1 Introduction
Vertebrates have well-conserved reproductive systems including certain brain nuclei (Peter, 1983), circulating steroids (Demski, 1984), and key neurotransmitter types (Kah et al., 2007). The brain integrates information about external and internal conditions and controls reproduction via release of gonadotropin-releasing hormone 1 (GnRH1) produced in the preoptic/hypothalamic region. The GnRH1 decapeptide stimulates pituitary release of the gonadotropins, luteinizing hormone (LH) and follicle-stimulating hormone (FSH), which in turn activate gonadal production of gametes and steroid hormones. Numerous neurochemicals including steroids, serotonin, dopamine, glutamate, GABA, gonadotropin-inhibiting hormone and GnRH1 itself can influence GnRH1 neuron activity (reviewed in Van der Kraak (2009). Recently, the neuropeptide kisspeptin has been added to this list (Clarkson et al., 2008), but the brain areas involved in processing kisspeptin signals remain incompletely described, as do the potential roles of environmental impacts on kisspeptin signaling.
Kisspeptins, by binding to the G-protein coupled receptor Kiss1r, serve as critical upstream regulators of GnRH1 function (Clarkson et al., 2008). The kisspeptins are a set of peptides, 10 to 54 amino acids in length, that originate from two related propeptides and end with a C-terminal RF-amide domain. To date, kisspeptins have been identified in fish (van Aerle et al., 2008), amphibians (Biran et al., 2008), reptiles (Lee et al., 2009), and mammals (Popa et al., 2008). The critical role of kisspeptin signaling during reproductive development was discovered in humans suffering from hypophysiotrophic hypogonadism, a condition in which gonads fail to develop at puberty. Kiss1r was formerly called GPR54, but the use of lower case italics kiss1r to refer to the gene and capitalized Kiss1r for the protein has now been adopted by fish biologists (Akazome et al., 2010; Gottsch et al., 2009). The failure of the gonads to grow was caused by a mutation in Kiss1r (Seminara et al., 2003). Further demonstrating the necessity of kisspeptin signaling for puberty, two groups (d'Anglemont de Tassigny et al., 2007; Seminara et al., 2003) were able to disrupt GnRH function by disrupting genomic DNA encoding kisspeptin or its receptor in mice. Taken together, recent findings have revealed kisspeptins and Kiss1r as key regulators of reproduction.
Kisspeptins can exert their effects wherever there are receptors. In all organisms studied to date, GnRH1 neurons express Kiss1r and respond to kisspeptins by increasing gonadotropin release (Oakley et al., 2009). Furthermore, kisspeptins bind Kiss1r and cause target GnRH1 neurons to strongly depolarize as well as increase GnRH1 release, as demonstrated in ewes and female rhesus monkeys (Keen et al., 2008; Messager et al., 2005). Kisspeptins also cause changes in gene expression in GnRH1 neurons, including increased gnrh1 mRNA expression and increased Fos levels (Irwig et al., 2004). Kisspeptin effects on GnRH neurons also cause downstream effects on the pituitary, including increased LH (Navarro et al., 2004) and FSH release. The direct effects of kisspeptin on GnRH1 neurons demonstrate the essential role of Kiss1r expressed on these neurons. Kisspeptin, like GnRH1, may also act outside the Brain- Pituitary-Gonad (BPG) axis. To understand the role(s) of kisspeptin in relation to other regulatory pathways, we need to know where it acts, which requires localizing its receptors. GnRH receptor localization in the brain has identified the sites where GnRH can mediate key neuromodulatory roles for GnRH peptides (Chen and Fernald, 2006). Similarly, localizing kiss1r will reveal sites of potential action and provide insight into its possible roles.
Teleost fishes have a wide variety of reproductive systems and respond strongly to environmental signals, making them useful for analysis of inputs to hypophysiotropic GnRH1 neurons (Elizur, 2009). Several diploid fish species have two kisspeptin genes, kiss1 and kiss2, with Kiss2 peptide more effective at stimulating LH and FSH release (Felip et al., 2008). It is well known that the BPG axis regulates development as well as seasonal timing of reproduction in fish. In several teleost species including zebrafish (Danio rerio), fathead minnow (Pimephales promelas), and Nile tilapia (Oreochromis niloticus) (Biran et al., 2008; Filby et al., 2008; Martinez-Chavez et al., 2008), kiss1r mRNA expression increases at the onset of puberty. This matches what is found in mammals, where kisspeptins and Kiss1r regulate GnRH1 neurons through increased activity at puberty (Han et al., 2005). Like puberty, environmental inputs including food availability, annual cycles, and social interactions can regulate the reproductive system, suggesting that they may also impact kisspeptin signaling.
In many social species, the presence of dominant conspecifics suppresses reproductive maturation or competence of non-dominant animals. For example, subordinate male naked mole rats (Heterocephalus glaber) have smaller testes (Faulkes et al., 1991), and subordinate male brown rats (Rattus norvegicus) have decreased testosterone levels and testes size (Blanchard et al., 1995). Among fishes, dominant male siamese fighting fish Beta splendens (Leitz, 1987), stoplight parrotfish (Sparisoma viride) (Cardwell and Liley, 1991), mouthbrooding cichlids (Astatotilapia burtoni) (Francis et al., 1993), rainbow trout (Onchorhynchus mykiss) (Cardwell et al., 1996), and the cooperatively breeding cichlid (Neolamprologus pulcher) (Fitzpatrick et al., 2006) develop larger testes and produce more testosterone than their subordinate conspecifics. Therefore, the use of species with dramatic social regulation of reproductive capacity can provide important information on how kisspeptin signaling may be influenced by the social environment.
We used an African cichlid fish, Astatotilapia burtoni, to understand the effects of social status on Kiss1r. Male A. burtoni exist in two distinct social phenotypes: territorial (T) males that display vivid coloration and aggressive behaviors and comprise approximately 10–30% of the population in their native habitat in the shorepools of Lake Tanganyika, and non-territorial (NT), pale-colored males that make up the remaining 70–90% of the population (Fernald and Hirata, 1977). Reproductive maturation of NT males is inhibited by the presence of T males (Davis and Fernald, 1990; Francis et al., 1993). Subordinate A. burtoni, like subordinate males of rodent and fish species described above, have smaller testes than dominant individuals. In A. burtoni, social dominance also produces changes in hypothalamic GnRH1 neurons, including increased GnRH1 mRNA levels (White et al., 2002) and 50% increases in cell soma area (Davis and Fernald, 1990). To understand the mechanisms responsible for such dramatic plasticity we need to understand which types of neurons and molecules activate GnRH1 neurons in response to social cues.
Information from other species suggests kisspeptin is a likely candidate as a signaling molecule for regulation of GnRH1 activity. To identify the role of kiss1r we: 1) mapped the expression pattern of a kiss1r homolog in the brain of A. burtoni, and; 2) asked how social status affects kiss1r mRNA levels in dominant compared to subordinate males.
Materials and Methods
Animals
African cichlid fish, Astatotilapia burtoni, bred from wild-caught stock, were maintained under conditions simulating those of the natural environment (pH 7.8-8.2, temperature 27±1 °C, 12:12-hour light:dark cycle with full spectrum lighting). Fish were fed shortly after light onset with cichlid pellets and flakes (AquaDine, Healdsburg, CA). All work was performed in compliance with the animal care and use guidelines of the Stanford University Administrative Panel on Laboratory Animal Care.
Male fish (82-94 mm in standard length) were placed in pairs to establish social dominance and behavioral and physiological differences between T and NT males. Two pairs of size- matched T males (n = 11 pairs) from different aquaria were introduced with three females to a new experimental aquarium, identical to their former aquarium except that a perforated divider separated the tank into two equal sections. One pair of males and three to four females were housed on each side of the divider. Fish on one side of a tank could interact chemically and visually, but not physically, with those in the adjacent side of the tank. Within minutes of being transferred to the aquaria, one male on each side asserted his prior T status and the other became NT. Fish were kept for 4 weeks in this new condition to suppress the reproductive axis in the subordinate males (Fox et al., 1997; White et al., 2002). Gonadosomatic index (ratio of gonad mass to body mass) was calculated for all males sacrificed and was significantly higher (two sample t-test, t = 3.856, p = 0.0011, n = 10T, 11NT) in T males than in NT males after excluding one significant outlier, a T male with extremely high GSI measurement (Grubbs's test: p < 0.01). Pairing and behavioral procedures and tissue collection followed protocols described in detail by Korzan et al (2008).
Cloning kiss1r from A. burtoni cDNA
A partial sequence of kiss1r was amplified by polymerase chain reaction from A. burtoni cDNA derived from mRNA isolated from whole brain tissue. Primers were based on conserved regions of kiss1r from a related cichlid fish, Oreochromis niloticus (GenBank accession number BAD34454.1): sense: AACTCCTGTCACGATGTACTCCTCC; antisense: AAACTGTCCCTATCCTTCTTATGT. The reaction products were purified, subcloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and sequenced (Sequetech, Mountain View, CA).
Sequence information from these original clones was used to design gene-specific primers for 5′ and 3′ RACE (rapid amplification of cDNA ends) and subsequently to clone the full-length cDNAs. 5′ RACE and 3′ RACE cDNA were prepared according to the manufacturer's instructions (SMART RACE cDNA Amplification Kit, BD Biosciences, USA) using Advantage 2 polymerase mix (BD Biosciences, USA). Specific primers based on partial A. burtoni sequences of kiss1r were used in combination with the 5′ or 3′ universal (UPM) and nested universal (NUP) primers to amplify the cDNA.
kiss1r sequence and phylogenetic analysis
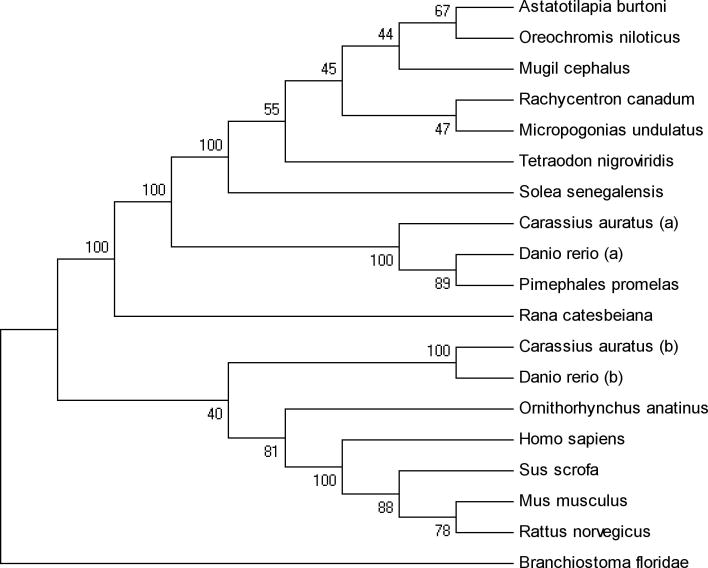
The sequence of A. burtoni Kiss1r (GQ860302) was aligned with those of other species (Clustal W; http://www.ch.embnet.org/software/ClustalW.html), identical amino acids were identified, and phylogenetic analyses were performed to situate A. burtoni Kiss1r among available known sequences. For Kiss1r sequences that were available in GenBank only as DNA, we translated the DNA sequence to protein sequences. The transmembrane regions of the Kiss1r protein sequences were aligned, and a phylogenetic tree was generated by neighbor-joining methods and bootstrap values after 500 replications were calculated using MEGA4.1 (Tamura et al., 2007). Species names and GenBank accession numbers for the Kiss1r homologous sequences are:
Oreochromis niloticus: BAD34454.1; Mus musculus: AAK83236.1; Homo sapiens: AAK83235.1; Rattus norvegicus: AAD19664.1; Rachycentron canadum: ABG82165.1; Micropogonias undulates: ABC75101.1; Solea senegalensis: ABW96362.1; Mugil cephalus: ABG76790.1; Kiss1ra Carassius auratus: ACK77792.1; Kiss1rb Carassius auratus: ACK77793.1; Kiss1ra Danio rerio: NP_001099149.1; Kiss1rb Danio rerio: NP_001104001.1; Rana catesbeiana: ACD44939.1; Sus scrofa: NP_001038089.1; Pimephales promelas: ABV45419.1; Tetraodon nigroviridis: CAG06231.1. The outgroup for this tree was a predicted Kiss1r homolog from the Florida lancet, Branchiostoma floridae: XP_002246008.1.
kiss1r distribution in A. burtoni tissues
To determine the distribution of kiss1r in A. burtoni tissues, PCR was performed on tissue samples from adult A. burtoni (whole brain, olfactory bulbs, pituitary, retina, saccule, gill, kidney, spleen, stomach, intestine, liver, skeletal muscle, heart, testis, ovary, telencephalon, hypothalamus and preoptic area, midbrain, cerebellum, hindbrain, and spinal cord). Tissues were removed, rapidly frozen on dry ice, and homogenized; total RNA was isolated and DNAase- treated using RNeasy kits (Qiagen Inc., Valencia, CA). Approximately 1 μg total RNA for each sample was used for making cDNA with an iScript reverse transcriptase kit (Bio-Rad Laboratories, Hercules, CA). cDNA was diluted 1:20 in nuclease-free water for RT-PCR.
Advantage 2 Polymerase (Clontech, Mountain View, CA) was used for PCR reactions, following Advantage 2 PCR protocol. PCR was carried out on an iCycler thermocycler (Bio-Rad Laboratories, Hercules, CA) using the following reaction conditions: 3 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. PCR products were electrophoretically run alongside Trackit 1kb DNA ladder on a 1% agarose gel containing ethidium bromide and then photographed. Negative controls included running the same procedure without any cDNA tissue template, or without RT, both of which produced no reaction products. The following primers were used to amplify A. burtoni kiss1r (GQ860302): sense: GGAACAAACTTCTCTCTAGGAAGACACGA, antisense: TGTTAAATGATCGGGATCAGTTCACC, giving a product of size 1093 bp. As a positive quality control, A. burtoni-specific primers for the housekeeping gene actin (CN469235) sense: CGCTCCTCGTGCTGTCTTC, antisense: TCTTCTCCATGTCATCCCAGTTG, were used to amplify a 179 bp PCR product in the same samples.
Localization of kiss1r mRNA using in situ hybridization
In situ hybridization followed standard procedures as modified by our laboratory (Burmeister and Fernald, 2005; Grens et al., 2005). Brains were flash frozen in Tissue-Tek OCT compound (Ted Pella, Redding, CA) inside Peel-A-Way plastic molds (Polysciences, Inc., Warrington, PA) on dry ice and stored at -80°C. Brain tissue was sectioned coronally in three series at 14 μm using a Microm HM 550 cryostat (Thermo Scientific, Waltham, MA) and mounted on charged glass slides (VWR) that were then stored at -80°C until use. Slides were brought to room temperature, fixed for 10 minutes in 4% paraformaldehyde in phosphate buffered saline (PBS), rinsed twice for 3 minutes each in PBS, immersed in 0.1 M triethanolamide (TEA) buffer for 3 minutes, acetylated in 0.25% acetic anhydride in 0.1 M TEA for 10 minutes, rinsed twice for 3 minutes each in 2× sodium citrate sodium chloride (SSC) buffer, dehydrated in an ethanol series, and air dried. 35S radioactively labeled probes complementary to A. burtoni kiss1r were diluted to 5 × 106 cpm/ml, and DIG probes complementary to A. burtoni gnrh1, gnrh2, and gnrh3 were diluted to 1 ng/ml in hybridization solution (Sigma-Aldrich, St. Louis, MO) supplemented with 1 g/ml dithiothreitol (DTT). Preheated probe mix was added to each slide and then slides were coverslipped and immersed overnight in a 60°C mineral oil bath. After removing slides from the mineral oil, residual oil was removed by immersion in chloroform. Probe and coverslips were removed in two rinses of 4× SSC, and slides were then washed with 2× SSC with DTT. To detect the GnRH probe, slides were incubated in anti-DIG-peroxidase primary antibody (Roche, Indianapolis, IN), amplified (Tyramide Signal Amplification kit, NEN Life Sciences, Boston, MA), and stained using digoxigenin. Finally, slides were dehydrated in ethanol, air dried, dipped in nuclear emulsion diluted 1:1 in water (NBT-2; Eastman Kodak, Rochester, NY), air dried again, and stored in a light-tight box at 4°C for 3-4 weeks. Following development, slides were stained with cresyl violet, dehydrated in an ethanol/xylene series, and coverslipped.
We used sense versions of the probe to test the specificity of our in situ hybridization, and in no case was signal seen above background. Brain nuclei were identified using published atlases from A. burtoni (Burmeister et al., 2009; Fernald and Shelton, 1985) and sea bass (Cerda-Reverter and Peter, 2003).
In situ hybridization quantitative analysis
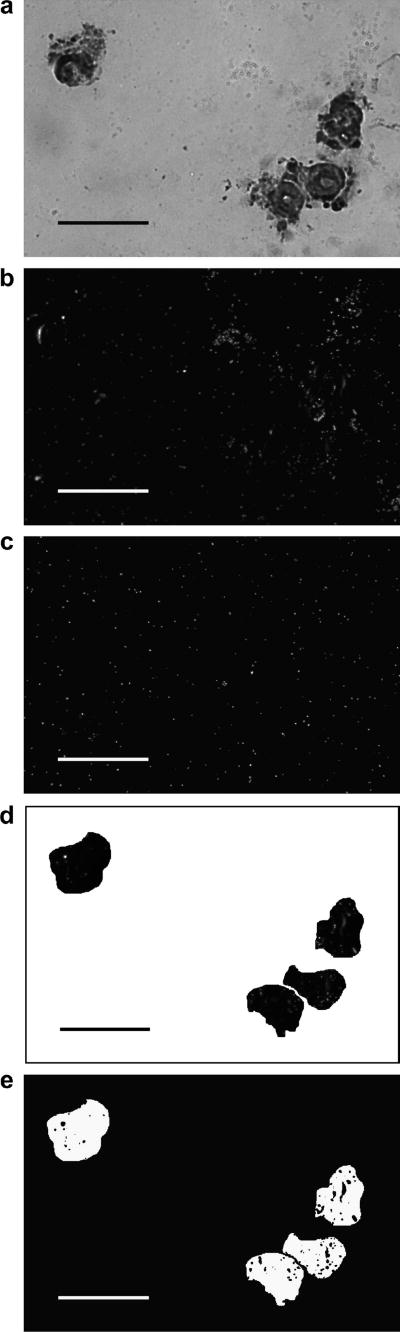
For each brain (5 NT; 4 T), all sections containing GnRH1 neurons and all sections containing GnRH3 neurons were photographed (Zeiss Axioscope; 40× objective). Photomicrographs were captured digitally as uncompressed TIFF files (SPOT camera; Diagnostic Instruments, Sterling Heights, MI). After staining, a series of 3 images was used to quantify kiss1r staining within DIG-labeled GnRH1 and GnRH3 neurons. Brightfield microscopy (Fig. 1a) of coronal sections revealed the cell bodies stained with DIG using the GnRH probe. A darkfield image (Fig. 1b) with the same field of view as each brightfield image was also photographed (Darklite Illuminator; Micro Video Instruments, Avon, MA). Darkfield images revealed some clusters of high silver grain density, reflecting higher radioactive probe binding by kiss1r mRNA. To account for background levels of silver grain signal for each section containing GnRH1 or GnRH3 neurons, another darkfield photograph was also taken of a region of the slide adjacent to the brain section of interest (Fig. 1c). The silver grains in this region were scattered and present at low density, reflective of background levels of activation of the emulsion. To measure silver grain density specifically in the GnRH1 and GnRH3 neurons, neurons were selected from the darkfield image (PhotoShop 7; Adobe, San Jose, CA), using the brightfield image as a guide. This resulted in a grayscale image of only darkened GnRH1 or GnRH3 neurons (Fig. 1d) with light silver grains.
Figure 1.
A series of images used in the analysis of kiss1r mRNA content in one representative section of A. burtoni brain. All scale bars represent 50 μm.
a. Brightfield microscopy of coronal sections revealed the cell bodies stained with DIG using the GnRH1 probe.
b. Darkfield reflected-light image was taken for the same field of view as in 1a. Silver grain clusters reveal kiss1r probe concentration.
c. Background image of a region adjacent to the brain section of interest.
d. Darkfield image of the GnRH1 neurons from Fig. 1a
e. Segmented version of image from Fig. 1d with non-grain and grain regions distinguished and made binary, i.e. black and white. Silver grains are seen as black spots on a white background. Images similar to Fig. 1d were used for subsequent analysis (Fig. 7).
To quantify the silver grain labeling kiss1r in GnRH1 and GnRH3 neurons, images were analyzed using ImageJ software (Rasband, 1997-2009). Images were displayed with contrast inverted so that the silver grains are seen as black spots on a white background. Silver grain images were segregated from background using a K means clustering plugin (http://ij-plugins.sourceforge.net/index.html) (Number of clusters: 4, Cluster center tolerance: 0.0010, Randomization seed: 48). ImageJ was then used to convert the image to binary (Fig. 1e). Using the “Analyze Particle” function, cell area was measured and recorded using these parameters: Size pixelˆ2: 200-infinity and Circularity: 0.0 – 1.0. Silver grains were measured and recorded using the following parameters: Size pixelˆ2: 0-100 and Circularity: 0.0 – 1.0.
For all darkfield images, we recorded the total number and total area of silver grains, the total number and total area of cells, and the fraction of the area containing cells that also contained silver grains. Total number and total area of silver grains were also recorded for the control darkfield image (as seen in Fig. 1c.) The camera captured 1.92 × 104 pixels per image, so to make background silver grain amounts comparable to cell silver grain amounts we divided cell area (in units of pixels) by 1.92 × 104. To account for background levels of silver grains in each brain section, we subtracted a factor incorporating background grains amount and cell area from the measured grain amounts as follows:
Normalized Grain Number = Total Grain Number – (Background Grain Number)*(Cell Pixels/1.92 × 104)
This procedure gave a metric of silver grain number for the GnRH1 and GnRH3 neurons in each brain. To confirm the accuracy of our automated grain quantification, the number of grains was also counted manually for comparison. From images displayed on a computer monitor (Dell Inspiron LCD), the number of grains in each image was counted manually for 41 section images and the product-moment correlation coefficient of the manual and automated count was r = 0.92 (p < 0.00001). Normalized Grain Number values were compared between T and NT males and between GnRH1 and GnRH3 neurons using t-Tests.
Quantitative RT-PCR (qRT-PCR)
kiss1r mRNA levels were measured by quantitative reverse transcription-PCR (qRT-PCR) with the MyIQ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Animals were sacrificed by rapid cervical transection, and their brains removed and frozen in Qiagen lysis buffer for whole brain analysis. Total RNA free from genomic DNA contamination was extracted (Qiagen RNeasy Plus Mini and Micro Kits, Qiagen, Valencia, CA), and 1 μg total RNA for each sample was used to make cDNA by reverse transcription (iScript cDNA kit, Bio-Rad Laboratories, Hercules, CA). cDNA was diluted 1:20 in nuclease-free water for qRT-PCR.
Quantitative RT-PCR primers were designed with Vector NTi (Invitrogen, Carlsbad, CA) and PrimerQuest (Integrated DNA Technologies, Coralville, IA) software to be complementary to A. burtoni kiss1r (GQ860302), g3pdh (AF123727) or 18S rRNA (U67333). All qRT-PCR primers were designed to avoid dimers or hairpin structures and to have similar melting temperatures (ca. 60°C) and GC content (ca. 50% of residues). Primers for g3pdh and 18SrRNA were identical to those used previously for A. burtoni (Zhao and Fernald, 2005). The following primers were used to amplify A. burtoni kiss1r (GQ860302): sense: CGTGACAGTCTACCCCCTGAA, antisense: TCCAAATGCAAATGCTGACAA, giving a product of size 77 bp.
qRT-PCR was performed in 30-μL duplicate reactions with 1× IQ SYBR Green Supermix (Bio-Rad Laboratories), 0.25 μM of each primer, and 2.5 ng/μL cDNA (RNA equivalent). PCR parameters were as follows: 5 min at 95 °C followed by 40 cycles of 30 sec at 95 °C, 30 sec at 60 °C, and 30 sec at 72 °C, followed by melt curve analysis. Each amplicon was specifically amplified, as demonstrated by single peaks in melt curves for each reaction product. Fluorescence data from MyIQ were processed using Real-time PCR Miner software (Zhao and Fernald, 2005) to calculate threshold cycle number (CT) and amplification efficiency for each sample. CT values were below 30 for all reactions. Comparison between T and NT whole brains revealed no differences in expression levels of the housekeeping genes g3pdh (t-Test: paired two sample for means; p = 0.452; t stat = −0.796; n = 8 per group) and 18S rRNA (t-Test: paired two sample for means; p = 0.326; t stat = −1.057; n = 8 per group), indicating they are appropriate reference genes for this study. To normalize kiss1r gene concentration, kiss1r values for each sample were divided by the geometric mean of g3pdh and 18S rRNA concentration values. Mean normalized kiss1r values were compared between T and NT males using paired t-Tests.
Results
Kiss1r sequence and phylogenetic analysis
Using primers designed based on the Oreochromis niloticus kiss1r cDNA sequence, we amplified cDNA from the Astatotilapia burtoni library and cloned it (TOPO vector). The A. burtoni Kiss1r transmembrane protein sequence was highly homologous (99.7% identical) with the O. niloticus sequence and also homologous with Kiss1r in other species (Fig. 2). The A. burtoni form was 99.0% identical with the mullet, Mugil cephalus, 97.9% identical with the cobia, Rachycentron canadum; 97.2% identical with the Atlantic croaker, Micropogonias undulates; and 97.2% identical with the Senegalese sole, Solea senegalensis. The sequence of A. burtoni Kiss1r is more similar to that of the Kiss1ra gene in the goldfish Carassius auratus and zebrafish Danio rerio, than to the Kiss1rb paralog in these two species (Fig. 2).
Figure 2.
Phylogenetic tree of Kiss1r predicted amino acid sequences shows fish and tetrapod lineages, with the lancet, Branchiostoma floridae, as an outgroup. The transmembrane region of the translated protein sequence of A. burtoni Kiss1r was aligned with known Kiss1r sequences, and the phylogenetic tree was generated by neighbor-joining methods using MEGA4.1. Calculated bootstrap values (after 500 replications) are shown (MEGA4.1).
kiss1r distribution in A. burtoni tissues
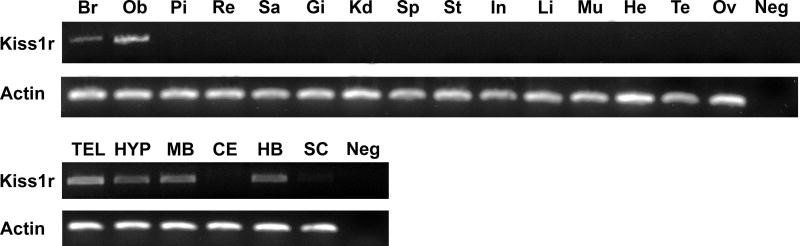
We analyzed expression of kiss1r mRNA in several tissues using RT-PCR (Fig. 3). Of the tissues examined, only the brain and olfactory bulb showed levels of kiss1r mRNA that were detectable by our assay. In the brain, the telencephalon, hypothalamus (including the preoptic area), midbrain, and hindbrain expressed kiss1r, but the cerebellum and spinal cord did not show detectable levels. No kiss1r mRNA was detectable in pituitary, retina, saccule of the inner ear, gill, kidney, spleen, stomach, intestine, liver, skeletal muscle, heart, testis, or ovary.
Figure 3.
kiss1r mRNA transcripts are present in several A. burtoni tissues and brain regions. Semiquantitative gel analysis of PCR products from cDNA reverse transcribed using RNA from whole brain (Br), olfactory bulbs (Ob), pituitary (Pi), retina (Re), saccule (Sa), gill (Gi), kidney (Kd), spleen (Sp), stomach (St), intestine (In), liver (Li), skeletal muscle (Mu), heart (He), testis (Te), ovary (Ov), telencephalon (TEL), hypothalamus and preoptic area (HYP), midbrain (MB), cerebellum (CE), hindbrain (HB), and spinal cord (SC). Negative control (Neg) column is the PCR without template tissue cDNA. Upper rows are PCR products obtained with A. burtoni kiss1r-specific primers, and lower rows are products from actin-specific primers from the same tissue samples.
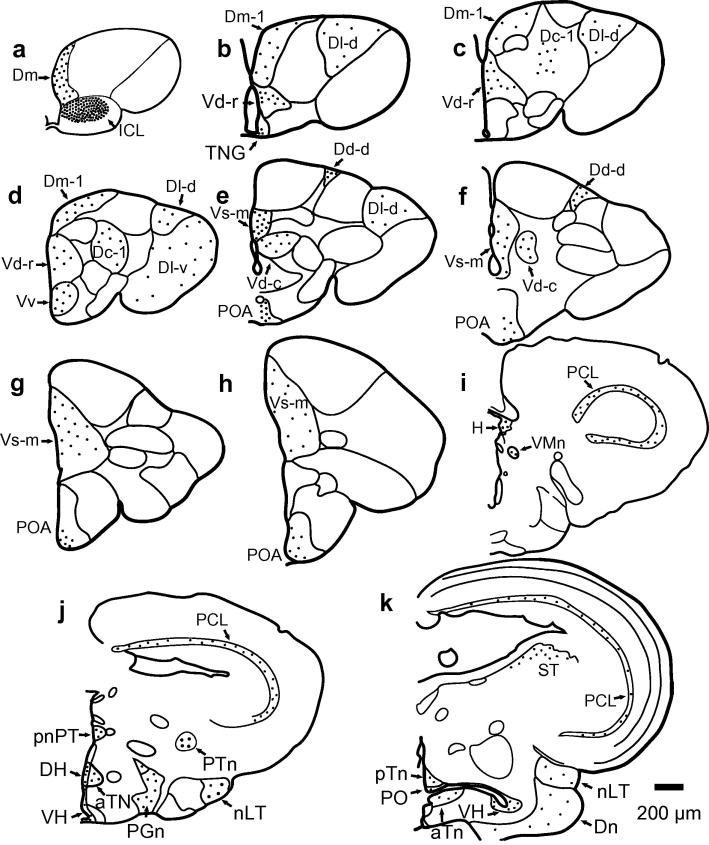
kiss1r localization in the brain
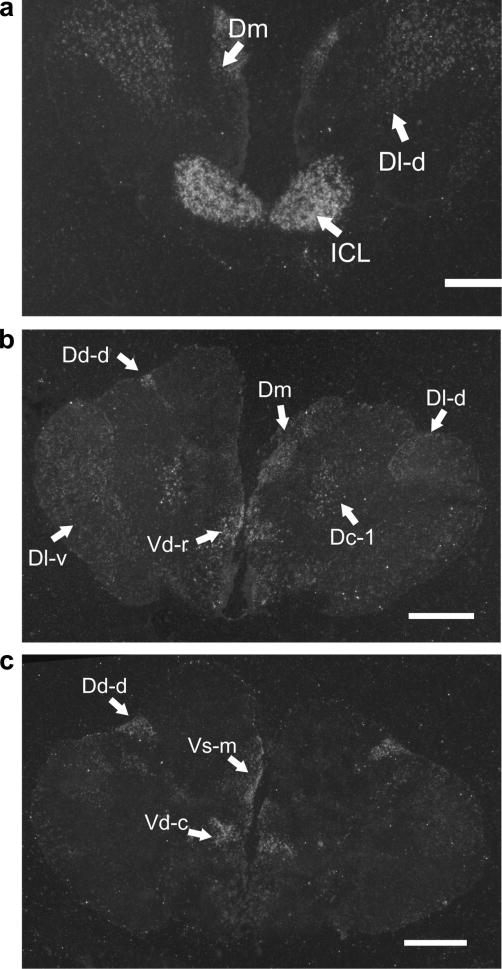
To identify which nuclei in the A. burtoni brain express kiss1r mRNA, we probed coronal brain sections with an S35 labeled RNA probe. kiss1r mRNA hybridized in several regions throughout the brain in both territorial and non-territorial male A. burtoni (Figs. 4 and 5). Strong expression was observed throughout the entire internal cellular layer (ICL) of the olfactory bulb. Telencephalic nuclei (Burmeister et al., 2009) containing kiss1r mRNA detectable by the in situ probe in the dorsal telencephalon (D) included the terminal nerve ganglion (TNG), part 1 of medial division of D (Dm-1), ventral subdivision of lateral division of D (Dl-v), dorsal subdivision of lateral division of D (Dl-d), part 1 of central division of D (Dc-1), and dorsal subdivision of dorsal division of D (Dd-d). In the ventral telencephalon, kiss1r mRNA was detected in medial part of Vs (Vs-m), rostral part of dorsal division of V (Vd-r), caudal part of dorsal division of V (Vd-c), ventral division of V (Vv). In the diencephalon, kiss1r signal was found in the preoptic area (POA), periventricular organ of the posterior tuberculum (pnPT), habenular nucleus (H), ventromedial thalamic nucleus (VMn), posterior thalamic nucleus (PTn, located slightly more rostrally than shown previously (Fernald and Shelton, 1985)),dorsal hypothalamus (DH), ventral hypothalamus (VH), diffuse nucleus of the inferior lobe (Dn), anterior tuberal nucleus (aTn), paraventricular organ (PO), posterior tuberal nucleus (pTn), preglomerular nucleus (PGn), and nucleus of the lateral torus (nLT). In the mesencephalon, kiss1r signal was found in the torus semicircularis (ST), and periventricular cell layer of the tectum. In the rhombencephalon, kiss1r signal was found in the raphe nucleus, reticular formation, and vagal lobe.
Figure 4.
In situ hybridization shows kiss1r mRNA localization in the brain. Photographs of 14 μm rostral to caudal coronal sections of A. burtoni brain sections were taken using darkfield illumination. Scale bars represent 1 mm.
Figure 5.
Illustration of kiss1r mRNA expression in olfactory bulb, telencephalon, diencephalon, and mesencephalon. Dots indicate location and relative density of silver grains. (a-h) Illustration of kiss1r expression pattern in the olfactory bulb and telencephalon of A. burtoni (Brain outlines from Burmeister et al., 2009). (i-k) Illustration of kiss1r expression pattern in the diencephalon and mesencephalon of A. burtoni (Brain outlines from Fernald and Shelton, 1985). Scale bar represents 200 μm.
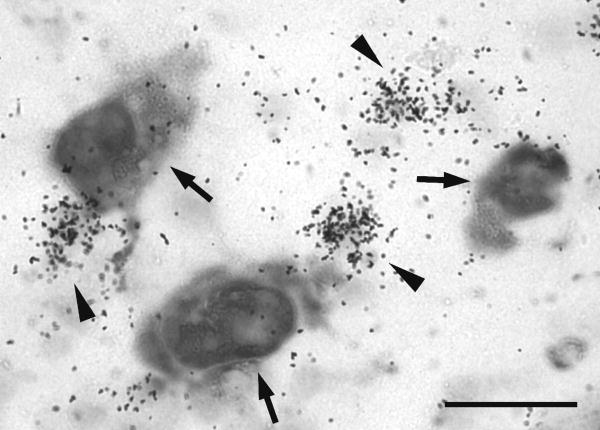
GnRH1 and GnRH3 neurons both expressed kiss1r mRNA. No GnRH2 neurons were observed to express kiss1r mRNA above background levels. Additionally, we discovered a population of cells in the POA that expressed kiss1r mRNA but not GnRH1 (Fig. 6). Because these cells were not stained with DIG, we did not quantify silver grains in this population as we did for the GnRH1 neurons. Nonetheless, we observed that this silver grain staining was more densely clustered than that seen in GnRH1 neurons although the identity of these cells remains unknown.
Figure 6.
Some cells in the preoptic area express kiss1r but not gnrh1. Photograph in the preoptic area of a non-territorial male stained for gnrh1 mRNA with DIG (large cells) and kiss1r mRNA with 35S-labeled probe (small black dots). GnRH1 neurons are indicated with arrows. Clusters of silver grains representing non-GnRH1 neurons in the POA are indicated by arrowheads. Scale bar represents 50 μm.
Abundance of Kiss1r mRNA in the brain as a function of social status
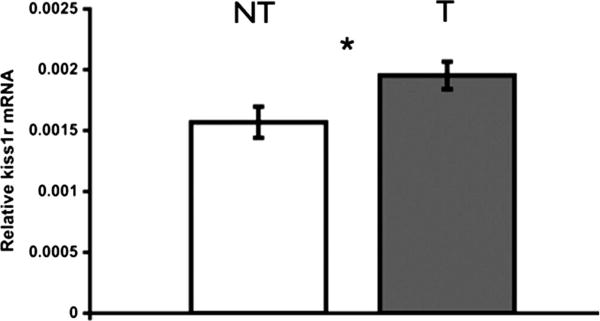
Kiss1r mRNA levels measured by qRT-PCR in the whole brain of males differed with social status. Territorial males had higher Kiss1r mRNA levels compared to non-territorial males (two sample t-test, t = −2.412, p = 0.0467, n = 8 per group) (Fig. 7).
Figure 7.
Territorial males (T) have higher levels of kiss1r mRNA in the brain compared to non-territorial males (NT) (paired two sample t-test, p = 0.0467, n = 8T, 8NT). Data shown are whole brain mean ± SE transcript levels relative to the geometric mean of two housekeeping genes (g3pdh and 18srrna).
Comparison of GnRH1 and GnRH3 neuron kiss1r expression
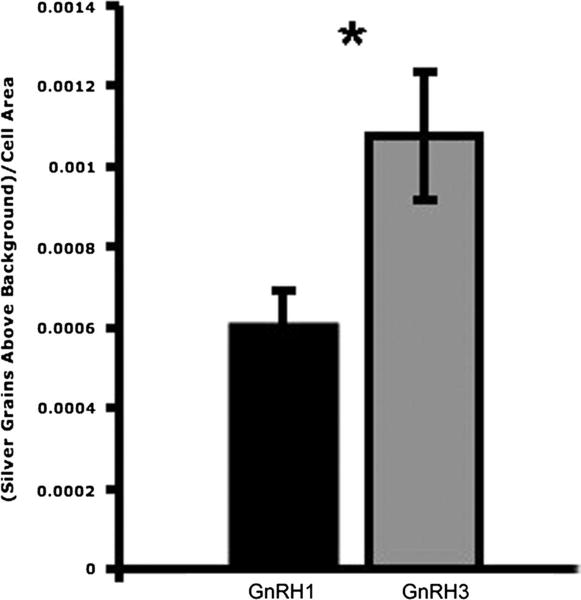
From a series of darkfield and brightfield images, kiss1r expression was quantified in both GnRH1 neurons in the POA and GnRH3 neurons in the forebrain using ImageJ software (Fig. 1). kiss1r levels in GnRH1 neurons of T males (n = 6) and NT males (n = 5) were measured from darkfield images of silver grains following autoradiographic in situ hybridization, and no significant difference was found between T and NT males (two sample t-test, t = −0.650; n = 5T, 5NT, p = 0.534). There was also no significant difference in kiss1r expression in GnRH3 neurons between T males and NT males (two sample t-test, t = 0.631, p = 0.545, n = 6T, 4NT). A significantly higher density of kiss1r silver grains was found in GnRH3 neurons compared to GnRH1 neurons (two sample t-test, t = −2.325, p = 0.034, n = 9) (Fig. 8).
Figure 8.
Significantly more kiss1r was found in GnRH3 neurons compared to GnRH1 neurons (two sample t-test, t = −2.325, p = 0.034, n = 9). Silver grains on cells from T and NT males were counted using ImageJ and normalized by subtracting background silver grain density, as illustrated in Fig. 1.
Discussion
These experiments confirm expression of kiss1r mRNA in GnRH1 and GnRH3 neurons, and they reveal a fairly extensive distribution of kiss1r throughout the brain, suggesting widespread effects of kisspeptin. We found expression in novel cell populations, including cells in the ICL of the olfactory bulb, non-GnRH1 cells in the POA, and cells in several nuclei of the telencephalon, diencephalon, mesencephalon, and rhombencephalon. Furthermore, we identify higher kiss1r mRNA levels in the whole brain in dominant males compared to social subordinates, which suggests a role for kisspeptin receptor in mediating the reproductive potentiation that characterizes dominant male A. burtoni.
Kiss1r Sequence and Phylogeny
Multiple kiss1r genes have been identified in several fish species, including zebrafish (Biran et al., 2008), medaka (Lee et al., 2009), and goldfish (Li et al., 2009), as well as in platypus and clawed frog. Mechaly et al. (2010) suggest that one form of kiss1r has been lost in fishes of the orders Pleuronectiformes and Tetraodontiformes. Genome sequences of fugu, pufferfish and three-spined stickleback reveal only one kiss1r gene. It seems likely that cichlids, belonging to the Order Perciformes, may also have only one kiss1r gene, but establishing this possibility as fact requires further confirmation. The predicted Kiss1r protein sequence identified in A. burtoni is phylogenetically more closely related to the Kiss1ra paralogs and the Kiss1r found in Rana catesbeiana, a tetrapod, than to the Kiss1rb paralogs. The identification of a single kiss1r gene in several teleosts and tetrapods is consistent with a gene duplication in cyprinids, including C. auratus and D. rerio, that is not found in the tetrapod or other teleost lineages. Since we know that the actinopterygian lineage did experience a duplication, our tree is consistent with current schema about vertebrate evolution (Hoegg et al., 2004). Multiple kiss1r splice variants do exist in several fish species, however, including Senegalese sole (Solea senegalensis) (Mechaly et al., 2009) and Atlantic halibut (Hippoglussus hippoglossus) (Mechaly et al., 2010). Thus, the possibility remains that multiple splice forms may also exist in A. burtoni.
kiss1r distribution in A. burtoni tissues
kiss1r mRNAs are expressed in different tissues in different species. For example, in zebrafish, Danio rerio, kiss1ra is expressed mostly in the brain and gonads (Biran et al., 2008). D. rerio kiss1rb, on the other hand, is expressed in brain and pituitary as well as spleen, gills, kidney, intestine and pancreas, muscle, and adipose tissue. The A. burtoni Kiss1r protein is more closely related in sequence to D. rerio Kiss1ra than to D. rerio Kiss1rb. The expression pattern of A. burtoni kiss1r, however, is more similar to D. rerio kiss1rb. This discrepancy between sequence and expression pattern may suggest that the D. rerio Kiss1r genes have diverged in their functions throughout the body since the duplication event occurred in the D. rerio lineage.
kiss1r mRNA localization in the brain
In situ hybridization staining revealed kiss1r mRNA in GnRH1-expressing neurons of the POA, consistent with previous work showing that GnRH1 neurons express Kiss1r in mice (Messager et al., 2005), rats (Irwig et al., 2004), and Nile tilapia (Parhar et al., 2004). This result supports the hypothesis that kisspeptin could regulate GnRH1 directly in teleosts, particularly A. burtoni. Our results also show that kiss1r mRNA is located throughout the brain and not exclusively in GnRH1 neurons. Filby et al (2008) used qRT-PCR on macrodissected brain regions of the fathead minnow, Pimephales promelas, and found high expression of kiss1r in olfactory bulb, telencephalon and preoptic area, optic tectum, and a combined region that included hypothalamus, thalamus, and midbrain tegmentum. Our data provide important new information about where kiss1r expression occurs within specific neuroanatomically-defined nuclei in the teleost brain.
Expression of kiss1r in olfactory bulb suggests a role for kisspeptin in chemical communication or other olfactory roles. We found that the internal cellular layer (ICL) of the olfactory bulb in A. burtoni strongly expresses kiss1r (Figs. 4 and 5). The olfactory bulb in teleosts, including cichlids, is essential for many kinds of behavior, including feeding, homing, reproduction, territoriality, and predator avoidance (reviewed by Hara et al. (1993)). In particular, pheromonally-mediated reproductive behavior is controlled by mitral cells, whose axons form the lateral part of the medial olfactory tract (Hamdani, 2007; Satou et al., 1984). The small, densely packed granule cells of the ICL inhibit mitral cells in the mitral cell layer (MCL) (Satou, 1990; Satou et al., 1984). The granule cells are thought to mediate a form of lateral inhibition that may help distinguish closely related stimuli (Laberge and Hara, 2001). Inputs come, for example, from corticotrophin releasing factor (CRF), whose receptors are expressed in ICL cells (Van Pett et al., 2000); neuropeptide Y (NPY), which increases electrical activity of the olfactory epithelium in hungry axolotls (Ambystoma mexicanum) (Mousley et al., 2006); and GnRH3 neurons in the terminal nerve ganglion (Eisthen et al., 2000). Other reproductive neuropeptides are received by the olfactory bulb in A. burtoni. For example, GnRH-immunoreactive fibers project to the olfactory bulb, and GnRH-R2, the receptor type expressed by GnRH-releasing neurons in the brain, is also localized in the olfactory bulb (Chen and Fernald, 2006). Olfaction is thought to be important for reproduction in male A. burtoni, as their olfactory system responds to steroid cues in the water (Cole and Stacey, 2006; Robison et al., 1998), and they respond behaviorally to olfactory cues, increasing their courtship when chemicals from female conspecifics are present (Caprona, 1974). The high levels of kiss1r mRNA in the ICL suggest that kisspeptin could modulate responsiveness or precision of olfactory input by influencing granule cells' activity.
Kisspeptin signaling in the forebrain may also influence memory. We found kiss1r mRNA in the telencephalon in both Dorsolateral (Dl) and Dorsomedial (Dm) nuclei. In rats, kiss1r mRNA is expressed in hippocampus and amygdala, functionally similar to Dl and Dm, respectively (Lee et al., 1999). Dl lesions in goldfish (Carrasius auratus) have shown this region to be necessary for learning tasks that have temporal and emotional components (Portavella et al., 2002). Experiments indicated that goldfish Dm is involved specifically in learning tasks involving an aversive component. More recent work, however, suggests that goldfish do use Dm for spatial memory of extra-maze cues (Saito, 2006). The localization of Kiss1r in the forebrain suggests future functional studies to test the possibility that kisspeptin signaling is involved with spatial and emotional memory pathways in fish.
Finding non-GnRH1 kiss1r neuron populations in the POA raises the question of other potential roles for kisspeptin in neuroendocrine pathways. Other neuropeptidergic neurons besides GnRH1-expressing cells in the POA of A. burtoni have been linked to social status and endocrine changes. Somatostatin (SS), for example, a neuropeptide involved in inhibition of somatic growth, is expressed in a population of neurons in the POA (Hofmann and Fernald, 2000). These SS neurons are responsive to social status in male A. burtoni, increasing in size in territorial males compared to non-territorial males. Arginine vasotocin (AVT) is expressed exclusively by a population of neurons in the POA and hypothalamus of A. burtoni (Greenwood et al., 2008), and neurons in the anterior POA contain higher AVT mRNA levels in territorial males than in non-territorial males. Since GnRH1, AVT, and SS neurons in the POA interact, it may be that kisspeptin also has a gonadotropin-releasing effect through some intermediate signaling pathway that reaches GnRH1 cells indirectly. Alternatively, Kiss1r in non-GnRH1 POA neurons may serve non-gonadotropin-releasing roles, for instance in modulating behavioral circuits or regulating other endocrine pathways.
Abundance of Kiss1r mRNA in the brain as a function of social status
Because of similarities between GnRH1 neuron responses to puberty and to the transition from subordination to social dominance, cellular phenomena discovered in the context of social dominance may shed light on mechanisms occurring during puberty. Similar to changes seen during sexual maturation, social ascension in male A. burtoni leads to increased gnrh1 mRNA levels as well as dramatic changes in GnRH1 neuron morphology (Fernald, 2009), raising the possibility of parallel mechanisms between social transition and sexual maturation. In other species, including both fish and mammals, levels of kiss1r gene expression increase around the time of puberty. Levels of kiss1r are greater in dominant A. burtoni T males compared to non- dominant NT males, and males can switch between social statuses in both directions. If the increase we have shown in kiss1r expression is accompanied by increases in kisspeptin signaling to GnRH1 neurons or increased transcription of gnrh1, then reversible changes in kiss1r levels could lead to the known GnRH1 plasticity associated with social transition. Since we identified a difference between T and NT males in whole brain kiss1r levels, but not in levels of kiss1r found in GnRH1 neurons, the population(s) of cells in the brain that increase kiss1r expression along with social dominance requires future study.
Many factors known to be important for activation of reproduction can influence kisspeptin signaling. Kisspeptin levels are modulated in diverse species by internal states including pregnancy (Roa et al., 2006), nutrition (Forbes et al., 2009; Wu et al., 2009), and circadian rhythms (Robertson et al., 2009) as well as by environmental signals including, in seasonal breeders, photoperiod length (Clarke et al., 2009; Simonneaux et al., 2009). Several types of stressors can impact kiss1 and kiss1r expression. For example, in female rats, three stressors that decrease LH pulse frequency (restraint, hypoglycaemia, and lipopoplysaccarhide) decreased kiss1r mRNA levels (Kinsey-Jones et al., 2009). Our study suggests that social cues may also play an important role in kisspeptin signaling in fish and potentially in other species that rely on kisspeptin for development and activation of gonadotrophs and the reproductive axis.
Acknowledgments
The authors extend thanks to Chun-Chun Chen, Geetanjali Chakraborty, April Zhang, and Helen McCurdy for technical assistance and to Katharine Mach, Julie Desjardins, and two anonymous reviewers for valuable comments on this manuscript. This work was funded by NIH NS 34950 to R.D.F., F32NS061431 to K.P.M., and 5F32MH074222 to W.J.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Akazome Y, Kanda S, Okubo K, Oka Y. Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain. Journal of Fish Biology. 2010;76(1):161–182. doi: 10.1111/j.1095-8649.2009.02496.x. [DOI] [PubMed] [Google Scholar]
- Biran J, Ben-Dor S, Levavi-Sivan B. Molecular identification and functional characterization of the kisspeptin/kisspeptin receptor system in lower vertebrates. Biol Reprod. 2008;79(4):776–786. doi: 10.1095/biolreprod.107.066266. [DOI] [PubMed] [Google Scholar]
- Blanchard DC, Spencer RL, Weiss SM, Blanchard RJ, McEwen B, Sakai RR. Visible burrow system as a model of chronic social stress: behavioral and neuroendocrine correlates. Psychoneuroendocrinology. 1995;20(2):117–134. doi: 10.1016/0306-4530(94)e0045-b. [DOI] [PubMed] [Google Scholar]
- Burmeister SS, Fernald RD. Evolutionary conservation of the egr-1 immediate-early gene response in a teleost. J Comp Neurol. 2005;481(2):220–232. doi: 10.1002/cne.20380. [DOI] [PubMed] [Google Scholar]
- Burmeister SS, Munshi RG, Fernald RD. Cytoarchitecture of a Cichlid Fish Telencephalon. Brain Behav Evol. 2009;74(2):110–120. doi: 10.1159/000235613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprona MDCd. The Effect of Chemical Stimuli from Conspecifics on the Behavior of Haplochromis burtoni (Cichlidae, Pisces) Cellular and Molecular Life Sciences (CMLS) 1974;30(December):1394–1395. doi: 10.1007/BF01919654. [DOI] [PubMed] [Google Scholar]
- Cardwell JR, Liley NR. Androgen Control of Social Status in Males of a Wild Population of Stoplight Parrotfish, Sparisoma viride (Scaridae) Hormones and Behavior. 1991;25:1–18. doi: 10.1016/0018-506x(91)90035-g. [DOI] [PubMed] [Google Scholar]
- Cardwell JR, Sorensen PW, Kraak GJVD, Liley NR. Effect of Dominance Status on Sex Hormone Levels in Laboratory and Wild-Spawning Male Trout. General and Comparative Endocrinology. 1996;101:333–341. doi: 10.1006/gcen.1996.0036. [DOI] [PubMed] [Google Scholar]
- Cerda-Reverter JM, Peter RE. Endogenous melanocortin antagonist in fish: structure, brain mapping, and regulation by fasting of the goldfish agouti-related protein gene. Endocrinology. 2003;144(10):4552–4561. doi: 10.1210/en.2003-0453. [DOI] [PubMed] [Google Scholar]
- Chen CC, Fernald RD. Distributions of two gonadotropin-releasing hormone receptor types in a cichlid fish suggest functional specialization. J Comp Neurol. 2006;495(3):314–323. doi: 10.1002/cne.20877. [DOI] [PubMed] [Google Scholar]
- Clarke IJ, Smith JT, Caraty A, Goodman RL, Lehman MN. Kisspeptin and seasonality in sheep. Peptides. 2009;30(1):154–163. doi: 10.1016/j.peptides.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, d'Anglemont X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. Journal of Neuroscience. 2008;28(35):8691–8697. doi: 10.1523/JNEUROSCI.1775-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TB, Stacey NE. Olfactory responses to steroids in an African mouth-brooding cichlid, Haplochromis burtoni(Gunther) Journal of Fish Biology. 2006;68(3):661–680. [Google Scholar]
- d'Anglemont de Tassigny X, Fagg LA, Dixon JP, Day K, Leitch HG, Hendrick AG, Zahn D, Franceschini I, Caraty A, Carlton MB, Aparicio SA, Colledge WH. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci U S A. 2007;104(25):10714–10719. doi: 10.1073/pnas.0704114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MR, Fernald RD. Social control of neuronal soma size. J Neurobiol. 1990;21(8):1180–1188. doi: 10.1002/neu.480210804. [DOI] [PubMed] [Google Scholar]
- Demski LS. The Evolution of Neuroanatomical Substrates of Reproductive Behavior: Sex Steroid and LHRH-Specific Pathways Including the Terminal Nerve. American Zoologist. 1984;24(3):809–830. [Google Scholar]
- Eisthen HL, Delay RJ, Wirsig-Wiechmann CR, Dionne VE. Neuromodulatory effects of gonadotropin releasing hormone on olfactory receptor neurons. J Neurosci. 2000;20(11):3947–3955. doi: 10.1523/JNEUROSCI.20-11-03947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizur A. The KiSS1/GPR54 system in fish. Peptides. 2009;30(1):164–170. doi: 10.1016/j.peptides.2008.08.018. [DOI] [PubMed] [Google Scholar]
- Faulkes CG, Abbott DH, Jarvis JUM. Social suppression of reproduction in male naked mole-rats, Heterocephalus glaber. J Reprod Fert. 1991;91:593–604. doi: 10.1530/jrf.0.0910593. [DOI] [PubMed] [Google Scholar]
- Felip A, Zanuy S, Pineda R, Pinilla L, Carrillo M, Tena-Sempere M, Gomez A. Evidence for two distinct KiSS genes in non-placental vertebrates that encode kisspeptins with different gonadotropin-releasing activities in fish and mammals. Mol Cell Endocrinol. 2008 doi: 10.1016/j.mce.2008.11.017. [DOI] [PubMed] [Google Scholar]
- Fernald RD. Social Regulation of Reproduction: What Changes and Why? Hormones, Brain and Behavior. 2. Maryland Heights, MO: Elsevier; 2009. pp. 683–691. [Google Scholar]
- Fernald RD, Hirata NR. Field study of Haplochromis burtoni: habitats and co-habitant. Environmental Biology of Fishes. 1977;2(December):299–308. [Google Scholar]
- Fernald RD, Shelton LC. The organization of the diencephalon and the pretectum in the cichlid fish, Haplochromis burtoni. J Comp Neurol. 1985;238(2):202–217. doi: 10.1002/cne.902380207. [DOI] [PubMed] [Google Scholar]
- Filby AL, van Aerle R, Duitman J, Tyler CR. The kisspeptin/gonadotropin-releasing hormone pathway and molecular signaling of puberty in fish. Biol Reprod. 2008;78(2):278–289. doi: 10.1095/biolreprod.107.063420. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JL, Desjardins JK, Stiver KA, Montgomerie R, Balshine S. Male reproductive suppression in the cooperatively breeding fish Neolamprologus pulcher. Behavioral Ecology. 2006;17(1):25–33. [Google Scholar]
- Forbes S, Li XF, Kinsey-Jones J, O'Byrne K. Effects of ghrelin on Kisspeptin mRNA expression in the hypothalamic medial preoptic area and pulsatile luteinising hormone secretion in the female rat. Neurosci Lett. 2009;460(2):143–147. doi: 10.1016/j.neulet.2009.05.060. [DOI] [PubMed] [Google Scholar]
- Fox HE, White SA, Kao MH, Fernald RD. Stress and dominance in a social fish. J Neurosci. 1997;17(16):6463–6469. doi: 10.1523/JNEUROSCI.17-16-06463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis RC, Soma K, Fernald RD. Social regulation of the brain-pituitary-gonadal axis. Proc Natl Acad Sci U S A. 1993;90(16):7794–7798. doi: 10.1073/pnas.90.16.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottsch ML, Clifton DK, Steiner RA. From KISS1 to kisspeptins: An historical perspective and suggested nomenclature. Peptides. 2009;30(1):4–9. doi: 10.1016/j.peptides.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AK, Wark AR, Fernald RD, Hofmann HA. Expression of arginine vasotocin in distinct preoptic regions is associated with dominant and subordinate behaviour in an African cichlid fish. Proc Biol Sci. 2008;275(1649):2393–2402. doi: 10.1098/rspb.2008.0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens KE, Greenwood AK, Fernald RD. Two visual processing pathways are targeted by gonadotropin-releasing hormone in the retina. Brain Behav Evol. 2005;66(1):1–9. doi: 10.1159/000085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdani EH, Doving Kjell B. The functional organization of the fish olfactory system. Prog Neurobiol. 2007;82:80–86. doi: 10.1016/j.pneurobio.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Han SK, Gottsch ML, Lee KJ, Popa SM, Smith JT, Jakawich SK, Clifton DK, Steiner RA, Herbison AE. Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty. Journal of Neuroscience. 2005;25(49):11349–11356. doi: 10.1523/JNEUROSCI.3328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara TJ. Role of olfaction in fish behavior. In: Pitcher TJ, editor. Behaviour of teleost fishes. 1. New York: Chapman & Hall; 1993. [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59(2):190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hofmann HA, Fernald RD. Social status controls somatostatin neuron size and growth. J Neurosci. 2000;20(12):4740–4744. doi: 10.1523/JNEUROSCI.20-12-04740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwig MS, Fraley GS, Smith JT, Acohido BV, Popa SM, Cunningham MJ, Gottsch ML, Clifton DK, Steiner RA. Kisspeptin activation of gonadotropin releasing hormone neurons and regulation of KiSS-1 mRNA in the male rat. Neuroendocrinology. 2004;80(4):264–272. doi: 10.1159/000083140. [DOI] [PubMed] [Google Scholar]
- Kah O, Lethimonier C, Somoza G, Guilgur LG, Vaillant C, Lareyre JJ. GnRH and GnRH receptors in metazoa: a historical, comparative, and evolutive perspective. Gen Comp Endocrinol. 2007;153(1-3):346–364. doi: 10.1016/j.ygcen.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Keen KL, Wegner FH, Bloom SR, Ghatei MA, Terasawa E. An increase in kisspeptin-54 release occurs with the pubertal increase in luteinizing hormone-releasing hormone-1 release in the stalk-median eminence of female rhesus monkeys in vivo. Endocrinology. 2008;149(8):4151–4157. doi: 10.1210/en.2008-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey-Jones JS, Li XF, Knox AM, Wilkinson ES, Zhu XL, Chaudhary AA, Milligan SR, Lightman SL, O'Byrne KT. Down-regulation of hypothalamic kisspeptin and its receptor, Kiss1r, mRNA expression is associated with stress-induced suppression of luteinising hormone secretion in the female rat. J Neuroendocrinol. 2009;21(1):20–29. doi: 10.1111/j.1365-2826.2008.01807.x. [DOI] [PubMed] [Google Scholar]
- Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Brain Res Rev. 2001;36(1):46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Lee DK, Nguyen T, O'Neill GP, Cheng R, Liu Y, Howard AD, Coulombe N, Tan CP, Tang-Nguyen AT, George SR, O'Dowd BF. Discovery of a receptor related to the galanin receptors. FEBS Lett. 1999;446(1):103–107. doi: 10.1016/s0014-5793(99)00009-5. [DOI] [PubMed] [Google Scholar]
- Lee YR, Tsunekawa K, Moon MJ, Um HN, Hwang JI, Osugi T, Otaki N, Sunakawa Y, Kim K, Vaudry H, Kwon HB, Seong JY, Tsutsui K. Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology. 2009;150(6):2837–2846. doi: 10.1210/en.2008-1679. [DOI] [PubMed] [Google Scholar]
- Leitz T. Social Control of Testicular Steroidogenic Capacities in the Siamese Fighting Fish Betta splendens Regan. The Journal of Experimental Zoology. 1987;244:473–478. [Google Scholar]
- Li S, Zhang Y, Liu Y, Huang X, Huang W, Lu D, Zhu P, Shi Y, Cheng CH, Liu X, Lin H. Structural and functional multiplicity of the kisspeptin/GPR54 system in goldfish (Carassius auratus) J Endocrinol. 2009;201(3):407–418. doi: 10.1677/JOE-09-0016. [DOI] [PubMed] [Google Scholar]
- Martinez-Chavez CC, Minghetti M, Migaud H. GPR54 and rGnRH I gene expression during the onset of puberty in Nile tilapia. Gen Comp Endocrinol. 2008;156(2):224–233. doi: 10.1016/j.ygcen.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Mechaly AS, Vinas J, Murphy C, Reith M, Piferrer F. Gene structure of the Kiss1 receptor-2 (Kiss1r-2) in the Atlantic halibut: insights into the evolution and regulation of Kiss1r genes. Mol Cell Endocrinol. 2010;317(1-2):78–89. doi: 10.1016/j.mce.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Mechaly AS, Vinas J, Piferrer F. Identification of two isoforms of the Kisspeptin-1 receptor (kiss1r) generated by alternative splicing in a modern teleost, the Senegalese sole (Solea senegalensis) Biol Reprod. 2009;80(1):60–69. doi: 10.1095/biolreprod.108.072173. [DOI] [PubMed] [Google Scholar]
- Messager S, Chatzidaki EE, Ma D, Hendrick AG, Zahn D, Dixon J, Thresher RR, Malinge I, Lomet D, Carlton MB, Colledge WH, Caraty A, Aparicio SA. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc Natl Acad Sci U S A. 2005;102(5):1761–1766. doi: 10.1073/pnas.0409330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousley A, Polese G, Marks NJ, Eisthen HL. Terminal nerve-derived neuropeptide y modulates physiological responses in the olfactory epithelium of hungry axolotls (Ambystoma mexicanum) J Neurosci. 2006;26(29):7707–7717. doi: 10.1523/JNEUROSCI.1977-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro VM, Castellano JM, Fernandez-Fernandez R, Barreiro ML, Roa J, Sanchez-Criado JE, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology. 2004;145(10):4565–4574. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30(6):713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhar IS, Ogawa S, Sakuma Y. Laser-captured single digoxigenin-labeled neurons of gonadotropin-releasing hormone types reveal a novel G protein-coupled receptor (Gpr54) during maturation in cichlid fish. Endocrinology. 2004;145(8):3613–3618. doi: 10.1210/en.2004-0395. [DOI] [PubMed] [Google Scholar]
- Peter RE. Evolution of Neurohormonal Regulation of Reproduction in Lower Vertebrates. American Zoologist. 1983;23(3):685–695. [Google Scholar]
- Popa SM, Clifton DK, Steiner RA. The role of kisspeptins and GPR54 in the neuroendocrine regulation of reproduction. Annu Rev Physiol. 2008;70:213–238. doi: 10.1146/annurev.physiol.70.113006.100540. [DOI] [PubMed] [Google Scholar]
- Portavella M, Vargas JP, Torres B, Salas C. The effects of telencephalic pallial lesions on spatial, temporal, and emotional learning in goldfish. Brain Res Bull. 2002;57(3-4):397–399. doi: 10.1016/s0361-9230(01)00699-2. [DOI] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, Maryland: U. S. National Institutes of Health; 1997-2009. [Google Scholar]
- Roa J, Vigo E, Castellano JM, Navarro VM, Fernandez-Fernandez R, Casanueva FF, Dieguez C, Aguilar E, Pinilla L, Tena-Sempere M. Hypothalamic expression of KiSS-1 system and gonadotropin-releasing effects of kisspeptin in different reproductive states of the female Rat. Endocrinology. 2006;147(6):2864–2878. doi: 10.1210/en.2005-1463. [DOI] [PubMed] [Google Scholar]
- Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin- releasing hormone/luteinizing hormone surge. Endocrinology. 2009;150(8):3664–3671. doi: 10.1210/en.2009-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison RR, Fernald RD, Stacey NE. The olfactory system of a cichlid fish responds to steroidal compounds. Journal of Fish Biology. 1998;53:226–229. [Google Scholar]
- Saito KaSW. Deficits in acquisition of spatial learning after dorsomedial telencephalon lesions in goldfish. Behavioural Brain Research. 2006;172(2):187–194. doi: 10.1016/j.bbr.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Satou M. Synaptic organization, local neuronal circuitry, and functional segregation of the teleost olfactory bulb. Prog Neurobiol. 1990;34(2):115–142. doi: 10.1016/0301-0082(90)90004-z. [DOI] [PubMed] [Google Scholar]
- Satou M, Oka Y, Kusunoki M, Matsushima T, Kato M, Fujita I, Ueda K. Telencephalic and preoptic areas integrate sexual behavior in hime salmon (landlocked red salmon, Oncorhynchus nerka): results of electrical brain stimulation experiments. Physiol Behav. 1984;33(3):441–447. doi: 10.1016/0031-9384(84)90167-7. [DOI] [PubMed] [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349(17):1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- Simonneaux V, Ansel L, Revel FG, Klosen P, Pevet P, Mikkelsen JD. Kisspeptin and the seasonal control of reproduction in hamsters. Peptides. 2009;30(1):146–153. doi: 10.1016/j.peptides.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- van Aerle R, Kille P, Lange A, Tyler CR. Evidence for the existence of a functional Kiss1/Kiss1 receptor pathway in fish. Peptides. 2008;29(1):57–64. doi: 10.1016/j.peptides.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Van der Kraak GJ. Fish Neuroendocrinology. Elsevier; 2009. The GnRH System and the neuroendocrine regulation of reproduction. [Google Scholar]
- Van Pett K, Viau V, Bittencourt JC, Chan RK, Li HY, Arias C, Prins GS, Perrin M, Vale W, Sawchenko PE. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol. 2000;428(2):191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- White SA, Nguyen T, Fernald RD. Social regulation of gonadotropin-releasing hormone. J Exp Biol. 2002;205(Pt 17):2567–2581. doi: 10.1242/jeb.205.17.2567. [DOI] [PubMed] [Google Scholar]
- Wu M, Dumalska I, Morozova E, van den Pol A, Alreja M. Melanin-concentrating hormone directly inhibits GnRH neurons and blocks kisspeptin activation, linking energy balance to reproduction. Proc Natl Acad Sci U S A. 2009;106(40):17217–17222. doi: 10.1073/pnas.0908200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12(8):1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]