integrase. The G418-resistance titers for the Int(+) and Int(−) vector were 25,000 ± 5,000 infectious units (IU)/ml and 38 ± 7 IU/ml, respectively. (c) The DNA sequence surrounding the neo gene insertion site. The 533-bp PCR fragment was cloned into pBluescript by TA cloning and sequenced. The loxP site was boxed. (d) Southern blot analysis of YY2-derived clones containing site-specific gene insertion. G418-resistant clones derived from the integrase-deficient HIV7/lox-neo vector were picked and expanded. The genomic DNAs from six such clones were digested with either HindIII or EcoRI, separated on a gel, blotted and hybridized with a neo-specific probe. The expected fragment size for site-specific gene insertion is 2.2 kb with HindIII digestion and 2.1 kb with EcoRI digestion. HT1080 DNA and YY2 DNA digested with HindIII or EcoRI were also run on the same gel to serve as negative controls. CMV, cytomegalovirus; GFP, green fluorescence protein; HIV, human immunodeficiency virus; IE, immediate early; LTR, long-terminal repeat; Vpr, viral protein R.
To enrich for the clones with site-specific gene insertion, we generated the targeting vector from a packaging plasmid containing a mutation in the integrase gene.24 Because this mutation blocks normal HIV integration, G418-resistant cells should be derived preferentially via recombination with the Cre-loxP system. As shown in Figure 3b, a pool of only 35 G418-resistant colonies was sufficient to exhibit the expected PCR fragment whereas a pool of three colonies did not (lanes 8 and 9). Genomic Southern analysis of 15 randomly picked clones showed that six clones exhibited the expected 2.2- and 2.1-kb fragments after digestion with HindIII and EcoRI, respectively (Figure 3d). To ensure that the observed site-specific gene insertion is not YY2-specific, we established a pool of HT1080-derived clones containing randomly positioned loxP sites in the genome. Using the same approach described above, we showed site-specific neo gene insertion into these loxP sites (data not shown). The observed gene insertion event is therefore not dependent on a particular loxP site in the genome. Together, these results suggested that a transgene introduced via the targeting lentiviral vector could be inserted into multiple loxP sites present in the human genome and that the insertion depended on the presence of copackaged Vpr-Cre fusion proteins.
Discussion
Genotoxicity induced by random retro- and lentiviral vector integration raises serious safety concerns of applying these viral vectors in human gene therapy. The strategy described here takes advantage of the high efficiency of lentivirus for gene delivery, but bypasses the normal viral integration process and use instead the Cre/loxP system for site-specific gene insertion. Different strategies have been used before to introduce Cre into cells, including DNA transfection, protein transduction, and viral vector-mediated gene delivery.17,18,25,26,27 Many of these approaches led to stable integration of the cre gene into the host genome, and constitutive Cre expression was shown to cause detrimental effect to the host cell.25,26 Direct protein transduction avoids such a risk but is limited by the level of the introduced Cre protein. Our strategy of using the same lentiviral virion to deliver both the Cre protein and the gene of interest has at least two advantages. First, only copackaged Vpr-Cre proteins in the virion but not its gene are delivered to the target cells, thereby avoiding the problem of cell toxicity induced by stable Cre expression. Second, Vpr-Cre is in close association with the gene of interest, thereby increasing the probability of site-specific gene insertion once the target loxP site in the host genome is located by the lentiviral preintegration complex. This feature effectively lowers the threshold level of Cre required to facilitate efficient recombination between the two loxP sites in the targeting vector and in the host genome, respectively.
For Cre encapsidation, we capitalized on the unique property of HIV-1 Vpr. Among the key HIV-1 encoded proteins that can mediate nuclear import of the preintegration complex, Vpr has the unique capacity to serve as an efficient vehicle for the transportation of Vpr-linked proteins into the nucleus of infected cells.28,29,30 Vpr is packaged into virions at an higher efficiency than other HIV-1-encoded accessory proteins (Vif, Vpu, Nef) and is in a molar ratio of ~1:7 relative to Gag.31 Vpr is present at a level of ~100–200 copies/virion, a level potentially sufficient to mediate efficient site-specific gene insertion. The challenges for our approach are whether the Cre recombinase activity is compromised by protein fusion and whether Vpr-Cre is encapsidated into virions at sufficient levels to mediate gene insertion. Our data demonstrated that Vpr-Cre fusion maintained significant level of the recombinase activity and was packaged efficiently into virions. We chose to fuse Cre to the C-terminus of Vpr based on several considerations; the amino terminal α-helical region of Vpr is considered to be most important for nuclear localization and virion packaging.32 Fusion to the N-terminus of Vpr may therefore disrupt these functions. The fact that the C-terminus of Vpr is not required for packaging is consistent with this hypothesis.32 Although the functional role of the N-terminus of Cre is still unclear, the region is not well conserved among members of the tyrosine recombinase family and can be modified without significant loss of its activity.33,34 Based on these considerations, fusion between the C-terminus of Vpr and the N-terminus of Cre is therefore most likely able to succeed in directing site-specific gene insertion with our strategy. As the recombinase activity of Vpr-Cre is not as efficient as that of wild-type Cre, introduction of additional mutations into Vpr-Cre to increase its recombinase activity may further improve the efficiency of site-specific gene insertion. As direct protein–protein fusion may cause structural disturbance and result in reduced protein activity, insertion of a linker between Vpr-Cre may also improve the Cre enzymatic activity.35,36
Our strategy for site-specific gene insertion requires two recombination events. The first event involves recombination between the two loxP sites in the targeting vector, permitting the generation of a circular DNA substrate for gene insertion. This step is important as the presence of only a single loxP site in the linear targeting vector DNA genome, when recombined with the loxP site in the host genome, can lead to chromosome breakage. Transduction of YY2 cells with the targeting vector resulted in the emergence of G418-resistant colonies even though the neo gene in the vector was not directly linked to a promoter. As these vectors possess functional integrase and lentiviral vectors preferentially integrate into introns of active genes,6 most of these colonies may result from normal lentiviral integration and neo gene expression from an adjacent cellular promoter. Because several potential splice sites are present upstream from the neo gene in the vector, splicing of the RNA initiated from the cellular promoter could result in the generation of a mature transcript encoding functional neomycin phosphotransferase. Consistent with this hypothesis, the number of G418-resistant colonies was dramatically reduced when integrase-deficient vector was used for transduction (Figure 3b). In such a case, ~40% of the G418-resistant colonies were the result of site-specific gene insertion. Whether the efficiency for site-specific gene insertion can be enhanced further by an increase at the level of the copackaged Vpr-Cre or its recombinase activity remains to be determined.
Although the wild-type loxP sequence is absent from the human genome, many so-called pseudo-loxP sites in the human genome could serve as potential sites for gene targeting. An analysis by Thyagarajan et al. indicated that at least four pseudo-loxP sites could support Cre-mediated gene insertion and excision in human cells.19 The recombination efficiency among these four sites varied with the site at Xp22 having an efficiency similar to wild-type loxP whereas the site at 5p15 having a 100-fold lower efficiency. The site in Xp22 therefore may serve as a potential target for the insertion of a therapeutic gene to reduce the risk of insertional mutagenesis. This site is mapped in an intergenic region in X chromosome and is located ~138 kb away from the transcription start site of a nearest cellular gene that codes for short stature homeobox isoform b (SHOXb). Whether this site at Xp22 can serve as an efficient target for gene insertion and whether such insertion alters the expression of SHOXb will be evaluated in future studies. Directed molecular evolution has also been applied to create Cre recombinase variants that recognize a new DNA target sequence called loxH on human chromosome 22 but not the wild-type loxP sequence.37 Such an approach allows the design of Cre variants that recognize additional pseudo-loxP sites in the human genome. Using the strategy described here, incorporating these Cre variants should allow direct gene insertion into the cognate pseudo-loxP site. The efficiency for gene targeting in our study remains between 1/1,000 and 1/2,000. Since the excision event is kinetically favored with the Cre-loxP system,38,39 an inserted gene can readily be deleted in the presence of Vpr-Cre. Mutant loxP sites favoring insertion over excision have been identified with bacterial screening systems.38 These approaches can be applied to create mutant loxP sites that favor gene insertion instead of excision, thereby increasing the gene targeting efficiency. Contrary to Cre, the other site-specific recombination system, the phage φC31 recombinase, favors unidirectional gene insertion that would occur at higher frequencies than the reversible gene insertion directed by Cre.40,41 As the lentiviral strategy described here requires two recombination events to insert a transgene into the host genome (Figure 3a), it may not be compatible with the phage φC31 system to mediate site-specific gene insertion. Finally, the approach of Vpr fusion may be similarly applied to incorporate other proteins such as zinc-finger containing nucleases to enhance gene targeting by homologous recombination.
Materials and Methods
Plasmids. To construct the vpr-cre fusion gene, the vpr gene was PCR amplified from pCMV-HIV-123 with the following primers:
vpr-s: 5′-GCGCATACCGCTCGAGATGGAACAAGCCCCAAGAAGAC-3′
vpr-r: 5′-TTTGGTGTACGGTCAGTAAATTGACCAGGATCTACTGGCTCCATTTCT-3′
The cre gene was PCR amplified from pBS185 with the following primers:
cre-s: 5′-GCAAGAAATGGAGCCAGTAGATCCTGGTCCAATTTACTGACCGACAC-3′
cre-r: 5′-CGCGTACGCGTACGCGTATGGCGAATTCCTAATCGCCATCTTTCCAAGCAG-3′
A second PCR amplification was carried out with the annealed products from the first two PCRs using vpr-s and cre-r as the primers. The PCR product was cloned into pBluescript SK(−) to generate pBS-vpr-cre for sequencing. To construct pCMV-vpr-cre, the VSV-G gene in pCMV-G42 was replaced with the 1.3-kb fragment containing the vpr-cre fusion gene from pBS-vpr-cre. The targeting vector pHIV7/lox-neo includes in its backbone a 1.8-kb fragment containing the hygromycin-resistant gene controlled by the 3-phosphoglycerate kinase (pgk) promoter. A 1.4-kb fragment containing neo gene preceded by a loxP site was inserted into a unique Xba1 site in the 3′ LTR to generate pHIV7/lox-neo. Plasmid pC-HelpIN− encoding a defective integrase was provided by Dr J. Reiser.24 Plasmid pC-HelpIN− Δvpr harboring a defective vpr gene was created by fill-in of EcoRI-digested pC-HelpIN− followed by religation. The retroviral vector, pCCGLΔEn, used to establish YY1 and YY2 cells has a 740-bp fragment containing the GFP gene flanked by loxP sites. Expression of the GFP gene is controlled by an internal CMV IE promoter. The retroviral vector, pCG-β-gal, contains the GFP gene flanked by loxP sites followed by the β-gal gene. Expression of the GFP-β-gal cassette is initiated from the 5′LTR. Plasmid plox-geo was constructed by isolating a 3.9-kb fragment containing the geo gene from pSAβgeo43 and inserting it into pdLox-2 containing a multiple cloning site flanked by two loxP sites.
Cell lines. Cell lines used for this study were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Cells resistant to antibiotics were selected at a concentration of 400 µg/ml of either G418 or hygromycin. To establish YY1, HT1080 cells were transduced with CCGLΔEn. YY1 was isolated after three rounds of FACS for GFP expression. YY2 was established by transfecting YY1 cells with the cre-expression plasmid pBS185. YY2 was isolated by limiting dilution after three rounds of FACS for GFP− cells. To establish HT1080/Gβ cells, HT1080 cells were transduced with vector CG-β-gal. A clone expressing high levels of GFP was isolated by FACS and limiting dilution.
Lentiviral vector production and transduction. Lentiviral vectors were generated by transient transfection of 293T cells with the vector construct, the indicated packaging plasmid and pCMV-G.42 For the production of Vpr-Cre-containing vectors, pCMV-vpr-cre was also included in the transfection mixture. The transfection solution was replaced after 14–16 hours with fresh culture medium-containing 10 mmol/l sodium butyrate, and the vector was harvested after additional 24 hours. For cell transduction, the vector was applied at different dilutions to the cells in the presence of 8 µg/ml polybrene. Antibiotics selection or FACS analysis was performed 48 hours after transduction. To determine the efficiency of gene targeting, YY2 cells were transduced with HIV7/lox-neo containing Vpr-Cre and selected in hygromycin-containing medium. As this vector contains the hygromycin-resistance gene under the control of the pgk promoter, the number of hygromycin-resistant colony represents the total infectious units applied. The same vector preparation was also used to transduce YY2 cells and selected in G418-containing medium. Various numbers of G418-resistant colonies were pooled and subjected to PCR analysis of gene targeting events. The ratio between the number of G418-resistant colony required to detect the gene targeting event and the total infectious vector titer represents the efficiency of gene targeting.
Western blot analysis. Virions harvested and purified through a 20% sucrose cushion were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis. Immunoblotting was performed using a rabbit anti-Vpr polyclonal antibody (dilution 1:2,500) obtained from NIH AIDS Research and Reference Reagent Program or a rabbit anti-Cre polyclonal antibody (dilution 1: 100,000) obtained from Novagen (Madison, WI). A horse radish peroxidase-conjugated polyclonal goat anti-rabbit immunoglobulin G (dilution 1: 100,000) from Amersham-Pharmacia (Piscataway, NJ) was used as a secondary antibody. Chemiluminescent detection was performed using ECL-Plus detection reagents from Amersham-Pharmacia.
The Cre activity assay. To assay for the Cre enzymatic activity, 293T cells were transiently transfected with pCMV-vpr-cre and pCG-β-gal. Cre-expression plasmid, pBS185,18 was similarly transfected to serve as a positive control. Cells were lysed 48 hours later in RIPA buffer containing a mixture of protease inhibitors (Roche Applied Sciences, Alameda, CA) and the supernatant was collected after a brief centrifugation. The β-gal enzymatic activity was measured using the Tropix Galacto-Star chemiluminescent reporter gene assay system from Applied Biosystems (Foster City, CA). The BCA Protein Assay Reagent Kit (Pierce, Rockford, IL) was used to determine the protein concentration and to normalize the cell extract used in each assay. The in vitro recombination assay was performed according to Sauer.44 Briefly, purified virions containing either Vpr-Cre or Vpr were incubated in the Cre reaction mixture (1.25 mmol/l Tris-HCl, pH 7.5/1.67 mmol/l NaCl/10 mmol/l MgCl2) with 0.2 µg of the GFP plasmid at 30 °C for 30 minutes in the presence or the absence of 0.25% NP40. Recombination was determined by PCR amplification using a pair of primers with the following sequences:
CMV IE promoter: 5′-CGGGACTTTCCAAATTCGTAACAAC-3′
GFP gene: 5′-CTGAAGCACTGCACGCCGTAG-3′.
Analysis of site-specific gene insertion by PCR and Southern blot. The DNA isolation kit from Gentra Systems (Minneapolis, MN) was used for the purification of genomic DNA. For the Southern blot analysis, HindIII- or EcoRI-digested DNA was separated on 1% agarose gel and blotted. The filter was hybridized with a 777-bp neo-specific probe prepared by PCR amplification using a pair of primer with the following sequences:
neo-r: 5′-GTCAAGAAGGCGATAGAAGG-3′
neo-s: 5′-TATGACTGGGCACAACAGAC-3′
To detect site-specific gene insertion, 400-ng genomic DNA was denatured at 94 ºC, annealed at 60 ºC, and elongated at 72 ºC with HotStarTaq DNA polymerase (Qiagen, Valencia, CA) for 35–40 cycles. Site-specific gene insertion was detected by PCR amplification using primer pairs with the following sequences:
CMV1: 5′-CGGGACTTTCCAAAATGTCGTAAC-3′
CMV2: 5′-GGAGGTCTATATAAGCAGAGCTC-3′
CMV3: 5′-TAGAAGACACCGGGACCGATCC-3′
neo1: 5′-CTGCGTGCAATCCATCTTGTTC-3′
neo2: 5′-GGCAAGAAAGCCATCCAGTTTAC-3′
neo3: 5′-GTCTAGCTATCGCCATGTAAGC-3′
The sequences for the PCR primers to amplify the vectors are as follows:
Vec1: 5′-GACCTGAAAGCGAAAGGGAA-3′
Vec2: 5′-ACTGCGAATCGTTCTAGCTC-3′
Acknowledgments
We thank Dr B. Sauer for providing pBS185 and Dr J. Reiser for providing pC-HelpIN−. This work was funded in part by the NIH grant P01A146030.
REFERENCES
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA., and, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32 Database issue:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. Gene therapy. β-thalassemia treatment succeeds, with a caveat. Science. 2009;326:1468–1469. doi: 10.1126/science.326.5959.1468-b. [DOI] [PubMed] [Google Scholar]
- Inoue N, Izui-Sarumaru T, Murakami Y, Endo Y, Nishimura J, Kurokawa K, et al. Molecular basis of clonal expansion of hematopoiesis in 2 patients with paroxysmal nocturnal hemoglobinuria (PNH) Blood. 2006;108:4232–4236. doi: 10.1182/blood-2006-05-025148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL., and, Kossev P. Molecular genetics of benign tumors. Cancer Invest. 2002;20:362–372. doi: 10.1081/cnv-120001182. [DOI] [PubMed] [Google Scholar]
- Fedele M, Battista S, Manfioletti G, Croce CM, Giancotti V., and, Fusco A. Role of the high mobility group A proteins in human lipomas. Carcinogenesis. 2001;22:1583–1591. doi: 10.1093/carcin/22.10.1583. [DOI] [PubMed] [Google Scholar]
- Odero MD, Grand FH, Iqbal S, Ross F, Roman JP, Vizmanos JL, et al. Disruption and aberrant expression of HMGA2 as a consequence of diverse chromosomal translocations in myeloid malignancies. Leukemia. 2005;19:245–252. doi: 10.1038/sj.leu.2403605. [DOI] [PubMed] [Google Scholar]
- Katz RA, Merkel G., and, Skalka AM. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- Holmes-Son ML, Appa RS., and, Chow SA. Molecular genetics and target site specificity of retroviral integration. Adv Genet. 2001;43:33–69. doi: 10.1016/s0065-2660(01)43003-3. [DOI] [PubMed] [Google Scholar]
- Holmes-Son ML., and, Chow SA. Integrase-lexA fusion proteins incorporated into human immunodeficiency virus type 1 that contains a catalytically inactive integrase gene are functional to mediate integration. J Virol. 2000;74:11548–11556. doi: 10.1128/jvi.74.24.11548-11556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Zhu K, Segal DJ, Barbas CF, 3rd, Chow SA. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., and, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., and, Henderson N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- Thyagarajan B, Guimarães MJ, Groth AC., and, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- Popov S, Rexach M, Zybarth G, Reiling N, Lee MA, Ratner L, et al. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka MA, Koepp DM, Silver PA., and, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmi M, Glynn J, Jin MJ, Jolly DJ, Yee JK., and, Chen ST. Requirements for efficient production and transduction of human immunodeficiency virus type 1-based vectors. J Virol. 1999;73:1828–1834. doi: 10.1128/jvi.73.3.1828-1834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam PY, Li S, Wu J, Hu J, Zaia JA., and, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- Mochizuki H, Schwartz JP, Tanaka K, Brady RO., and, Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A, Brandon EP, Kootstra N, Gage FH., and, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc Natl Acad Sci USA. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP., and, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Nolden L, Edenhofer F, Haupt S, Koch P, Wunderlich FT, Siemen H, et al. Site-specific recombination in human embryonic stem cells induced by cell-permeant Cre recombinase. Nat Methods. 2006;3:461–467. doi: 10.1038/nmeth884. [DOI] [PubMed] [Google Scholar]
- BouHamdan M, Kulkosky J, Duan LX., and, Pomerantz RJ. Inhibition of HIV-1 replication and infectivity by expression of a fusion protein, VPR-anti-integrase single-chain variable fragment (SFv): intravirion molecular therapies. J Hum Virol. 2000;3:6–15. [PubMed] [Google Scholar]
- Yao XJ, Kobinger G, Dandache S, Rougeau N., and, Cohen E. HIV-1 Vpr-chloramphenicol acetyltransferase fusion proteins: sequence requirement for virion incorporation and analysis of antiviral effect. Gene Ther. 1999;6:1590–1599. doi: 10.1038/sj.gt.3300988. [DOI] [PubMed] [Google Scholar]
- Cavrois M, De Noronha C., and, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- Müller B, Tessmer U, Schubert U., and, Kräusslich HG. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J Virol. 2000;74:9727–9731. doi: 10.1128/jvi.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XJ, Subbramanian RA, Rougeau N, Boisvert F, Bergeron D., and, Cohen EA. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal α-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T., and, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D., and, Scocca JJ. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HL, Yao XQ, Xue C, Wang Y, Xiong XH., and, Liu ZM. Increasing the homogeneity, stability and activity of human serum albumin and interferon-α2b fusion protein by linker engineering. Protein Expr Purif. 2008;61:73–77. doi: 10.1016/j.pep.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Cao Y, Marks JD, Marks JW, Cheung LH, Kim S., and, Rosenblum MG. Construction and characterization of novel, recombinant immunotoxins targeting the Her2/neu oncogene product: in vitro and in vivo studies. Cancer Res. 2009;69:8987–8995. doi: 10.1158/0008-5472.CAN-09-2693. [DOI] [PubMed] [Google Scholar]
- Buchholz F., and, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- Thomson JG, Rucker EB, 3rd, Piedrahita JA. Mutational analysis of loxP sites for efficient Cre-mediated insertion into genomic DNA. Genesis. 2003;36:162–167. doi: 10.1002/gene.10211. [DOI] [PubMed] [Google Scholar]
- Araki K, Araki M., and, Yamamura K. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe HM., and, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe HM, Wilson SE., and, Smith MC. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- Burns JC, Friedmann T, Driever W, Burrascano M., and, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G., and, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Meth Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]