Abstract
The leader gene approach is a data mining method based on the systematic search for genes involved in a specific process and their ranking according to the number of interconnections with the other genes identified. The genes with the strongest interconnections are termed leader genes, since they may be supposed to play an important role in the process. The potential of malignant progression of OLP to oral squamous cell carcinoma (OSCC) is still not completely clear. In this study, the leader gene approach is applied to investigate the association between OLP and OSCC at a molecular level. Results were integrated with those obtained in an experimental analysis (see paper 1 of this series). Genes involved in OLP and OSCC were identified by systematic queries to dedicated databases. Interconnections among identified genes were calculated and given a confidence value using STRING database. Leader genes were identified by clustering genes according to their interconnections. This theoretical analysis shows that OLP and OSCC share two leader genes: TP53 and CDKN1A, involved in the PI3K signalling events mediated by AKT pathway. This finding and those obtained in the experimental analysis suggest the possible involvement of some key genes/proteins LCK, PIK3CA, BIRC5, TP53 and CDKN1A in the malignant progression from OLP to OSCC. Moreover, these findings support the role of some molecular pathways, namely IL2 signalling events mediated by PI3K, PI3K signalling events mediated by AKT, and, possibly, Aurora A signalling in the association between OLP and OSCC.
Keywords: computational biology, gene interactions, genomics, oral lichen planus, oral carcinoma
Background
Oral lichen planus (OLP) is a T-cell-mediated chronic inflammatory oral mucosal disease of unknown etiology [1]. The histological lesions are characterized by a sub-epithelial inflammatory infiltrate, mainly constituted by T lymphocytes and focused to the basal keratinocytes [1]. OLP lesions are characterized by a higher degree of cell turnover than healthy tissue: the proliferation, the maturation and the apoptosis of basal keratinocytes require a fine regulation at a genomic level [2]. From these observations, OLP may be seen as a complex multifactorial disease[2]. The World Health Organization defined OLP as a premalignant condition, with a possible progression to oral squamous cell carcinoma (OSCC) [1,2]. The progression from OLP to OSCC is a relatively rare event: although frequencies ranging from 0.4 to 6.5% have been reported, most studies suggest that the malignant progression rate is around 1%[2,3]. The molecular mechanisms determining the possible malignant development in OLP lesions are still not completely elucidated [3]. Further research is therefore advocated to clarify the molecular aspects of this potential relationship, including the identification of potential prognostic markers at a genomic and proteomic level [4].
Bioinformatics and data-mining can play a central role in the analysis and interpretation of genomic and proteomic data [5,6]. In particular, these disciplines may be useful to further clarify the pathophysiology of complex diseases, which are characterized by various biologic pathways, dependent upon the contribution of a large number of genes forming complex networks of interactions [5, 6]. Interactions between genes may be direct (physical interactions between the proteins, confirmed by experimental techniques, such as NMR or crystallography) or indirect (involvement in the same metabolic pathway or co-expression in different conditions) [7].
Recently, a data-mining method, defined as the “Leader Gene approach” have been proposed [5, 6]. This mining algorithm is based on the systematic search for the genes involved in a given process; the interaction among these genes are then calculated and genes are ranked according to the number and the confidence of all experimentally established interactions [5, 6], as derived from free Web-available databases, such as STRING (Search Tool for the Retrieval of Interacting Genes, Heidelberg, Germany) [7]. Genes in the highest rank are defined as “leader genes”, since they can be preliminarily supposed to play an important role in the analyzed process [5,6]. These genes may become potential targets for a targeted experimentation, which may be simpler than mass-scale molecular genomics and, at the same time, powerful [5,6]. The Leader Gene approach was applied to different cellular processes and pathological conditions, such as the human T lymphocyte cell cycle, human kidney transplant and periodontitis[5,6]; the results were integrated with a targeted experimental analysis, to draw an overall picture of these processes [8]. Recently, the Leader Gene approach was applied to infer the epidemiologically-established correlation between human periodontitis and type-2 diabetes [6]. Overall, the results showed that periodontitis and diabetes share four leader genes and that all leader genes are linked in a complex map of interactions. This finding may suggest an important role of leader genes in the association between these diseases; leader genes may be supposed, at least preliminarily, to act as hubs in the interaction map[5,6]. Leader genes may therefore become promising targets for a dedicated experimentation. In this data-mining ab-initio study, we apply the leader gene approach to preliminarily investigate the molecular mechanisms underlying the potential association between OLP and OSCC. The theoretical results are integrated with some experimental findings, recently obtained by mean of immunohistochemistry and tissue micro-array on patient biopsies.
Methodology
General architecture
The data mining algorithm followed is represented in Figure 1 [5,6]. First, a preliminarily set of genes with an established role in a specific process is identified by an iterative search of large-scale gene databases (PubMed, GeneBank, Geneatlas, Genecards), using several keyword-based searches and the official HGNC nomenclature. Second, the preliminary set of genes is expanded using the STRING database (version 8.2), excluding textminingderived interactions, to identify genes linked to those playing an established role in the process under study, and therefore potentially involved in it [7]. Only interactions with a confidence score ≫0.9, as given by STRING, are considered. Results are then filtered to discard false positives via a keyword-based query in PubMed, until no new genes are retrieved. Third, the interactions between all the genes identified are mapped using STRING. This database gives a combined association score to each gene-gene interaction, representing its strength. The combined association scores referring to each single gene are then summed to obtain a weighted number of links (WNL). Fourth, genes are clustered (hierarchical and Kmeans algorithms) according to their WNL. The genes belonging to the highest class are defined as leader genes. The other genes are defined as class B genes, class C genes, and so on. The differences in WNL among different classes are compared by ANOVA test (alpha value = 0.001).
Figure 1.
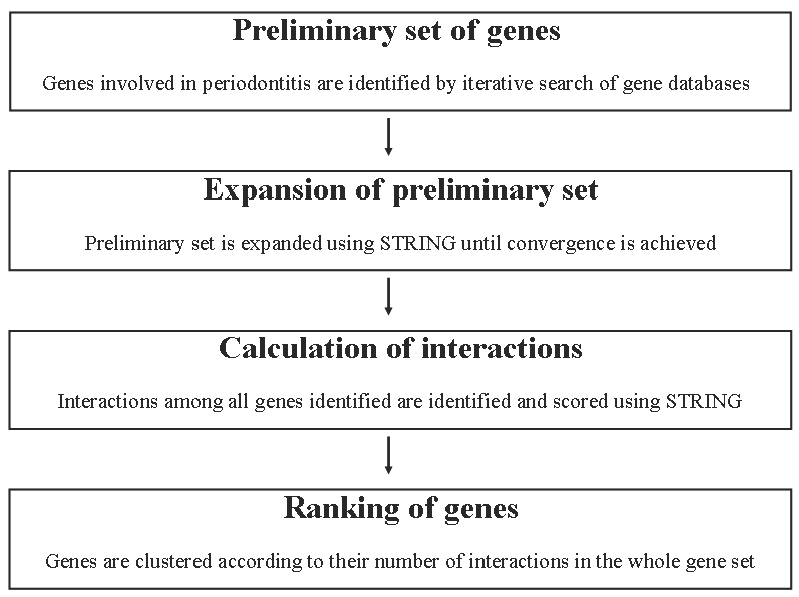
Flow chart of the leader gene approach (reproduced from [6]).
Leader genes in OLP and OSCC
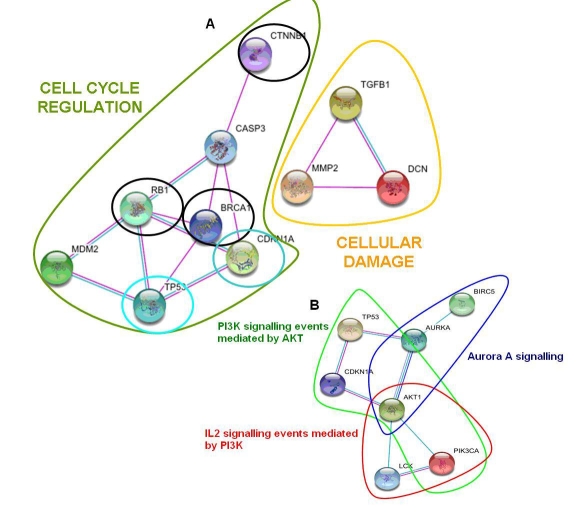
The above-described leader gene algorithm is applied to both OLP and OSCC. The resulting leader genes are compared. Research was last updated on 2th November 2009. Overall, 48 human genes were identified as involved or potentially involved in OLP; 7 of them were classified as leader genes (p≪0.001 for their WNL vs other classes). In total, 208 human genes resulted as involved or potentially involved in OSCC; 5 of them were classified as leaders (p≪0.001 for their WNL vs other classes). Table 1 summarizes the established or putative role of all leader genes. OLP and OSCC share 2 leader genes: TP53 and CDKN1A. All leader genes involved in the control of cell cycle are closely interacting (Figure 2a).
Figure 2.
Maps of interactions, according to STRING software [7]. The lines connecting single genes represent physical interactions between proteins, confirmed by various experimental methods (magenta lines), correlation in gene expression experiments (dark blue) or the involvement in the same metabolic pathway (light blue). A) Interactions among leader genes in human oral lichen planus (OLP) and in human oral squamous cell carcinoma (OSCC). Leader genes in OLP involved in cell cycle regulation are circled in green; leader genes in OLP potentially determining cellular damage are circled in yellow. Leader genes in OSCC are circled in black. Leader genes shared between OLP and OSCC are circled in blue. B) Interactions among the leader genes (TP53 and CDKN1A) shared between OLP and OSCC and some key genes (LCK, PIK3CA and BIRC5) recently showed as playing a role in the malignant progression from OLP to OSCC. Molecular pathways and further genes (AKT1 and AURKA) linking the above-mentioned ones are also shown.
Pathways characterization and comparison with experimental data
PI3K signalling pathway may play a role in the pathogenesis of OSCC [9]. The involvement of leader genes in this pathway has been checked in the Pathway Interaction Database (PID). Both shared leader genes and MDM2 are involved in PI3K signalling mediated by ATK1 pathway. The results of the data-mining process are compared and integrated, using STRING and PID databases, with the results of a recent experimental study, which identified LCK, PIK3CA and survivin (BIRC5) as potential biomarkers of the progression from OLP to OSCC. These genes are strongly interconnected with the two shared leader genes between OLP and OSCC, TP53 and CDKN1A (Figure 2b). The interactions seem to be mediated by AKT1 gene; three molecular pathways are involved in this network, namely
IL2 signalling events mediated by PI3K
PI3K signalling events mediated by AKT, and
Aurora A signalling, including AURKA gene
Discussion
We applied the Leader Gene approach, a previously-validated datamining algorithm [5,6,8], to obtain a preliminary evaluation of the potential correlation between OLP and OSCC at a genomic level. Overall, two leader genes, i.e. those with the strongest interconnections among all genes involved in OLP or OSCC, were shared between these conditions: TP53 and CDKN1A. All the other leader genes involved in cell cycle were close interconnecting. These findings might suggest that leader genes may play an important role in the association between OLP and OSCC: even a small variation in the expression, in the sequence or in the regulation of these genes might have an impact on several other genes [8]. Moreover, the shared leader genes can be supposed, at least preliminarily, to act as hubs in the interaction map [5, 6]. Recently, the Leader Gene approach was applied to formulate new hypotheses on the correlation between periodontitis and diabetes: results showed that 4 leader genes were shared between the two diseases, while all the remaining leader genes were strongly linked [6]. In the present analysis, only two genes were linked; this may suggest, at least preliminarily, that the association between OLP and OSCC is weaker than that between periodontitis and diabetes. This hypothesis is in line with the rates of malignant progression from OLP to OSCC and those referring to periodontitis-diabetes comorbidity [2,3,6]. Data-mining was completely conducted abinitio [6]. A blind search might represent a proof of the validity of this approach, if the retrieved results are in line with current knowledge. After the ab-initio identification of leader genes, scientific literature was mined to verify the role of leader genes in OLP and OSCC.
Evidence of the role of leader genes in OLP and in OSCC
All the identified leader genes are actually playing a role in OLP or in OSCC. For instance, MMP2, TGFB1 and DCN are involved in cellular damage and their expression is increased in OLP lesions [10]. Similar findings were observed for CASP3 [11]; moreover, this gene and MDM2 are regulators of cell cycle and apoptosis, [12, 13]. Leader genes in OSCC regulate cell cycle. A genome-wide profiling revealed the association of RB1 and BRCA1 with OSCC [14], while a reduced expression of CTNN1B in associated with aggressive OSCC[15]. TP53 and CDKN1A, the two shared leader genes, play a major role in the potential malignant progression. TP53 is associated to genome repair and pro-apoptotic signalling. An increase in TP53 expression is observed in OLP lesions [16]. Mutations in this gene are associated with progression to OSCC and the expression of TP53 protein correlates with poor prognosis in OSCC patients [16]. CDKN1A inhibition and misregulation frequently occur in OSCC [17].
Molecular pathways potentially involved between OLP and OSCC
The potential association between impairment in the regulation of PI3K signalling pathway mediated by AKT1 and the pathogenesis of OSCC has been recently suggested [9]. Both TP53 and CDKN1A are involved in this pathway[18,19], as well as another leader gene in OLP, MDM2 [20]. This finding may support the possible correlation between the two conditions. Recent findings, obtained in OLP and OSCC patients, have shown that the overexpression of three key proteins (LCK, PIK3CA and BIRC5) may be associated to an high risk of malignant progression (Oluwadara, paper 1 of this series). These proteins form a complex network of direct and experimentally-established interactions with TP53 and CDKN1A. Such network includes two other genes (AKT1 and AURKA) and three different pathways: the PI3K signalling pathway mediated by AKT1, the IL2 signalling events mediated by PI3K, and the Aurora A signalling. These pathways may play a role in the malignant progression from OLP to OSCC. For instance, it may be very preliminary suggested that an impairment in IL2 signalling, which promotes T-cell proliferation and subsequent continuous T-cell signalling and activation, may be associated to a reduction in immune activity and to an enhancement in the malignant potential of OLP lesions.
Limitations
The present analysis presents several limitations. The first limitation is the lack of an experimental validation; however, like all datamining processes, the Leader Gene approach is only able to provide new well-grounded hypotheses, which may be either verified or discarded by a targeted experimentation, e.g. with microarrays or real-time polymerase chain reaction (RT-PCR) [5,6]. Another limitation is the possible bias related to database mining [5,6]. However, only strong and experimentally-established gene and protein interactions were considered [7]. These choices might limit, at least partially, a possible bias related to database mining.
Conclusion
This analysis could further confirm that data-mining of existing databases may represent a starting point to improve our knowledge about cellular processes and diseases, to formulate new hypotheses and to plan targeted experimentation [5,6]. In particular, the analysis of gene interaction maps and the ranking of genes according to their interconnections might help in identifying new targets for dedicated experimental analyses, which may confirm or discard each hypothesis and suggest potential risk factors and therapy targets [5,6]. The results of this study must be interpreted together with the findings obtained in an experimental analysis conducted on a large number of biopsies from OLP and OSCC patients (Oluwadara, paper 1 of this series). Overall, these studies suggest the possible involvement of some key genes and of the encoded proteins (LCK, PIK3CA, BIRC5, TP53 and CDKN1A) in the malignant progression from OLP to OSCC. Moreover, these findings support the role of some molecular pathways, namely IL2 signalling events mediated by PI3K, PI3K signalling events mediated by AKT, and, possibly, Aurora A signalling in the association between OLP and OSCC. Further details about these complex molecular networks will likely be provided by targeted DNA or protein microarrays, as previously suggested [4,5,6]. These analyses will be focused only on selected molecular targets and therefore will be much easier to interpret than mass-scale experiments. On this basis, further insights towards a better understanding of the molecular mechanisms underlying the potential progression from OLP to OSCC may be retrieved.
Supplementary material
Acknowledgments
We are grateful to Dr Victor Sivozhelezov (Nanoworld Institute, University of Genoa, Genoa, Italy) for useful discussion. The authors declare no conflict of interest directly relevant to this study.
Footnotes
Citation:Giacomelli et al., Bioinformation 4(6): 258-262 (2009)
References
- 1.Chiappelli F, Cajulis OS. Quintessence Int. 2004;35:223. [PubMed] [Google Scholar]
- 2.Lodi G, et al. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100:40. doi: 10.1016/j.tripleo.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 3.Cortès-Ramírez DA, et al. Med Oral Patol Oral Cir Bucal. 2009;14:E118. [PubMed] [Google Scholar]
- 4.Oluwadara O, Chiappelli F. Bioinformation. 2009;3:332. doi: 10.6026/97320630003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Covani U, et al. J Periodontol. 2008;79:1974. doi: 10.1902/jop.2008.080062. [DOI] [PubMed] [Google Scholar]
- 6.Covani U, et al. J Periodontol. 2009;80:1265. doi: 10.1902/jop.2009.080671. [DOI] [PubMed] [Google Scholar]
- 7.Jensen LJ, et al. Nucleic Acids Res. 2009;37(Database issue):D412. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giacomelli L, Nicolini C. J Cell Biochem. 2006;99:1326. doi: 10.1002/jcb.20991. [DOI] [PubMed] [Google Scholar]
- 9.Bian Y, et al. Cancer Res. 2009;69:5918. doi: 10.1158/0008-5472.CAN-08-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, et al. Head Neck. 2008;30:1237. doi: 10.1002/hed.20869. [DOI] [PubMed] [Google Scholar]
- 11.Tobon Arroyave SI, et al. Oral Diseases. 2004;10:173. doi: 10.1046/j.1601-0825.2003.00998.x. [DOI] [PubMed] [Google Scholar]
- 12.Woo M, et al. Genes Dev. 1998;12:806. doi: 10.1101/gad.12.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanda N, et al. J Oral Pathol Med. 2000;29:385. doi: 10.1034/j.1600-0714.2000.290804.x. [DOI] [PubMed] [Google Scholar]
- 14.Sparano A, et al. Laryngoscope. 2006;116:735. doi: 10.1097/01.mlg.0000205141.54471.7f. [DOI] [PubMed] [Google Scholar]
- 15.Cai ZG, et al. Chin Med J (Engl) 2008;121:1866. [PubMed] [Google Scholar]
- 16.Ogmundsdóttir HM, et al. J Oral Pathol Med. 2009;38:565. doi: 10.1111/j.1600-0714.2009.00766.x. [DOI] [PubMed] [Google Scholar]
- 17.González-Moles MA, et al. Arch Oral Biol. 2006;51:1093. doi: 10.1016/j.archoralbio.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Zhou B, et al. Nat Cell Biol. 2001;3:245. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 19.Abubaker J, et al. Mol Cancer. 2009;8:51. doi: 10.1186/1476-4598-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salcedo A, et al. EMBO J. 2006;25:4752. doi: 10.1038/sj.emboj.7601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.