Abstract
HIF-1 α (hypoxia inducible factor-1 α isoform) has been exploited as a target in cancer therapeutics. HIF-1 α is the isoform-2 of HIF-1 α subunit. It is a 735 residues long protein modeled in this study. The HIF-1 α is absolutely critical for continued survival of cancer cells as it is involved in the activation of glycolysis and it helps an oxygen-starved cell convert sugar to energy without using oxygen. It also initiates angiogenesis to bring in a fresh oxygen supply. HIF-1 α operates only in presence of free radicals. In the present study, five antioxidants, namely lycopene, ascorbic acid, α-tocopherol, curcumin and curcumin dipiperoyl ester which are potent scavengers of reactive oxygen species (ROS) have been docked to HIF-1 α modeled protein in order to assess their binding and consequently, their inhibitory activity. The binding energy score has been found to be in the order, curumin dipiperoyl ester > lycopene > curcumin > tocopherol > ascorbic acid. However, subsequent experiments should be designed to validate these observations.
Background
One of the important factors highlighted recently in cancer is the activation of glycolysis leading to angiogenesis [1] and cell proliferation, decreased apoptosis, cellular immortalization and invasion/metastasis. Novel molecules for cell proliferation are provided through glycolysis, which is activated by the well known factor HIF-1 α [2–4]. Transcription factor is a guide to many cancers by activating the transcription of many genes that code for proteins involved in several pathways which are closely related to cancer growth. The progressive survival of cancer cells depends on glycolytic energy through ATP generation [5–6] and the level of ATP is reduced remarkably under oxygen starved conditions in the absence of HIF-1 α [2]. In most common human cancers as well as in pre-neoplastic and pre-malignant lesions, such as colonic adenoma, breast ductal carcinoma HIF-1 α is over-expressed. It is reported that HIF-1 α expression may occur very early in carcinogenesis, before histological evidence of angiogenesis or invasion [3]. It has been suggested that HIF-1 α is a biomarker of carcinogenesis and for drug design, pharmaceutical companies use it as suitable target for cancer chemoprevention.
Studies suggest that HIF-1 α can only operate in the presence of free radicals and it does not work if antioxidants remove these free radicals [7]. They showed that while this protein was abundant in untreated cancer cells taken from mice, it disappeared in vitamin C-treated cells. It might be possible that antioxidants adopted two alternative pathways for reducing the activity of HIF-1 α, firstly, these directly remove the free radicals and make the survival of HIF-1 α difficult and another way may be that these antioxidants bind at the active site of the protein to inhibit its activity. HIF1 is a hetero-dimer composed of two subunits (an alpha and a beta). The beta subunit has been recognized as the aryl hydrocarbon receptor nuclear trans-locator (ARNT) [9]. Two alternative transcripts of hypoxia inducible factor-1gene encodes for different isoforms (NP_001521.1 and NP_851397.1). The HIF1 alpha is isoform 2 (NP_851397.1) and it is shorter with a distinct Cterminus, compared to isoform 1 as it lacks an alternate segment at the 3´ CDS region. Therefore, it is important to describe the mechanism of in induction by HIF-1 α. However, the protein databank (PDB) contains the NMR structure of cysteine/histidine-rich 1 (CH1) domain of p300 bound to the C-terminal transactivation domain of HIF-1α, which is smaller than the target receptor protein [8]. Hence, we employed threading technique to construct the model. We then used docking tools to evaluate the binding of anti-oxidants like lycopene, curcumin, α tocopherol and ascorbic acid. We also docked curcumin dipiperoyl ester for comparison as its bioavailability is enhanced remarkably to curcumin and it is a potent anticancer drug to curcumin [10].
Methodology
Target sequence
The 735 residue long protein sequence of Human hypoxia inducible factor 1 (NCBI_ID: NP_851397.1 (hypoxia-inducible factor 1, alpha subunit isoform 2) was downloaded from NCBI RefSeq database [12].
BLAST search
The target sequence for HIF-1 alpha was submitted to NCBI-Blast and searched against PDB [13] to select suitable structural templates.
Wurst server
Wurst server is used in the absence of a suitable homolog. The target sequence is aligned to more than 3000 templates using a dynamic programming algorithm with a structure and sequence features based score function [14].
ArchPRED
We used ArchPRED for loop region prediction [15].
SwissPdbViewer
We used swisspdbviewer to subsequently model the structure using the selected template and loop structures [16].
RAMPAGE server
The RAMPAGE server is used to validate the predicted model [17].
Discussion
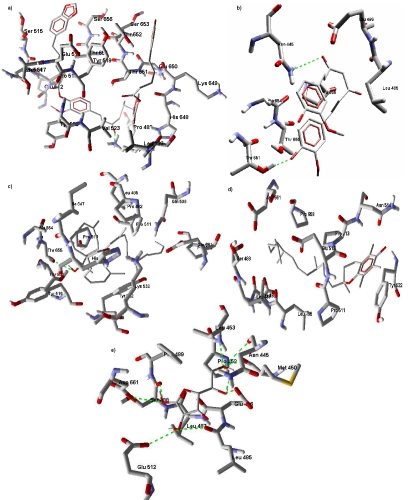
Ramachandran plot analysis of the modelled structure show 77.4% in favoured region, 14.1% in allowed region and 8.1% in outlier region of the plot. This modeled protein structure has five cavities detected using the molegro software with volumes 288.768 A3, 29.184 A3, 26.112 A3, 25.6 A3 and 19.968 A3. The cavity with volume 288.768 A3 shows good energy score for all the five antioxidants (lycopene, ascorbic acid, tocopherol, curcumin and dipiperoyl ester of curcumin) used in this study. The ranking of the anti-oxidants using the energy score is curcumin-dipiperoyl ester 〈 lycopene 〈 curcumin 〈 tocopherol 〈 ascorbic acid. Data shows curcumin dipiperoyl ester with the lowest energy score is most favourable. The five antioxidants used in the study exhibit hydrogen bond interaction with the interacting residues as shown in Figure 1. The analysis shows ascorbic acid with maximum H-bond interaction and tocopherol with minimum H-bond interactions. Table 1 (see supplementary material) shows the interacting properties of the anti-oxidants to the target protein. Curcumin also show low energy score which is close to lycopene. The energy score of curcumin molecule was changed by substituting it with dipiperoyl ester as reported eslwehere [11].
Figure 1.
HIF-1 α protein docked with (a) curcumin dipiperoyl ester; (b) curcumin; (c) lycopene; (d) tocopherol; (e) ascorbic acid
Conclusion
It is known that antioxidants inhibit the activity of HIF-1 α by scavenging free radicals. Data here show that the five antioxidants (lycopene, ascorbic acid, α- tocopherol, curcumin and curcumin conjugates with piperic acid) have similar mode of binding at the site of the hypoxia inducing factor-1 α with varying binding energy scores. Current analysis also shows the H-bonds interactions of these compounds with the active sites. However, subsequent experiments should be designed to validate these observations.
Supplementary material
Footnotes
Citation:Upadhyay et al, Bioinformation 4(6): 233-236 (2009)
References
- 1.Gatenby RA, Gillies RJ. Nat Rev Cancer. 2004;4:891. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Harris AL. Nat Rev Cancer. 2002;2:38. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 3.Schofield CJ, Ratcliffe. PJ. Nat Rev Mol Cell Biol. 2004;5:343. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 4.Xu RH, et al. Cancer Res. 2005;65:613. [PubMed] [Google Scholar]
- 5.Ramanathan A, et al. Proc Natl Acad Sci U S A. 2005;102:5992. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. Nat Rev Cancer. 2003;3:721. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 7.Gao P, et al. Cancer Cell. 2007;12:230. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman SJ, et al. Proc Natl Acad Sci U S A. 2002;99:5367. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hőpfl G, et al. Am J Physiol Regul Integr Comp Physiol. 2004;286:R608. doi: 10.1152/ajpregu.00538.2003. [DOI] [PubMed] [Google Scholar]
- 10.Mishra S, et al. Bioorganic & Medicinal Chemistry. 2005;13:1477. doi: 10.1016/j.bmc.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 11.Mishra S, et al. Free Radical Biology and Medicine. 2005;38:1353. doi: 10.1016/j.freeradbiomed.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Pruitt KD, et al. Nucleic Acids Res. 2007;35:D61. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephen F, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torda AE, et al. Nucleic Acids Res. 2004;32:532. doi: 10.1093/nar/gkh357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Fuentes N, et al. Nucleic Acids Res. 2006;34:173. doi: 10.1093/nar/gkl156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex N, Peitsch MC. Protein Data Bank Quarterly Newsletter. 1996;77:7. [Google Scholar]
- 17.Lovell SC, et al. Proteins: Structure, Function & Genetics. 2002;50:437. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.