Abstract
Studying the association between quantitative phenotype (such as height or weight) and single nucleotide polymorphisms (SNPs) is an important problem in biology. To understand underlying mechanisms of complex phenotypes, it is often necessary to consider joint genetic effects across multiple SNPs. ANOVA (analysis of variance) test is routinely used in association study. Important findings from studying gene-gene (SNP-pair) interactions are appearing in the literature. However, the number of SNPs can be up to millions. Evaluating joint effects of SNPs is a challenging task even for SNP-pairs. Moreover, with large number of SNPs correlated, permutation procedure is preferred over simple Bonferroni correction for properly controlling family-wise error rate and retaining mapping power, which dramatically increases the computational cost of association study.
In this paper, we study the problem of finding SNP-pairs that have significant associations with a given quantitative phenotype. We propose an efficient algorithm, FastANOVA, for performing ANOVA tests on SNP-pairs in a batch mode, which also supports large permutation test. We derive an upper bound of SNP-pair ANOVA test, which can be expressed as the sum of two terms. The first term is based on single-SNP ANOVA test. The second term is based on the SNPs and independent of any phenotype permutation. Furthermore, SNP-pairs can be organized into groups, each of which shares a common upper bound. This allows for maximum reuse of intermediate computation, efficient upper bound estimation, and effective SNP-pair pruning. Consequently, FastANOVA only needs to perform the ANOVA test on a small number of candidate SNP-pairs without the risk of missing any significant ones. Extensive experiments demonstrate that FastANOVA is orders of magnitude faster than the brute-force implementation of ANOVA tests on all SNP pairs.
Keywords: Association study, ANOVA test
1. INTRODUCTION
Quantitative phenotype association study analyzes genetic variation across a population in order to find the genetic factors underlying continuous phenotypes (such as height or weight). These phenotypes are often complex in the sense that they are likely due to the effects of multiple genes [6, 24]. The most abundant source of genetic variation is represented by single nucleotide polymorphisms (SNPs). A SNP is a DNA sequence variation occurring when a single nucleotide (A, T, G, or C) in the genome differs between individuals of a species. For inbred species, a SNP usually shows variation between only two of the four possible nucleotide types [12], which allows us to represent it by a binary variable. The binary representation of a SNP is also referred to as the genotype of the SNP.
Various statistics can be applied to measure the association between SNPs and the phenotypes of interest, among which ANOVA (analysis of variance) test is one of the standard statistic methods and has been routinely used in quantitative phenotype association study [18]. The goal of ANOVA test is to determine whether the group means are significantly different after accounting for the variances within groups. It accomplishes the comparison by decomposing the total variance in the data into within-group variance and between-group variance. If the between-group variance is sufficiently larger than the within-group variance, then the test concludes that there is significant (phenotypic) difference between the groups.
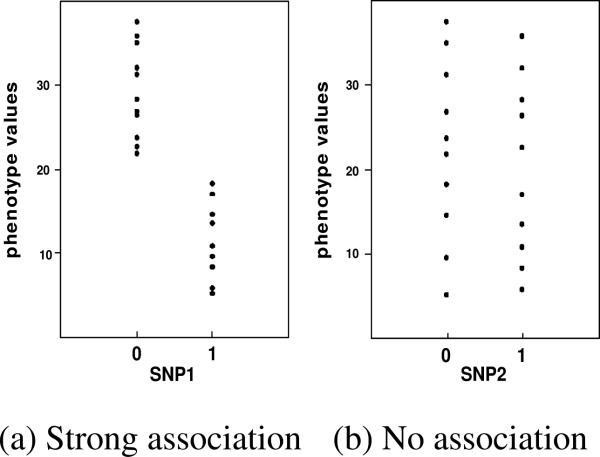
In the application of phenotype-SNP association study, the individuals’ phenotype values are grouped by the genotype of a SNP or a subset of SNPs. Figure 1(a) shows an example of strong association between a phenotype and a SNP. 0 and 1 on the x-axis represent the SNP genotype and the y-axis represents the phenotype. Each point in the figure represents an individual. It is clear from the figure that the phenotype values are partitioned into two groups with distinct means, hence indicating a strong association between the phenotype and the SNP. On the other hand, if the genotype of a SNP partitions the phenotype values into groups as shown in Figure 1(b), the phenotype and the SNP are not associated with each other.
Figure 1.
Examples of associations between a phenotype and two different SNPs
Recent advances in high-throughput techniques enable genotyping SNPs in genome-wide scale, resulting in large datasets containing thousands to hundreds of thousands of SNPs [1, 3]. The vast number of SNPs has posed great computational challenge to genome-wide association study. In order to understand the underlying biological mechanisms of complex phenotype, one needs to consider the joint effect of multiple SNPs simultaneously. Although the idea of studying the association between phenotype and multiple SNPs is straightforward, the implementation is nontrivial. For a study with total N SNPs, in order to find the association between n SNPs and the phenotype, a brute-force approach is to exhaustively enumerate all possible SNP combinations and evaluate their associations with the phenotype. The computational burden imposed by this enormous search space often makes the complete genome-wide association study intractable.
The computational challenge of genome-wide association study is further compounded by another well-known statistical problem – the multiple testing problem [14]. The multiple testing problem can be described as the potential increase in Type I error when statistical tests are performed multiple times. Let α be the Type I error for each independent test. If n independent comparisons are performed, the experimental-wise error α′ is given by
For example, when α = 0.05 and n = 20, α′ = 1 − 0.9520 = 0.64. We have 64% probability to get at least one spurious result. Determining the statistical significance of the association between the phenotype and SNPs is crucial. Bonferroni correction based on the assumption that all n tests are independent is too conservative for the genome-wise association studies since SNPs are often correlated. Alternatively, permutation procedure can be used and much preferred in association studies which automatically takes the correlation structure of SNPs into consideration.
The idea of permutation is to randomly permute the phenotype hundreds to thousands of times. For each permutated phenotype, the association analysis will be repeated. Then the null distribution of the test statistics is estimated and used to assess the statistical significance of the findings from the original phenotype. However, permutation test is very time-consuming since the test procedure needs to be performed in all permutations in order to find the threshold.
Algorithm development to support these large scale analysis is still in its infancy stage. Most existing work focuses on studying the association between the phenotype and SNP-pairs and can only handle a small number of SNPs. Given a pair of SNPs, the phenotype values can be partitioned into at most four groups by the genotype of the SNP-pair, i.e., 00, 01, 10, and 11. Since each SNP has a distinct location on the genome, the association study of a phenotype and SNP-pairs is also called two-locus association mapping. Important findings are appearing in the literature from studying the association between phenotypes and SNP-pairs [21, 22, 27].
Although the standard ANOVA test has been a valuable tool to find association between SNP-pairs and phenotype, it is usually not performed in genome-wide scale. This is due to the fact that the search space of two-locus association mapping in genome-wide scale prohibits an exhaustive search. Suppose that the dataset consists of N SNPs and the number of permutations is K. The total number of ANOVA tests is KN(N − 1)/2. Given a moderate number of SNPs N = 10, 000 and number of permutations K = 1, 000, the number of ANOVA tests is around 5 × 1010. Therefore, ANOVA test is often reserved for validating a small set of candidates identified by other methods [17, 25].
In this paper, we examine the computational aspect of ANOVA test. We present an efficient algorithm, FastANOVA, and show that the standard ANOVA test can be applied in genome-wide scale for two-locus association mapping even when the permutation procedure is needed. Unlike algorithms applying heuristics, FastANOVA is a complete algorithm, i.e., it guarantees to find the optimal solution, though it does not explicitly examine all possible SNP-pairs. In fact, a large portion of the SNP-pairs are pruned without the need of performing the tests. FastANOVA establishes an upper bound on the two-locus ANOVA test. The upper bound is the sum of two terms: one based on the ANOVA test between phenotype and a single SNP, and the other based on the pair-wise SNP genotype and the ordered phenotype values. This formulation of the upper bound allows the algorithm to calculate the bound for a large number of SNPs together, which enables fast candidate retrieval. Moreover, the intermediate results for calculating the second term of the upper bound is independent of phenotype permutations. Hence they only need to be computed once and can be reused in all permutations. Applying this bound, FastANOVA is able to identify SNP-pairs with significant ANOVA test values using only a small fraction of the time required by performing ANOVA test on all SNP-pairs. The principles developed in FastANOVA are also applicable to the general case of testing SNP subsets containing more than two SNPs.
2. RELATED WORK
The problem of phenotype-SNP association study has attracted extensive research interests and is an ongoing research area in biology and statistic communities. In this section, we give a briefly review of the related work from a computational point of view. Please refer to [4, 7, 11] for excellent surveys of existing work.
Under the assumption that the number of SNPs is limited, e.g., from tens to a few hundreds, exhaustive algorithms that explicitly enumerate all possible SNP combinations and evaluate their associations with the phenotype have been developed [16, 19]. These methods are not well adapted to genome-wide association study.
To avoid exhaustively enumerating the search space, a common approach is to break the problem into two steps [8, 10]. First, a subset of important SNPs are selected. Second, within the selected subset, the association between SNPs and the phenotypes are searched. These methods are not complete since the SNPs with weak marginal effects may not be selected in the first place. Genetic algorithm [5, 15] has been applied in finding SNP-pairs for quantitative phenotypes. These methods cannot guarantee to find the optimal solution.
Feature selection methods [13] have been proposed to address the problem of finding important SNPs. In feature selection, the selected feature subset usually contains features that have low correlation with each other but have strong correlation with the target feature. In the application of selecting SNPs, the goal is to select a subset of SNPs that can be used as proxies for all SNPs in the genome [9, 23]. The selected SNPs can then be used as the input SNPs in the association study. Apparently, these methods are also not complete.
3. TWO-LOCUS ANOVA TEST
Let (X1, X2, ···, XN} be the set of SNPs of M individuals (Xi ∈ {0, 1}, 1 ≤ i ≤ N) and Y = {y1, y2, ···, yM} be the quantitative phenotype of interest, where ym (1 ≤ m ≤ M) is the phenotype value of individual m.
For any SNP Xi (1 ≤ i ≤ N), we represent the F-statistic from the ANOVA test of Xi and Y as F(Xi, Y). For any SNP-pair (XiXj), we represent the F-statistic from the ANOVA test of (XiXj) and Y as F(XiXj, Y).
The basic idea of ANOVA test is to partition the total sum of squared deviations SST into between-group sum of squared deviations SSB and within-group sum of squared deviations SSW:
In our application of two-locus association study, Table 1(a) and Table 1(b) show the possible groupings of phenotype values by the genotypes of Xi and (XiXj) respectively.
Table 1.
Possible groupings of phenotype values by the genotypes of Xi and (XiXj)
| Xi = 1 | Xi = 0 |
|---|---|
| group A | group B |
| (a) Grouping of Y by Xi | |
| Xi = 1 | Xi = 0 | |
|---|---|---|
| Xj = 1 | group a1 | group b1 |
| Xj = 0 | group a2 | group b2 |
| (b) Grouping of Y by XiXj | ||
Let A, B, a1, a2, b1, b2 represent the groups as indicated in Table 1(a) and Table 1(b). We use SSB(Xi, Y) and SSB(XiXj, Y) to distinct the one locus (i.e., single-SNP) and two locus (i.e., SNP-pair) analyses. Specifically, we have
The F-statistics for ANOVA tests on Xi and (XiXj) are:
| (1) |
| (2) |
where g in Equation (2) is the number of groups that the genotype of (XiXj) partitions the individuals into. Possible values of g are 3 or 4, assuming all SNPs are distinct: If none of groups A, B, a1, a2, b1, b2 is empty, then g = 4. If one of them is empty, then g = 3.
Let be be the sum of all phenotype values. The total sum of squared deviations does not depend on the groupings of individuals:
Let be the sum of phenotype values in a specific group, and ngroup be the number of individuals in that group. SSB(Xi, Y) and SSB(XiXj, Y) can be calculated as follows:
Note that for any group of A, B, a1, a2, b1, b2, if ngroup = 0, then is defined to be 0.
The two-locus association mapping with permutation test is typically conducted in the following way [18].
First, for every SNP-pair (XiXj) (1 ≤ i < j ≤ N), the ANOVA test is performed and F(XiXj, Y) is recorded.
Second, a permutation test is performed to get a reference distribution in order to assess the statistical significance of previous findings. More specifically, a permutation Yk of Y is generated by sampling the phenotype Y without replacement. In other words, phenotype values are randomly assigned to individuals in the dataset with no single phenotype value being assigned to more than one individual. Let Y′ = {Y1, Y2, ···, YK} be the set of K permutations of Y. For each permutation Yk ∈ Y′, let FYk represent the maximum F-statistic value of all SNP-pairs, i.e.,
The distribution of {FYk|Yk ∈ Y′} is then used as the reference distribution for assessing the statistical significance of F(XiXj, Y) values found using the original phenotype Y: Given a Type I error threshold α, the critical value Fα is the αK-th largest value in {FYk|Yk ∈ Y′}. For example, suppose that α = 0.01 and K = 1000, then Fα is the 10th largest value in {FYk|Yk ∈ Y′}. The SNP-pair (XiXj) whose F-statistic value F(XiXj, Y) ≥ Fα is considered as significant at α.
Two computational problems need to be solved in this procedure. The first one is to find the critical value Fα for a given Type I error threshold α. The second one is to find all SNP-pairs (XiXj) whose F-statistics are greater than Fα. We formalize these two problems as follows.
Problem (1): Given the Type I error threshold α, find the critical value Fα, which is the αK-th largest value in {FYk|Yk ∈ Y′}.
Problem (2): Given the threshold Fα, find all SNP-pairs (XiXj) such that F(XiXj, Y) ≥ Fα.
A brute force approach to these two problems is to enumerate all SNP-pairs and find their F-statistics. In Problem (1), for each permutation Yk ∈ Y, all SNP-pairs need to be enumerated in order to find the maximum value FYk. In Problem (2), all SNP-pairs need to be enumerated to see if their test values are above the threshold Fα. Computationally, Problem (1) is more challenging, since the permutation number K can range form hundreds to thousands, which means the running time of finding the critical value Fα can be hundreds to thousands times longer than the running time of finding the significant SNP-pairs in Problem (2) using a brute-force search.
In the reminder of the paper, we first derive an upper bound on two-locus ANOVA test value and discuss how this upper bound enables an efficient ANOVA testing for a single phenotype. Then we show how this approach can be easily extended to handle the permutation procedure.
4. THE UPPER BOUND
4.1 Updating F-Statistic
Since the total sum of squared deviations does not change, from the calculation of F(Xi, Y) and F(XiXj, Y) (Equations (1) and (2)), we know that the relationship between these two tests only depends on the relationship between SSB(Xi, Y) and SSB(XiXj, Y). Next we show that SSB(XiXj, Y) can be updated from SSB(Xi, Y). For groups A, a1 and a2, let
Similarly, we have
Thus, SSB(XiXj, Y) can be updated using SSB(Xi, Y):
| (3) |
Note that if any one of {na1, na2, nA} is 0, then ΔA = 0. Similarly, if any one of {nb1, nb2, nB} is 0, then ΔB = 0.
Next, we develop an upper bound of SSB(XiXj, Y). We first show the derivation of an upper bound of ΔA. A similar idea can be applied to find an upper bound of ΔB.
4.2 Bounds of ΔA and ΔB
Let {ym|ym ∈ A} = {yA1, yA2, ···, yAnA} be the phenotype values in group A. Without loss of generality, assume that these phenotype values are arranged in ascending order, i.e.,
The derivative of ΔA with respect to Ta1 is:
Thus we have
We have the range of Ta1:
The maximum value of ΔA is attained when Ta1 = la1 or Ta1 = ua1, i.e.,
| (4) |
We use R1(XiXjY) to denote this upper bound.
Let {ym|ym ∈ B} = {yB1, yB2, ···, yBnB} be the phenotype values in group B. Without loss of generality, assume that these phenotype values are arranged in ascending order, i.e.,
Similarly, we can derive the bound on ΔB:
| (5) |
We use R2(XiXjY) to denote this upper bound. The symbols used in Inequalities (4) and (5) are summarized in Table 2.
Table 2.
Notations for the bounds on ΔA and ΔB
| Symbols | Formulas |
|---|---|
| R1 (XiXjY) | |
| R2 (XiXjY) |
From Equation (3), Inequalities (4) and (5), we have the overall upper bound on SSB(XiXj, Y):
Theorem 4.1. (Upper bound of SSB(XiXj, Y))
Property 4.2. The upper bound in Theorem 4.1 is tight.
The tightness of the bound is obvious from the derivation of the upper bound, since there exists some genotype of SNP-pair (XiXj) that makes the equality hold. For the same reason, we have the following property.
Property 4.3. The upper bound in Theorem 4.1 does not exceeds the total sum of squared deviations, i.e.,
5. THE FASTANOVA ALGORITHM
In this section, we show how our algorithm FastANOVA utilizes the upper bound in Theorem 4.1 to achieve efficient two-locus ANOVA testing. In Section 5.1, we describe the method for Problem (2) discussed in Section 3, that is, given the threshold Fα, find all SNP-pairs whose F-statistics are greater than Fα. Then in Section 5.2, we discuss how FastANOVA performs in permutation procedure, i.e., the scenario of Problem (2) in Section 3.
5.1 One Phenotype
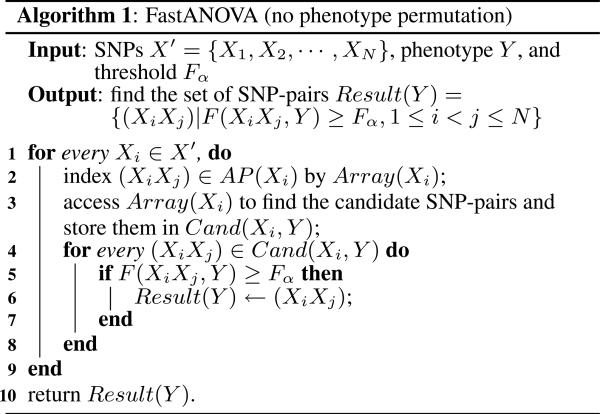
Given the threshold Fα, to find all SNP-pairs whose F-statistics are greater than Fα, a brute-force approach is to enumerate all SNP-pairs. To expedite this process, we employ the inequality in Theorem 4.1 to prune SNP pairs that will have no chance to pass the significance threshold Fα. From Equation (2), we know that finding SNP-pairs (XiXj) whose F-statistics F(XiXj, Y) ≥ Fα is equivalent to finding SNP-pairs satisfying
Theorem 4.1 suggests that we only need to compute the F-statistics for the SNP-pairs that satisfy:
We refer to these SNP-pairs as candidate SNP-pairs.
We now discuss how to apply the upper bound in Theorem 4.1 in detail. The set of all SNP-pairs is partitioned into non-overlapping groups such that the upper bound can be readily applied to each group. For every Xi (1 ≤ i ≤ N), let AP(Xi) be the set of SNP-pairs
For all SNP-pairs in AP(Xi), nA, TA, nB, TB and SSB(Xi, Y) are constants. Moreover, la1, ua1 are determined by na1, and lb1, ub1 are determined by nb1. Therefore, in the upper bound, na1 and nb1 are the only variables that depend on Xj and may vary for different SNP-pairs (XiXj) in AP(Xi).
Note that na1 is the number of 1's in Xj when Xi takes value 1, and nb1 is the number of 1's in Xj when Xi takes value 0. It is easy to prove That switching na1 and na2 does not change the F-statistic value and the correctness of the upper bound. This is also true if we switch nb1 and nb2. Therefore, without loss of generality, we can always assume that na1 is the smaller one between the number of 1's and number of 0's in Xj when Xi takes value 1, and nb1 is the smaller one between the number of 1's and number of 0's in Xj when Xi takes value 0. The following property specifies the values that na1 and nb1 can take. The proof is straightforward and omitted here.
Property 5.1. If there are m 1's and (M − m) 0's in Xi, then for any (XiXj) ∈ AP(Xi), the possible values that na1 can take are . The possible values that nb1 can take are .
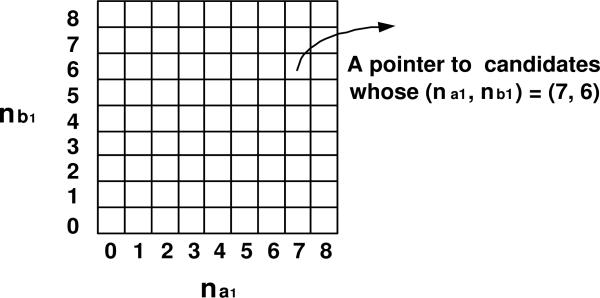
To efficiently retrieve the candidates, the SNP-pairs (XiXj) in AP(Xi) are grouped by their (na1, nb1) values and indexed in a 2D array, referred to as Array(Xi).
Example 5.2. Suppose that there are 32 individuals, and the genotype of Xi consists of half 0's and half 1's. Thus for the SNP-pairs in AP(Xi), the possible values of na1 and nb1 are {0,1,2, ···, 8}. Figure 2 shows the 9 × 9 array, Array(Xi), whose entries represent the possible values of (na1, nb1) for the SNP-pairs (XiXj) ∈ AP(Xi). The entries in the same column have the same na1 value. The entries in the same row have the same nb1 value. The na1 value of each column is noted beneath each column. The nb1 value of each row is noted left to each row. Each entry of the array is a pointer to the SNP-pairs (XiXj) ∈ AP(Xi) having the corresponding (na1, nb1) values.
Figure 2.
The index array Array(Xi) for efficient retrieval of the candidate SNP-pairs
Note that for a SNP-pair (XiXj) ∈ AP(Xi), na1 and na2 can be calculated faster than performing the two-locus ANOVA test. To obtain na1 and na2, we only need to count the numbers of 0's and 1's of Xj when Xi is equal to 0 and 1 respectively, which can be done by a linear scan of the M × 2 binary matrix consisting of the genotypes of Xi and Xj. In contrast, to calculate the F-statistic, we first need to scan the M × 3 binary matrix consisting of Xi, Xj and Y in order to find out how the phenotype values are grouped by the genotype of (XiXj). Then a constant time O(t) is required to compute the F-statistic.
Property 5.3. For any SNP Xi, the maximum number of the entries in Array(Xi) is .
The proof of Property 5.3 is straightforward and omitted here. In order to find candidate SNP-pairs, we scan all entries in Array(Xi) to calculate their upper bounds. Since the SNP-pairs indexed by the same entry share the same (na1, nb1) value, they have the same upper bound. In this way, we can calculate the upper bound for a group of SNP-pairs together. Note that for typical genome-wide association studies, the number of individuals M is much smaller than the number of SNPs N. Therefore, the additional cost for accessing Array(Xi) is minimal compared to performing ANOVA tests for all pairs (XiXj) ∈ AP(Xi).
Algorithm 1 describes the FastANOVA algorithm for finding the SNP-pairs whose F-statistics are greater than the threshold Fα. The inputs of FastANOVA include the N SNPs, the phenotype Y and the critical value Fα. For each Xi, FastANOVA first indexes (XiXj) ∈ AP(Xi) using Array(Xi). Then it retrieves the candidate SNP-pairs by accessing Array(Xi) and records them in Cand(Xi, Y). The candidates in Cand(Xi, Y) are then evaluated for their F-statistics. The candidates whose F-statistics are greater than or equal to Fα are reported by the algorithm.
|
5.2 Permutation Procedure
For multiple tests, permutation procedure is often used in genetic analysis for controlling family-wise error rate. For genome-wide association study, permutation is less commonly used because it often entails prohibitively long computation time. Our FastANOVA algorithm makes permutation procedure feasible in genome-wide association study.
Let Y′ = {Y1, Y2, ···, YK} be the K permutations of the phenotype Y. Following the idea discussed in Section 5.1, the upper bound in Theorem 4.1 can be easily incorporated in the algorithm to handle the permutations.
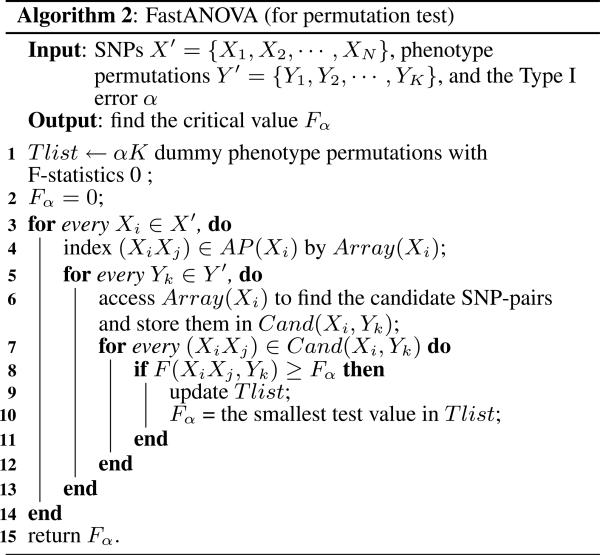
Property 5.4. For every SNP Xi, the indexing structure Array(Xi) is independent of the permuted phenotypes in Y′.
The correctness of this property relies on the fact that, for any (XiXj) ∈ AP(Xi), na1 and nb1 only depend on the genotype of the SNP-pair and thus remain constant for different phenotype permutations. Therefore, for each Xi, once we build Array(Xi), it can be reused in all permutations.
The FastANOVA algorithm for permutation test is described in Algorithm 2. The inputs include the N SNPs, K phenotype permutations, and the Type I error threshold α. The goal is to find the critical value Fα, which is the αK-th largest value in {FYk|Yk ∈ Y′}. Recall that FYk is the maximum F-statistic value for pheno-type Yk. We use Tlist to keep the αK phenotype permutations having the largest F-statistics found by the algorithm so far. Initially, Tlist contains K dummy phenotype permutations with test values 0. The smallest F-statistic value in Tlist, initially 0, is used as the threshold to prune the SNP-pairs. For each Xi, FastANOVA first indexes (XiXj) ∈ AP(Xi) using Array(Xi). Then it finds the set of candidate SNP-pairs Cand(Xi, Yk) by accessing Array(Xi) for every phenotype permutation Yk. The candidates in Cand(Xi, Yk) are then evaluated for their F-statistics. If a candidate's F-statistic value is greater than the current threshold, then Tlist is updated accordingly: If the candidate's phenotype Yk is not in the Tlist, then the phenotype in Tlist having the smallest F-statistic value is replaced by Yk. If the candidate's phenotype Yk is already in Tlist, we only need to update its corresponding F-statistic value to be the maximum value found for the phenotype so far. The threshold is also updated to be the smallest F-statistic value in Tlist.
Algorithm 2.
|
5.3 Complexity Analysis
In this section, we study the time and space complexities of the FastANOVA algorithm for permutation test. The complexity for a single phenotype can be analyzed in a similar way.
Time complexity: For each Xi, FastANOVA needs to index (XiXj) in AP(Xi). The complexity to build the indexing structure for all SNPs is O(N(N − 1)M/2). The worst case for accessing all Array(Xi) for all permutations is . Let represent the total number of candidates. The overall time complexity of FastANOVA is thus . The experimental results show that the overhead of building the indexing structures and accessing them for candidate retrieval are negligible when large permutation tests are needed. Note that the time complexity of the brute-force approach is O(KN(N − 1)M/2) = O(KN2M).
Space complexity: The total number of variables in the dataset, including the SNPs and the phenotype permutations, is N + K. The maximum space of the indexing structure Array(Xi) is . Note that for each SNP Xi, FastANOVA only needs to access one indexing structure, Array(Xi), for all permutations. Once the evaluation process for Xi is done for all permutations, Array(Xi) can be cleared from the memory. Therefore, the space complexity of FastANOVA is since . The space complexity is linear to the dataset size.
6. EXPERIMENTAL RESULTS
In this section, we present extensive experimental results on evaluating the performance of the FastANOVA algorithm. We show (1) the runtime comparison between FastANOVA and the brute-force approach under various experimental settings, (2) the punning effect of the upper bound, and (3) the relative computational cost of each component of FastANOVA. FastANOVA is implemented in C++. The experiments are performed on a 2.4 GHz PC with 1G memory running WindowsXP system.
Dataset: The SNP dataset used for the experiments is extracted from a set of combined SNPs from the 140k Broad/MIT mouse dataset [26] and 10k GNF [2] mouse dataset. This merged dataset has 156,525 SNPs for 71 individuals. The missing values in the dataset are imputed using NPUTE [20]. We use both real pheno-types and synthetic phenotypes in our experiments. The real phenotype data is available from the Jackson Lab [3].
6.1 Real Phenotypes
We use three real phenotypes in our experiments: cardiovascular (blood pressure), metabolism (water intake), and neurosensory (acoustic startle response). Table 3 shows the statistics of the geno-type datasets corresponding to the three phenotypes. The number of SNPs in the table indicates the number of unique SNPs in each genotype dataset.
Table 3.
Statistics of the genotype datasets
| cardiovascular | metabolism | neurosensory | |
|---|---|---|---|
| # individuals | 19 | 26 | 34 |
| # SNPs | 14,513 | 43,856 | 66,006 |
We first show the results on finding the critical value Fα, which is more time-consuming than finding the significance SNP-pairs given the critical value Fα for a single phenotype.
6.1.1 Finding critical value Fα
FastANOVA v.s. the brute-force approach
We compare FastANOVA with the brute-force approach under various experimental settings. Since the brute-force approach is very time-consuming, we use a moderate number of SNPs and permutations in the default setting in order to show the performance comparisons. The default setting is as follows: The Type I error threshold α = 0.01. The number of permutations is 100. The number of SNP is 10,000 for the two larger datasets of metabolism and neurosensory, and 2,900 for the cardiovascular SNP dataset. These experimental settings are chosen to demonstrate the performance gain and enhanced scalability offered by FastANOVA over the brute-force implementation. FastANOVA can handle much larger SNP panels and larger number of permutation tests. The performance of FastANOVA is expected to follow the same trends presented in the remainder of this section.
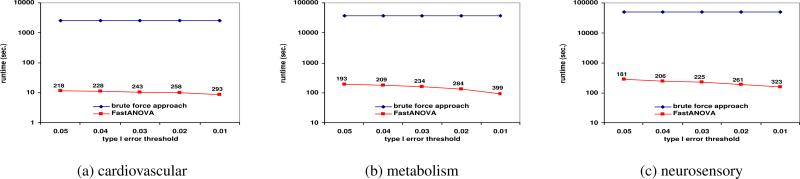
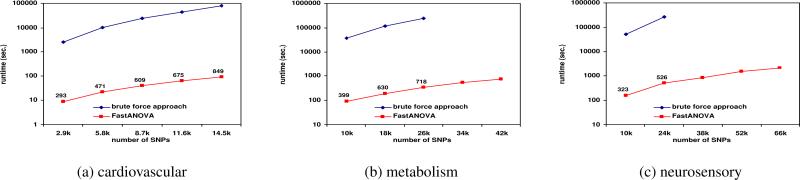
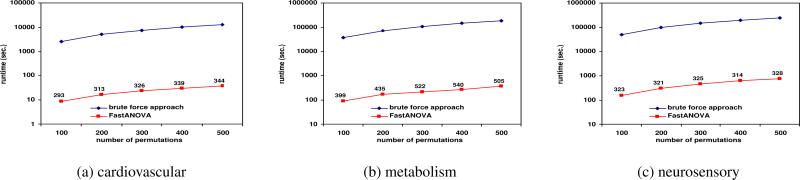
Figures 3, 4, and 5 show the running time comparison of FastANOVA and the brute-force approach on the three genotype phenotype datasets using different settings. The y-axis is in logarithm scale. The numbers above the runtime line of FastANOVA indicate the ratio of the runtimes of the brute-force approach over FastANOVA. We terminate the programs that have run over 72 hours without completion.
Figure 3.
Performance comparison between FastANOVA and the brute-force approach when varying Type I error thresholds
Figure 4.
Performance comparison between FastANOVA and the brute-force approach when varying the number of SNPs
Figure 5.
Performance comparison between FastANOVA and the brute-force approach when varying the number of permutations
Figure 3 shows the runtime comparison when varying the Type I error thresholds. For each dataset, the runtime of the brute-force approach does not change over different Type I error thresholds. The runtime of FastANOVA decreases as the threshold decreases. FastANOVA offers 218 fold speedup when α = 0.05 and 293 fold speedup when α = 0.01 on cardiovascular dataset. We can also ob serve a similar two-orders-of-magnitude speedup in the metabolism and neurosensory datasets. This is consistent with the pruning effect of the upper bound, which will be presented later in this section. In general, the lower the Type I error threshold, the more powerful the pruning effect, hence the faster the algorithm.
Figure 4 depicts the comparison of these two approaches when the number of SNPs changes. From these figures, it is clear that FastANOVA is about two orders of magnitude faster than the brute-force approach. The brute-force approach cannot finish in 72 hours when the number of unique SNPs is greater than 26k in the metabolism dataset and greater than 24k in the neurosensory dataset. We observe that the runtime ratio tends to increase (approaching three-orders-of-magnitude speedup) as the number of SNPs increases. This indicates that the performance gain of FastANOVA is even higher for larger SNP datasets.
Figure 5 shows the runtime comparison when the number of phenotype permutations changes. The runtime of the brute-force approach is linear with respect to the number of permutations. FastANOVA - is consistently two orders of magnitude faster than the brute-force approach. The performance gap increases as the number of permutations increases.
Pruning effect of the upper bound
Table 4 shows the percentage of SNP-pairs pruned under different experimental settings. Since the three datasets have different numbers of SNPs, the 1st to 5th rows in the category of “# SNPs” correspond to the settings from left to right on x-axis in each plot in Figure 4. Most SNP-pairs are pruned under all settings. Moreover, as the Type I error threshold decreases, the pruning ratio increases, which is consistent with runtime comparison shown in Figure 3. As the number of SNPs increases, the pruning ratio also increases. This is because, with more SNPs, the dynamic threshold used to prune the search space becomes higher. Hence a larger portion of SNPs are pruned. This is consistent with results shown in Figure 4. Note that from Table 4 we observe that the pruning ratio tends to remain steady when the number of permutations changes. However, we observe that the runtime ratio increases as the number of permutations increases. The reason for these two different trends will become clear after we show the results on the computational cost of each component of FastANOVA in the next subsection.
Table 4.
Pruning effects on cardiovascular, metabolism and neurosensory datasets when finding critical value Fα
| |
|
cardiovascular |
metabolism |
neurosensory |
|---|---|---|---|---|
|
α
|
0.05 | 99.881% | 99.724% | 99.701% |
| 0.04 | 99.907% | 99.758% | 99.751% | |
| 0.03 | 99.928% | 99.797% | 99.792% | |
| 0.02 | 99.949% | 99.877% | 99.853% | |
| 0.01 |
99.974% |
99.929% |
99.911% |
|
| # SNPs |
1st | 99.974% | 99.929% | 99.911% |
| 2nd | 99.991% | 99.985% | 99.979% | |
| 3rd | 99.996% | 99.996% | 99.997% | |
| 4th | 99.998% | 99.996% | 99.997% | |
| 5th |
99.998% |
99.993% |
99.998% |
|
| # Perm. | 100 | 99.974% | 99.929% | 99.911% |
| 200 | 99.966% | 99.935% | 99.917% | |
| 300 | 99.977% | 99.962% | 99.919% | |
| 400 | 99.977% | 99.961% | 99.914% | |
| 500 | 99.974% | 99.953% | 99.907% |
6.1.2 Finding significant SNP-pairs
In this subsection, we study the comparison between FastANOVA and the brute-force approach in finding significant SNP-pairs given a critical value Fα. Only the original phenotype (without permutations) is used in this procedure. We examine the detailed computation cost of each component of the FastANOVA algorithm. FastANOVA has three major components: building the indexing structure Array(Xi) for every SNP Xi, accessing Array(Xi) to find the candidate SNP-pairs, and performing ANOVA tests on these candidates.
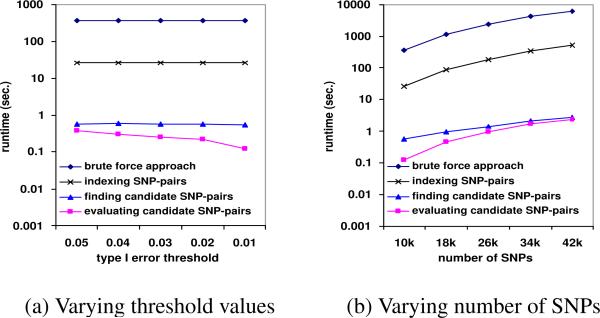
Due to space limitation, we only show the performance comparison on the metabolism dataset. Similar behaviors are also observed on the other two datasets. The default experimental setting is the same as before. Figure 6(a) and Figure 6(b) show the runtime of these three components when varying the Type I error threshold and number of SNPs in the dataset respectively. Since Fα is a function of α, in Figure 6(a), we plot the runtime with respect to α. In both figures, the three lines from the bottom show the runtime of these three components. The runtime of the brute-force approach is the top line. As we can see from these two figures, performing two-locus ANOVA tests on candidate SNP pairs is two to three orders of magnitude faster than performing such tests on all SNP-pairs. This is the benefit of the upper bound pruning since most SNP-pairs have been pruned and only a very small portion of candidates need to be evaluated for their F-statistics. The cost for accessing the indexing structures is also small, which demonstrates the efficiency of the method introduced in Section 5.1 for candidate retrieval. Among the three components of FastANOVA, the most time-consuming one is building the index structures. Yet, its runtime is only a small fraction of the runtime of performing the two-locus ANOVA tests on all SNP pairs. Note that, in permutation test, building the index structures is a one time cost. Once the index structures are built, they can be reused in all permutations. Therefore, the amortized overhead per permutation decreases when the number of permutations increases. This is why the pruning ratio remains steady as in Table 4 while the runtime ratio increases as in Figure 5 when the number of permutations increases.
Figure 6.
Runtime of each component of FastANOVA v.s. runtime of the brute-force approach in the process of finding significant SNP-pairs
6.1.3 Finding FYk for all permutations
Sometimes the users may be interested in finding FYk values of all phenotype permutations. In this way, the users can get the critical value Fα for any Type I error threshold α ranging from 0 to 1, without re-running the permutation tests for different thresholds. Recall that, given a set of phenotype permutations Y′ = {Y1, Y2, ···, YK}, FYk = max{F(XiXj, Yk)} 1 ≤ i < j ≤ N} is the maximum F-statistic value for permutation Yk. Fα is the αK-th largest value in {FYk|Yk ∈ Y′}. In this subsection, we show the pruning effect of the upper bound when it is applied to determine FYk for every Yk (1 ≤ k ≤ K). Note that in this case, for each permutation Yk, the dynamic threshold used to prune the search space is the largest F-statistic value of Yk identified by the algorithm so far.
Table 5 shows the pruning ratio of applying the upper bound to the three real phenotype datasets. The experimental setting is the same as the default setting before. As expected, the pruning ratios are slightly lower than those in Table 4, where smaller Type I error thresholds are used to prune the search space. However, the pruning ratios on all three datasets are still above 97%. Moreover, finding all FYk provides the advantage that we can get the Fα values for all possible values instead of just for a specific one.
Table 5.
Pruning effect on cardiovascular, metabolism and neurosensory datasets when finding for all permutations
| cardiovascular | metabolism | neurosensory |
|---|---|---|
| 97.865% | 97.844% | 98.061% |
6.2 Synthetic Phenotypes
To further study the performance of FastANOVA, we generate three synthetic phenotypes whose values follow three different distributions: uniform, standard normal, and standard exponential distribution. Our purpose is to study the pruning effect of the upper bound under different phenotype distributions. The default setting of the experiments in this subsection is as follows: #individuals = 32, #SNPs=10,000, #permutations=100, α = 0.01. There are 60,970 unique SNPs for these 32 individuals.
Table 6 shows the pruning ratio of FastANOVA under different settings using permutation test. In this table, we also include the pruning ratio when the number of individuals varies. We observe that the pruning effects are similar to that of real phenotypes, which indicates that the upper bound pruning is effective and insensitive to different phenotype distributions.
Table 6.
Pruning effect when finding critical value Fα using three synthetic phenotypes
| |
|
uniform |
normal |
exponential |
|---|---|---|---|---|
|
α
|
0.05 | 96.469% | 97.793% | 99.335% |
| 0.04 | 96.888% | 98.222% | 99.401% | |
| 0.03 | 97.695% | 98.631% | 99.502% | |
| 0.02 | 98.712% | 99.072% | 99.617% | |
| 0.01 |
99.605% |
99.506% |
99.737% |
|
| # SNPs |
10k | 99.605% | 99.506% | 99.737% |
| 22k | 99.864% | 99.814% | 99.924% | |
| 34k | 99.907% | 99.905% | 99.967% | |
| 46k | 99.928% | 99.889% | 99.965% | |
| 58k |
99.941% |
99.942% |
99.963% |
|
| # Perm. |
100 | 99.605% | 99.506% | 99.737% |
| 200 | 98.891% | 99.398% | 99.726% | |
| 300 | 98.897% | 99.072% | 99.780% | |
| 400 | 98.623% | 99.315% | 99.762% | |
| 500 |
98.709% |
99.199% |
99.759% |
|
| # indiv. | 28 | 99.756% | 99.695% | 99.893% |
| 30 | 99.422% | 99.577% | 99.880% | |
| 32 | 99.605% | 99.506% | 99.737% | |
| 34 | 99.073% | 99.289% | 99.773% | |
| 36 | 98.736% | 98.832% | 99.745% |
7. CONCLUSION AND FUTURE WORK
The large number of available SNPs poses great computational challenge to the genome-wide association study. To assess the significance of the findings, permutation test is usually required. These factors make the association study a very time-consuming process. Thus tools that can improve the efficiency of the association study are in demand.
In this paper we present an efficient algorithm, FastANOVA, for genome-wide two-locus ANOVA test. FastANOVA is a complete algorithm which guarantees to find the optimal solution. Experimental results demonstrate that FastANOVA is two to three orders of magnitude faster than the brute-force alternative. The efficiency of FastANOVA is gained from two sources. First, it utilizes an upper bound of the two-locus ANOVA test value to prune a majority of the SNP-pairs. Second, it identifies and reuses computation units that are independent of the phenotype and hence are invariant in permutation test. By eliminating redundant computation of these invariant units, FastANOVA is much more efficient than the brute-force method.
Even though FastANOVA is designed for two-locus association study of binary SNPs, the principles used in FastANOVA are general and applicable to the association study on SNP subsets containing more than two SNPs, and the heterozygous case where SNPs are encoded as {0, 1, 2}. In our future work, we will investigate how to apply these principles for association study considering joint effects of more than two SNPs and the heterozygous case.
8. ACKNOWLEDGMENTS
This research was partially supported by EPA grant STAR-RD832720, NSF grant IIS-0448392, and a Micosoft New Faculty Fellowship.
Footnotes
Categories and Subject Descriptors: H.2.8 [Database Applications]: Data Mining; J.3 [Life and Medical Sciences]: Biology and Genetics
General Terms: Algorithm, Performance
Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. To copy otherwise, to republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee.
9. REFERENCES
- 1. http://www.broad.mit.edu/
- 2. http://www.gnf.org/
- 3. http://www.jax.org/
- 4.Balding DJ. A tutorial on statistical methods for population association studies. Nature Reviews Genetics. 2006;7(10):781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 5.Carlborg O, Andersson L, Kinghorn B. The use of a genetic algorithm for simultaneous mapping of multiple interacting quantitative trait loci. Genetics. 2000;155(4):2003–2010. doi: 10.1093/genetics/155.4.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 7.Doerge RW. Multifactorial genetics: Mapping and analysis of quantitative trait loci in experimental populations. Nature Reviews Genetics. 2002;3:43–52. doi: 10.1038/nrg703. [DOI] [PubMed] [Google Scholar]
- 8.Evans DM, Marchini J, Morris AP, Cardon LR. Two-stage two-locus models in genome-wide association. PLoS Genet. 2006;2:e157. doi: 10.1371/journal.pgen.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halperin E, Kimmel G, Shamir R. Tag snp selection in genotype data for maximizing snp prediction accuracy. Proc. ISMB. 2005 doi: 10.1093/bioinformatics/bti1021. [DOI] [PubMed] [Google Scholar]
- 10.Hoh J, et al. Selecting snps in two-stage analysis of disease association data: a model-free approach. Ann. Hum. Genet. 2000;64:413–417. doi: 10.1046/j.1469-1809.2000.6450413.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nature Reviews Genetics. 2003;4:701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 12.Ideraabdullah FY, et al. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 2004;14(10a):1880–1887. doi: 10.1101/gr.2519704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Motoda H. Feature selection for knowledge discovery and data mining. Kluwer Academic Publishers; Boston: 1998. [Google Scholar]
- 14.Miller RG. Simultaneous Statistical Inference. Springer Verlag; New York: 1981. [Google Scholar]
- 15.Nakamichi R, et al. Detection of closely linked multiple quantitative trait loci using a genetic algorithm. Genetics. 2001;158(1):463–475. doi: 10.1093/genetics/158.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson MR, Kardia SL, Ferrell RE, Sing CF. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Research. 2001;11:458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohno Y, et al. Selective genotyping with epistasis can be utilized for a major quantitative trait locus mapping in hypertension in rats. Genetics. 2000;155:785–792. doi: 10.1093/genetics/155.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagano M, Gauvreau K. Principles of Biostatistics. Duxbury Press; Pacific Grove, CA: 2000. [Google Scholar]
- 19.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. American Journal of Human Genetics. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts A, McMillan L, Wang W, Parker J, Rusyn I, Threadgill D. Inferring missing genotypes in large snp panels using fast nearest-neighbor searches over sliding windows. Proc. ISMB. 2007 doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- 21.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 22.Scuteri A, et al. Genome-wide association scan shows genetic variants in the fto gene are associated with obesity-related traits. PLoS Genet. 3(7):2007. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sebastiani P, Lazarus R, Weiss ST, Kunkel LM, Kohane IS, Ramoni MF. Minimal haplotype tagging. Proc. Natl. Acad. Sci. USA. 2003;100(17):9900–9905. doi: 10.1073/pnas.1633613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segrè D, DeLuna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 25.Shimomura K, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11(6):959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- 26.Wade CM, Daly MJ. Genetic variation in laboratory mice. Nat. Genet. 2005;37:1175–1180. doi: 10.1038/ng1666. [DOI] [PubMed] [Google Scholar]
- 27.Weedon MN, et al. A common variant of hmga2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]