Abstract
Escherichia coli O157:H7, a food-borne pathogen, causes hemorrhagic colitis and the hemolytic-uremic syndrome. A putative virulence factor of E. coli O157:H7 is a 60-MDa plasmid (pO157) found in 99% of all clinical isolates and many bovine-derived strains. The well characterized E. coli O157:H7 Sakai strain (Sakai) and its pO157-cured derivative (Sakai-Cu) were compared for phenotypic differences. Sakai-Cu had enhanced survival in synthetic gastric fluid, did not colonize cattle as well as wild-type Sakai, and had unchanged growth rates and tolerance to salt and heat. These results are consistent with our previous findings with another E. coli O157:H7 disease outbreak isolate ATCC 43894 and its pO157-cured (43894-Cu). However, despite the essentially sequence identical pO157 in these strains, Sakai-Cu had changes in antibiotic susceptibility and motility that did not occur in the 43894-Cu strain. This unexpected result was systematically analyzed using phenotypic microarrays testing 1,920 conditions with Sakai, 43894, and the plasmid-cured mutants. The influence of the pO157 differed between strains on a wide number of growth/survival conditions. Relative expression of genes related to acid resistance (gadA, gadX, and rpoS) and flagella production (fliC and flhD) were tested using quantitative real-time PCR and gadA and rpoS expression differed between Sakai-Cu and 43894-Cu. The strain-specific differences in phenotype that resulted from the loss of essentially DNA-sequence identical pO157 were likely due to the chromosomal genetic diversity between strains. The O157:H7 serotype diversity was further highlighted by phenotypic microarray comparisons of the two outbreak strains with a genotype 6 bovine E. coli O157:H7 isolate, rarely associated with human disease.
Keywords: E. coli O157:H7, pO157, Phenotype Microarray, Phenotypic diversity
Introduction
Enterohemorrhagic Escherichia coli (EHEC) of the serotype O157:H7 is an important food-borne pathogen that causes hemorrhagic colitis (HC) and the hemolytic-uremic syndrome (HUS) in humans (Nataro and Kaper, 1998; O'Brien, 1998). Since the first outbreak of HC in 1982, E. coli O157:H7 has been associated with numerous outbreaks worldwide. The largest disease outbreak (~8,000 confirmed cases) occurred in Sakai City, Japan in 1996 and the E. coli isolate responsible is referred to as ‘Sakai’. E. coli O157:H7 is estimated to cause approximately 73,400 infections and over 60 deaths each year in the United states (Mead et al., 2000). Cattle are the major reservoir for E. coli O157:H7 and the most common source for the outbreaks in the United States (Grauke et al., 2002; Naylor et al., 2003). Healthy cattle carry E. coli O157:H7 transiently and sporadically without pathological symptoms and the rectoanal junction (RAJ) mucosa is the principal colonization site in the bovine gastrointestinal tract (GIT) (Naylor et al., 2003; Grauke et al., 2002; Cray and Moon, 1995; Wallace, 1999; Lim et al., 2007). Comparisons of E. coli O157:H7 isolates from clinical and bovine sources show genotypic diversity (Kim et al., 1999; Noller et al., 2003; Noller et al., 2003) and recent analysis reveals a specific lineage of E. coli O157:H7 associated with the most severe human disease (Manning et al., 2008). Further categorization of E. coli O157:H7 isolates by Shiga toxin (Stx)-encoding bacteriophage insertion sites designates genotypes 1 and 3 as most common among clinical isolates and genotypes 5 to7 as bovine-biased subtypes (Besser et al., 2007). The mechanism(s) underlying virulence potential among the E. coli O157:H7 is not understood but outbreak and bovine-biased strains likely represent two ends of the spectrum.
One putative virulence factors of E. coli O157:H7 is an F-like 60-MDa plasmid (pO157) found in 99% of all clinical isolates (Schmidt et al., 1996; Yoon and Hovde, 2008). Large plasmids, similar to pO157 (70–200 kb), are also found in most EHEC isolates from humans and animals (Nataro and Kaper, 1998). These plasmids carry hemolysin genes and genes that influence bacterial adhesion. Also epidemiological study suggests that there is a correlation between these large plasmids and the progression of diarrhea to HUS (Caprioli et al., 2005). The complete sequence of two pO157 from E. coli O157:H7 ATCC 43895 (43895) and Sakai (Sakai) are determined, but among the 100 putative genes, only 19 have been characterized (Makino et al., 1998; Burland et al., 2002; Lim et al., 2007). These include potential virulence factors, such as an enterohemolysin (ehxA), a type II secretion apparatus (etpC to –O), a serine protease (espP), a catalase-peroxidase (katP), a potential adhesion (toxB), attaching and effacing gene-positive conserved fragments (ecf), and a Cl esterase inhibitor (stcE) (Yoon and Hovde, 2008). Although, several studies show that pO157 genes contribute to both bacterial adherence in cell culture, survival in vitro, and colonization and persistence in cattle (Tatsuno et al., 2001; Lim et al., 2007), the biological significance of pO157 has not been fully demonstrated.
Our previous work compares outbreak E. coli O157:H7 strain ATCC 43894 (43894) and an isogenic pO157-cured strain (43894-Cu, previously called 277) (Lim et al., 2007). The 43894-Cu strain is more resistant to acid and bile and survives passage through the bovine GIT better than wild-type 43894, but does not colonize cattle at the RAJ mucosa as well as wild-type 43894. Many proteins, related to survival in salvage conditions are differentially expressed between 43894 and 43894-Cu. Among them, tryptophanase and glutamate decarboxylase isozymes are both increased by deletion of the pO157, suggesting that the pO157 regulates some chromosomal genes.
To determine if the established role of the pO157 in strain 43894 was similar in another well characterized E. coli O157:H7, we analyzed its role the E. coli O157:H7 Sakai. The pO157 from 43894 was sequenced and compared to the known sequences of the pO157 from Sakai. The wild-type and the pO157-cured mutant strains of ATCC 43894 and Sakai were compared for growth/survival in 1,920 conditions using high throughput phenotypic microarray technology (PM) and Sakai and Sakai-Cu were tested for (i) survival in acidic conditions, (ii) survival and persistence in cattle following an oral challenge of bacteria, (iii) growth rates and tolerance to salt and heat, (vi) antibiotic susceptibilities and motilities, and (v) gene expression related to acid resistance and motility.
Materials and Methods
Bacterial strains, media, and growth conditions
Bacterial strains used in this study are listed in Table 1. Southern-blot hybridization with a probe for the pO157-specific hemolysin gene confirmed loss of the pO157. All bacteria were grown in Luria-Bertani (LB) media (pH 7.5) unless otherwise indicated. d-Sorbitol MacConkey agar supplemented with 0.1 mg/ml 4-methylumbelliferyl-β-d-glucuronide (MUG), 50 µg/ml cefixime, 2.5 µg/ml potassium tellurite, and 40 µg/ml vancomycin (SMAC-CTVM) was used to culture E. coli O157:H7 strains from bovine samples as previously described (Rice et al., 2003). Trypticase soy broth (TSB; BBL/Becton Dickinson, Detroit, MI) was used for enrichment culture of bovine samples, acid resistance assays, and heat tolerance assays. M9 minimal medium containing 0.4% glucose was used for growth and salt tolerance assays.
Table 1.
Description of bacterial strains used in this study.
| Strains | Description | Stx | Geno- typea |
Origin | Source or reference |
|---|---|---|---|---|---|
| 43894 | Wild-type E. coli O157:H7 ATCC 43894 |
1&2 | 3b | Human isolate from outbreak associated with hamburger meat in Michigan, USA, 1982 |
ATCC |
| 43894-Cu (277) |
Isogenic mutant of 43894 with pO157 deletion |
1&2 | pO157 was cured using a mini plasmid incompatible with pO157d |
(Sheng et al., 2006) | |
| Sakai | Wild-type E. coli O157:H7 |
1&2 | 3 | Human isolate from outbreak associated with white radish sprout in Osaka, Japan, 1996 |
(Tatsuno et al., 2001) |
| Sakai-Cu | Isogenic mutant of Sakai with pO157 deletion |
1&2 | pO157 was cured using a mini plasmid incompatible with pO157d |
(Tatsuno et al., 2001) | |
| WSU180 | Wild-type E. coli O157:H7 |
1&2 | 6c | Bovine isolate from a dairy heifer in WA, USA, 2003 |
(Rice et al., 2003) |
Stx-encoding bacteriophage insertion site genotypes (Besser et al., 2007)
genotype 3, a principle genotype of human clinical isolates
genotype 6, a bovine-biased genotype
Plasmid deletion was confirmed by Southern-blot hybridization with a pO157-specific gene probe and chromosomal DNA integrity was confirmed by PFGE
Sequencing of pO157
The pO157 DNA was prepared using the Qiagen Plasmid Maxi Kit (Qiagen, Hilden, Germany) from an overnight culture of 43894. The crude pO157 was purified by a cesium chloride/ethidium bromide ultracentrifugation (Sambrook and Russell, 2001). The pO157 DNA was sequenced using the conventional shotgun sequencing approach adopting standard protocols developed previously (Wood et al., 2001; Hendrickson et al., 2004). Briefly, a small 2.5–3.5 kb insert library was generated by blunt-end ligation into the pUC19 vector. Random clones were picked directly into a freezing medium and the DNA was prepared by rolling circle amplifications protocols per the manufacturer’s recommended (Amersham Pharmacia Biotech, Piscataway, NJ). The plasmid DNA thus prepared was end-sequenced using 1/32nd BDT v3.1 (ABI) reaction chemistry and the sequencing reactions were run on 3730 XL capillary sequencers per the standard protocols. The data was processed and assembled using PHRED/PHRAP software tools and the assembled sequences were displayed using CONSED software tools. The gaps between the two contigs were sequenced using primers based on the sequences of both end of two contigs. The pO157 sequence of 43894 was compared with the published sequence of ATCC 43895 and Sakai using the web-based align program (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi).
Genotyping by multiplex PCR
Stx-encoding bacteriophage insertion site genotyping was determined as described by Besser et al. (Besser et al., 2007). Briefly, two multiplex PCR reactions each with three different PCR primer sets (one for stx1, yehVL, and wrbAR and the other for stx2, yehVR, and wrbAL) were performed. The following thermocycler parameters were used: 1 cycle of pre-denaturation (95 °C, 5 min); 35 cycles of denaturation (95 °C, 30 s), annealing (58 °C, 45 s), and extension (72 °C, 2 min); and 1 cycle of final extension (72 °C, 10 min). The PCR products were analyzed by gel electrophoresis using 1.2% agarose and 0.5x TBE buffer and ethidium bromide staining.
Acid resistance
Acid resistance in synthetic bovine gastric fluid (SGF) was determined as described previously with minor modifications (Lim et al., 2007). SGF was prepared with 0.15% bovine bile and adjusted to pH 2.0. Bacterial cells were grown at 37 °C in LB broth, inoculated into 10 ml of fresh SGF (108 CFU/ml), and incubated at 37 °C without shaking. Samples were cultured by direct plating on LB agar after 0.5, 1, 2, 3, and 4 h to determine the numbers of surviving cells.
Bovine oral challenge
All procedures involving animals were performed under guidelines approved by the University of Idaho Animal Care and Use and Biosafety Committees. Four 8- to 9-month old Holstein steers were used in a co-challenge method as described previously (Lim et al., 2007). Briefly, steers received a single dose of a bacterial mixture containing 1.0 × 1010 CFU Sakai and 1.0 × 1010 CFU Sakai-Cu. RAJ mucosal swab (RAMS) samples were collected into 3 ml TSB from each steer twice a week and plated directly onto SMAC-CTVM agar. After 18 h incubation at 37 °C, sorbitol- and MUG-negative colonies were confirmed to be O157 by latex agglutination (Pro-Lab Diagnostics, Toronto, Canada). The O157 colonies were differentiated as Sakai or Sakai-Cu by PCR with primers for the pO157-specific gene, ecf1 (Lim et al., 2007).
Bacterial growth
Growth rates of bacterial strains were monitored with a Bio-Tek Power Wave XS reader and KCjunior software. Bacteria were grown in LB broth at 37° C or in M9 minimal medium at 25 °C. After 1:1000 dilutions into fresh LB broth or M9 minimal medium, cells were inoculated to a 96-well plate and incubated with continuous shaking. Optical densities at 600 nm were measured every hr for 24 h for the plates incubated at 37 °C or for 48 h for the plates incubated at 25 °C.
Heat and salt tolerance
Bacteria grown in LB broth at 37° C for 18 h were diluted to 105 CFU/ml and 10 µl of diluted cultures were transferred into 1 ml of fresh TSB prewarmed to 55 °C or to 1 ml of M9 minimal medium containing 2.5 M NaCl (15%). Cells were exposed to 55 °C for 1 h or to 2.5 M NaCl at 25 °C for 24 h. The number of surviving cells was determined by plating on LB agar.
Antibiotic susceptibility test
Antibiotic susceptibility was performed by the standard Kirby-Bauer method (Bauer et al., 1999) using 28 different antibiotic disks (BBL/Becton Dickinson): Azithromycin (AZM), Carbenicillin (CB), Cefixime (CFM), Cefotaxime (CTX), Cefpodoxime (CPD), Cephalothin (CF), Chloramphenicol (C), Clindamycin (CC), Cloxacillin (CX), Colistin (CL), Erythromycin (E), Levofloxacin (LVX), Lincomycin (L), Lomefloxacin (LOM), Nafcillin (NF), Nalidixic acid (NA), Nitrofurantion (F/M), Norfloxacin (NOR), Penicillin (P), Sparfloxacin (SPX), Spectinomycin (SPT), Tetracycline (TE), Tilmicosin (TIL), Tobramycin, Trimethoprim (TMP), Trimethoprim/Sulfamethoxazole (SXT), Trovafloxacin (TVA), and Vancomycin (VA). The average diameter of two independent experiments was used for comparison.
Motility assay
Single colonies from cells grown on LB agar were stabbed into 0.3% soft agar with a sterilize toothpick and incubated at 37° C for 8 h or at 24° C for 18 h, respectively. The diameter of the motility halo was measured after incubation.
Real-time PCR
Bacterial cells were grown in M9 minimal media and collected by centrifugation at 3,000 × g at 4° C. Total RNA was isolated with the RNeasy kit (Qiagen) and cDNA was synthesized using Superscript II reverse transcriptase and random hexamers according to the manufacturer's guidelines. The Primer Express program version 2.0 (Applied Biosystems, Foster City, CA) was used to design primers for gadA, gadX, rpoS, fliC, and fliD. The expression level of the16S rRNA gene was used for normalization of results. The real time PCR reaction was performed using the ABI Prism 7500 real-time PCR system (Applied Biosystems) with a SYBR green I master mix. Results of reactions were analyzed using sequence detector systems software version 1.2.2 (Applied Biosystems).
Phenotypic Microarray (PM)
The PM analysis was performed by Biolog Inc. (Hayward, CA). The PM assay consists of twenty 96 well PM panels (PM 1–20) and tests bacterial growth/survival in 1,920 conditions. PM1 to PM8 tested for utilization of metabolites including a particular carbon, nitrogen, phosphate, or sulfur source, PM9 tested for growth in various osmolytes, PM 10 tested for growth in different pH conditions, and PM 11 through PM 20 tested for sensitivities to antibiotics and chemicals. Briefly, single colonies from each strain, grown on agar plates, were suspended in inoculating fluid containing a patented redox dye. Bacterial cell suspensions were transferred into PM panels and incubated in the OmniLog incubator reader. Bacterial growth/survival was assessed by the color change from the reduction of the redox dye and the color intensity was measured for 24 h. Wells without substrate that theoretically result in no signal were used as negative controls in each PM panel. Data were analyzed with Omnilog-PM software from Biolog.
Statistical analysis
The Student’s t test or one-way ANOVA were used to determine differences among E. coli O157:H7 strains.
Results and Discussion
To determine genotypic differences among the wild-type E. coli O157:H7 strains used in this study, Stx-encoding bacteriophage insertion site analysis was performed as described by Besser et al. (Besser et al., 2007). E. coli O157:H7 WSU180, a bovine isolate was used as an additional strain tested to analyze phenotypic diversity among wild-type strains. The two outbreak-associated strains, 43894 and Sakai, were genotype 3 and the bovine isolate, WSU180, was genotype 6 (Table 1). Genotype 3 is one of the principle genotypes among human clinical isolates and genotype 6 is a bovine-biased genotype, rarely associated with human disease (Besser et al., 2007). The method used to delete pO157 did not alter the chromosome of 43894 or Sakai. Previously we described that curing pO157 from 43894 did not damage chromosomal DNA (Lim et al., 2007). No differences in Sakai-Cu chromosomal DNA compared to wild-type DNA were detected following analysis by standard restriction digestion and DNA fragment separation using pulse-field gel electrophoresis (data not shown).
The 43894 pO157 was sequenced for comparison with the known Sakai pO157. The pO157 is considered a putative virulence factor of E. coli O157:H7 and is relatively well conserved, but genetic diversity (heterogeneities) are observed (Zhang et al., 2006; Wu et al., 2008). There are also slight variations in the size of pO157 among E. coli O157:H7 isolates ranging from 92 to 104 kb (Schmidt et al., 1996). The full sequence of pO157 from 43895 (92,077 bp) and Sakai (92,721 bp) were previously determined (Hayashi et al., 2001; Burland et al., 2002). It is known that there is high homology between 43894 and the sequenced 43895 (Burland et al., 2002), but for detailed comparison, the pO157 from 43894 was sequenced. The shotgun sequencing of pO157 from 43894 yielded two large contigs of 83,505 and 7,057 bases for a combined size of 90,645 bases. The sequence was identical to the pO157 from 43895 (data not shown). However comparison with the pO157 from Sakai showed that the Sakai pO157 possessed an extra 644 bp encoding one hypothetical gene. This extra gene is located between the genes for resolvase and transposase (IS629) in Sakai pO157 and shows 84% sequence homology in many cloning vectors. This suggests that the extra gene is likely not related to virulence mechanisms or structural functions.
Acid resistance may be important for bacterial survival, particularly for passage through the acidic stomach or for survival in foods with low pH (Lin et al., 1996). Diversity in acid resistance among wild-type E. coli O157:H7 strains is well known (Benjamin and Datta, 1995). To characterize acid resistance and the role of the plasmid among the strains in this study, the ability to survive in conditions with acid and bile were examined.
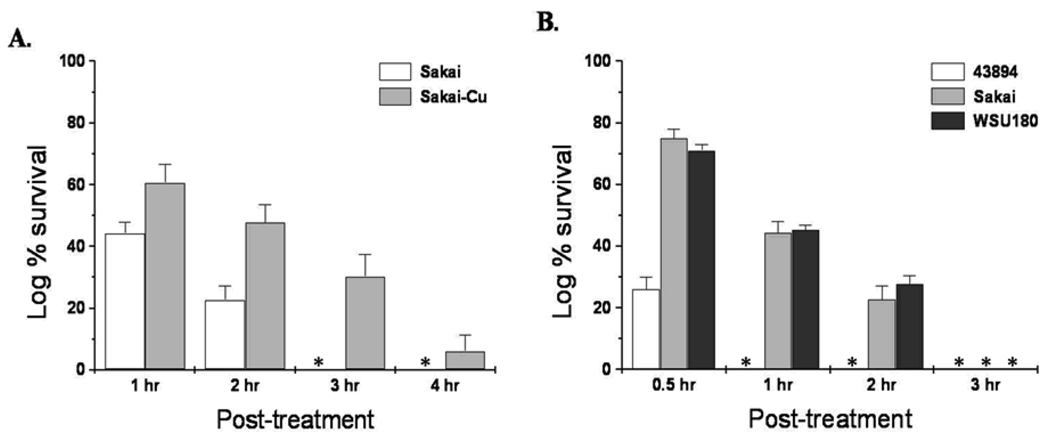
Our previous study shows that the removal of pO157 from 43894 confers better survival of this strain in SGF, pH 2 (Lim et al., 2007). To test for a similar phenotypic change in Sakai, the isogenic mutant Sakai-Cu was compared with Sakai wild-type for survival in SGF (pH 2). Both Sakai and Sakai-Cu survived well in SGF for 1 and 2 h, but Sakai-Cu survived better than Sakai (P < 0.05) at both time points and after 3 h incubation when no wild-type Sakai survived, some Sakai-Cu survived (Fig. 1A). Sakai-Cu was more resistant to synthetic bovine gastric fluid than wild-type Sakai. Thus, for both Sakai and 43894, the deletion of the pO157 resulted in increased acid resistance (Lim et al., 2007). We compared the acid resistance of Sakai-Cu and 43894-Cu in SGF (pH 2). During the first 2 h, ~20% more Sakai-Cu survived than 43894-Cu and by 4 h no 43894-Cu were recovered, but some Sakai-Cu survived (data not shown). We found varied acid resistance between wild-type 43894 and wild-type Sakai. To characterize this difference, three wild-type strains, 43894, Sakai, and WSU180 were compared in a survival assay using SGF at pH 2 to mimic the conditions in the bovine gastric abomasum. After 30 min, all three strains survived, but 43894 showed significantly lower survival compared to Sakai or WSU180 (P < 0.05). After 1 h in SGF, no surviving 43894 were recovered, but both Sakai and WSU180 survived well and similarly after 1h (P = 0.98) and after 2 h (P = 0.13). No bacterial strain survived after 3 h in the SGF (Fig. 1B).
Figure 1.
The survival of E. coli O157:H7 strains in acidic conditions with bile. Comparison of wild-type E. coli O157:H7 Sakai (Sakai) and an isogenic pO157-cured strain (Sakai-Cu) (A) and comparison of three wild-type E. coli O157:H7 strains (B). Bacteria were exposed to SGF, pH 2.0, for 4 h (A) or 3 h (B) at 37° C. The numbers of surviving bacteria at the times shown were determined by direct plate count on LB agar. Asterisks indicate no growth.
These results support our previous hypothesis that genes on pO157 influence acid resistance by regulation of chromosomal genes (Lim et al., 2007). Three basic systems for acid resistance in E. coli have been determined, but they are complex and the regulation and interplay of the acid resistance systems are not fully understood (Foster, 2004).
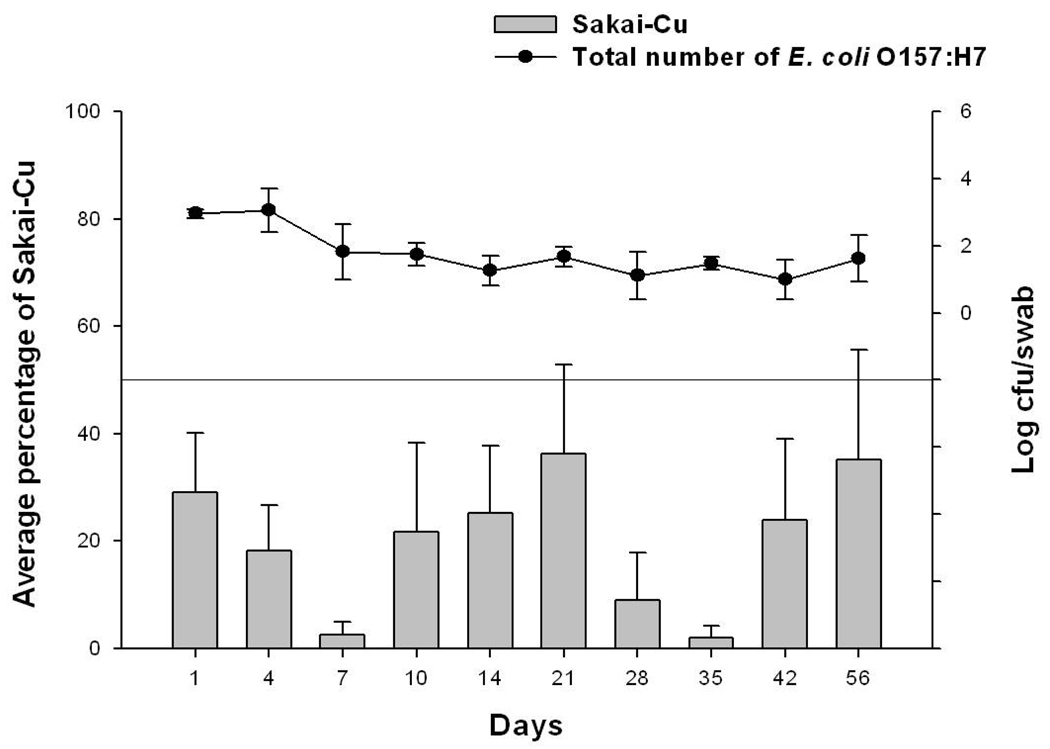
Even though 43894-Cu has an advantage over wild-type 43894 in surviving passage through the abomasum (bovine gastric stomach), we showed previously, that 43894-Cu did not colonize cattle as well as wild-type 43894 at the RAJ mucosa (Sheng et al., 2006; Lim et al., 2007). We hypothesized that pO157 in 43894 might affect gene expression(s) related to colonization or persistence through regulation of chromosomal DNA. Here, we investigated the difference between Sakai and Sakai-Cu for the ability of colonization and persist in cattle. An oral bacterial challenge method was used so that both the ability to survive passage through bovine GIT and the ability to colonize and persist at the RAJ mucosa could be assessed. Four Holstein steers received a single dose of equal numbers of both Sakai and Sakai-Cu, and were monitored for carriage of each bacterial strain by RAMS direct culture. All cattle carried the bacteria for the 56 day duration of the trial with values near 102 CFU/swab (Fig. 2). The average number of Sakai-Cu recovered from RAMS samples were lower at every sampling day compared to the number of wild-type Sakai (data not shown) so that the average percentage of the Sakai-Cu isolates was lower than wild-type (Fig. 2.) and the differences were significant on days 1, 4, 7 and 35 (P < 0.05). This finding is in agreement with our previous results with 43894, which indicate that the pO157 is a colonization factor for bacteria in cattle (Sheng et al., 2006; Lim et al., 2007).
Figure 2.
The effect of the pO157 on survival of E. coli O157:H7 in cattle following oral administration of the bacteria. Four steers were given a single oral dose of 1.0 × 1010 CFU containing both Sakai and Sakai-Cu. RAMS samples were cultured by direct plating onto SMAC-CTVM and sorbitol-negative MUG-negative colonies were confirmed to be the O157 serotype by latex agglutination. Isolates from each sample were sub-cultured and differentiated as Sakai or Sakai-Cu by PCR. Line height on each day represents the average number of total E. coli O157:H7 recovered from the steers expressed as log CFU/swab. Bar height represents the average percentage of Sakai-Cu in the total number of E. coli O157.
However, unlike 43894-Cu that did not persist in cattle as long as wild-type 43894, Sakai-Cu persisted for the duration of the experiment. This difference between the two pO157-cured strains (43894-Cu and Sakai-Cu) might be due to differences in the inherent acid resistance of the two parental wild-type strains (43894 and Sakai) or other chromosomal differences. All E. coli O157:H7 strains have similar fundamental chromosomal backbones but there is genetic diversity among strains (Wu et al., 2008). Recently Vanaja et al. reported that virulence factors including the pO157 genes showed increased expression in E. coli O157:H7 isolates from clinical genotype 1 compared to those from bovine genotype 5 (Vanaja et al., 2009). Both genotypes possess the pO157 but the expression levels of genes on pO157 were different. Therefore even though the pO157 genomic sequences are identical, the effect(s) of pO157 may differ among E. coli O157:H7 strains due to differences in chromosomal DNA sequences or gene expression.
One acid resistance mechanism in E. coli O157:H7 is the rpoS-dependent system in which rpoS regulates not only acid resistance, but also heat- and salt-tolerance (Cheville et al., 1996). Previously we showed that the enhanced acid resistance in 43894-Cu is linked to the synthesis of glutamate decarboxylase, but that there was no difference in growth, heat- or salt-tolerance between 43894 and 43894-Cu (Lim et al., 2007). Growth of Sakai and Sakai-Cu were indistinguishable in rich LB broth or in M9 minimal media at 37 °C (24 h) or 24 °C (48 h) (data not shown). Also, heat- and salt-tolerance assays showed no significant difference (P > 0.05) between Sakai and Sakai-Cu (data not shown). Thus, as in 43894 (Lim et al., 2007), the pO157 in Sakai did not influence growth kinetics or heat- and salt-tolerance.
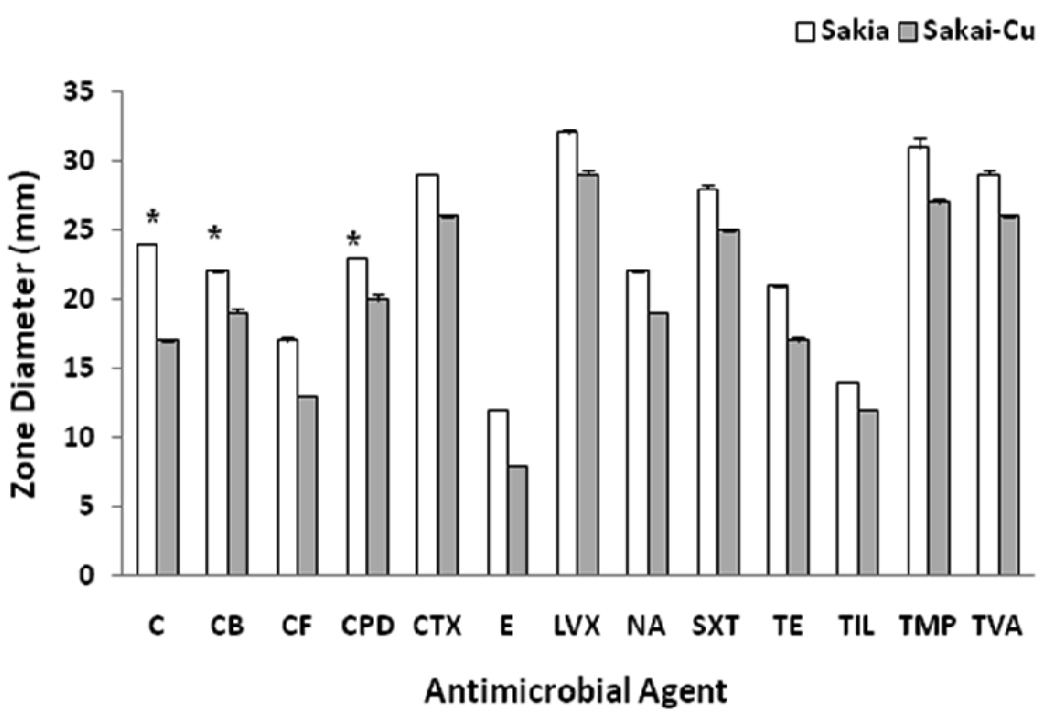
We compared strains for susceptibility to 28 different antibiotics. Sakai-Cu was more resistant than wild-type Sakai strain to 13 different antibiotics (Fig. 3). Among these 13 antibiotics, the susceptibility to chloramphenicol (C) and cefpodoxime (CPD) was changed from susceptible to intermediate, and to carbenicillin (CB) was changed from intermediate to the resistant category. Chloramphenicol is a bacteriostatic antibiotic and prevents protein systhesis. Cefpodoxime and carbenicillin are resistant to β-lactamase and prevent cell wall synthesis. Other antibiotics for which Sakai and Sakai-Cu had differential resistance include β-lactam, quinolone, and tetracycline antibiotics. There was no common mechanism among the antibiotic resistance/sensitivities influenced by the pO157. This result was not consistent with the previous findings with 43894-Cu. No antibiotic susceptibility differences were found between 43894 and 43894-Cu (Lim et al., 2007).
Figure 3.
Comparison of Sakai and Sakai-Cu for antibiotic susceptibility. Sakai and Sakai-Cu were streaked on Muller-Hinton agar and the antibiotic disks were placed on the plate. After incubation at 37°C for 18 h, the diameter of the growth inhibition zone was measured. The full names of the antibiotics are described in the text. Asterisks indicate differences from susceptible to intermediate (C and CPD) or intermediate to resistant (CB).
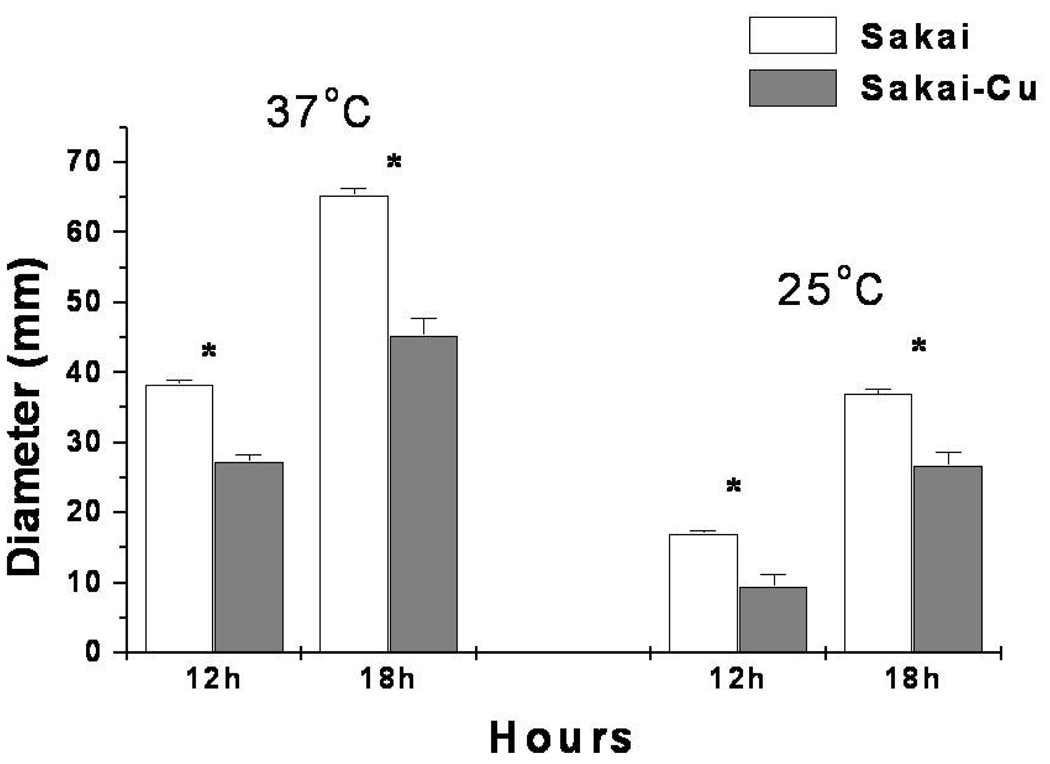
The observations that Sakai-Cu survived SGF and had increased resistance to various antibiotics compared to wild-type suggested that plasmid loss affected membrane structure. Since motility can be associated with a variety of membrane functions, we examined Sakai and Sakai-Cu for growth on standard swarm plates. As shown in Fig. 3, at 37 °C, Sakai-Cu swarmed to 27 and 65 mm in diameter after 12 h and 18 h incubation, respectively. This was ~30% less that the distances measured for wild-type Sakai movement (P < 0.05). Sakai-Cu showed a similar decreased motility at 25 °C (Fig. 4; P < 0.05). There was no significant difference in motility between 43894 and 43894-Cu (data not shown).
Figure 4.
Comparison of Sakai and Sakai-Cu for motility. Sakai and Sakai-Cu were stabbed into 0.3% motility agar and incubated for 18 h at 37°C and 25°C. The diameter of the zone of bacterial growth was measured. Asterisks indicate significant differences.
The expression of more than 50 genes is required for the motility phenotype and some of these are regulated by environmental signals (Sperandio et al., 2002). Low pH down-regulates motility via the H-NS protein (Soutourina a et al., 2002). Therefore, there might be a link between the increased acid resistance and reduced motility phenotypes that occur with the deletion of pO157 in Sakai.
To correlate relative gene expression with increased acid resistance and reduced motility phenotypes, quantitative real-time PCR (qRT-PCR) was conducted. The relative expression of three genes related to acid resistance, gadA, gadX, and rpoS, and two genes involved in flagella production, fliC and flhD, were compared between wild-type and pO157-cured strains. All experiments were performed at least in triplicate and ΔCt data was normalized to 16S rRNA as an internal control. There was a significant difference in gadA (4.84 ± 0.57 and 0.67 ± 0.61) and rpoS (3.12 ± 0.65 and 1.74 ± 0.53) expression between 43894 and 43894-Cu in the ΔCt value, respectively (P < 0.05). Therefore, we concluded that the expression of GadA was increased in 43894-Cu and it might be related to RpoS expression which is a well-known global regulator. However, although differences in expression of fliC and flhD in 43894 and differences in expression between Sakai and Sakai-Cu were measured among all five genes tested, these differences were not statistically significant (data not shown). It may be that PM was a more sensitive method to detect differences than qRT-PCR or that more complex mechanisms were involved in the phenotypic changes after the loss of pO157 in Sakai.
Among the assays for growth, acid resistance, survival and colonization of the bovine GIT, motility, heat and salt tolerance, and antibiotic susceptibility, the loss of pO157 showed effects on phenotypes that included similar and dissimilar effects between E. coli O157:H7 strains. To efficiently compare phenotypic changes associated with the deletion of pO157, we used a high throughput PM which tests 1,920 cellular phenotypes simultaneously (Bochner et al., 2001; Zhou et al., 2003). Mukherjee et al. used this technique to identify outbreak E. coli O157:H7 isolates associated with contaminated spinach, as unique in utilization of N-acetyl-D-galactosamine and showed that this strain phenotype could be a biomarker for a survival advantage associated with the food or with enhanced pathogenicity (Mukherjee et al., 2008).
The phenotypes of E. coli O157:H7 strains were analyzed and all PM results are shown in Supplement Table 1. Phenotypic differences between wild-type and pO157-cured strains are shown in Table 2 (43894 vs 43894-Cu) and Table 3 (Sakai vs Sakai-Cu). There was no difference in the utilization of carbon sources; however, interestingly, growth of 43894-Cu was increased compared to 43894 on nitrogen sources including tryptophan (Trp), tyrosine (Tyr), or phenylalanine (Phe). This suggests that pO157 in 43894 influences either the cleavage of these dipeptides, the metabolism of the corresponding amino acids, or both. Previously we reported that 43894-Cu had increased tryptophanase expression that may be related to the utilization of Trp (Lim et al., 2007). The metabolisms and transports of the aromatic amino acids are closely related and indicate that all may be influenced similarly by the O157 plasmid. The differences between Sakai and Sakai-Cu were mostly due to changes in chemical sensitivities, as shown in Table 3. When differences were detected Sakai-Cu grew better than Sakai in most chemical stress conditions but not in vancomycin, hygromycin B, Nitrofurazone, poly-L-lysine, tinidazole, orinidazol, or aparamycin. These results were consistent with the antibiotic susceptibility tests in which Sakia-Cu was more resistant than wild-type (Fig. 3). The PM assay detected differential growth between 43894 and 43894-Cu in the presence of several antibiotics (Table 3, Chemical section) for which no differences were detected by the antibiotic disk susceptibility tests. This may be due to the antibiotic concentrations tested. The discs use clinically relevant concentrations while each antibiotic was tested in the PM assay using four different concentrations. Most differences measured by the PM assay were at either the lowest or the highest antibiotic concentration (Supplement Table 1). This result showed that the pO157 in Sakai influenced various chemical sensitivities. To find the mechanisms of various phenotypic changes, a more extensive analysis with each chemical substrate is needed.
Table 2.
Phenotypic differences between wild-type E. coli O157:H7 (43894) and the isogenic pO157-cured strain (43894-Cu)
| Mode of Action | Chemical | 43894 | 43894-Cu |
|---|---|---|---|
| Nitrogen source | L-Arginine, L-Phenylalanine, L-Tryptophan | − | + |
| D-Lysine, D-Galactosamine | + | − | |
| L-Serine | ++ | − | |
|
Peptide Nitrogen source |
Ala-His | ++ | − |
| Gly-His, His-Asp, His-Leu, Ile-Phe, Ile-Trp, Ile-Tyr, Leu- Asp, Leu-Ile, Leu-Leu, Leu-Phe, His-Glu, His-His, Ile-Leu, Leu-His, Leu-Tyr, Met-Tyr, Phe-Met, Phe-Tyr, Trp-Val, Tyr- lle, Tyr-Val, Leu-Leu-Leu |
− | + | |
| Leu-Val, Met-Ile, Met-Leu, Met-Val, Phe-Ile, Trp-Leu, Trp- Tyr, Tyr-Leu, Tyr-Tyr, Val-Ile, Val-Leu, Gly-Gly-Phe |
− | ++ | |
| Leu-Trp, Trp-Lys, Trp-Phe, Trp-Trp | − | +++ | |
| Lys-Leu, Lys-Ser, Lys-Thr | + | − | |
| Phe-Trp, Thr-Leu, Val-Asp | + | +++ | |
| Phosphorous source | Thiophosphate, Dithiophosphate | − | ++ |
| Cysteamine-S-Phosphate | + | − | |
| Pyrophosphate | + | +++ | |
| Sulfur source | L-Cysteine | + | +++ |
| pH, deaminase | pH 9.5 + L-Aspartic Acid | − | + |
| Chemicals | Cefazolin, Cefoxitin, Piperacillin, Alexidine, Diamide, Chloroxylenol, Josamycin, Disulphiram, Phenethicillin, Orphenadrine |
− | + |
| Amikacin, Bleomycin, 1-Chloro-2,4-Dinitrobenzene, Methyltrioctylammonium, Chloride, Harmane, Ceftriaxone |
− | ++ | |
| Colistin, Tetracycline, Penimepicycline, 2-Nitroimidazole, Lithium Chloride, Blasticidin S, Tolylfluanid |
+ | − | |
| Streptomycin, Rifamycin SV | + | +++ | |
| Chromium Chloride | ++ | − | |
| Chloramphenicol | +++ | + | |
bacteria grew the same or less than the negative control which was a well without substrate
bacteria grew 1- to 2-fold more than the negative control
bacterial grew 2- to 3-fold more than the negative control
bacteria grew >3-fold more than the negative control
Table 3.
Phenotypic differences between wild-type E. coli O157:H7 (Sakai) and the isogenic pO157-cured strain (Sakai-Cu)
| Mode of Action | Chemical | Sakai | Sakai-Cu |
|---|---|---|---|
| Peptide Nitrogen source | Leu-Glu, Gly-Gly-D-Leu | + | − |
| Met-Lys | − | ++ | |
| Nutritional supplement | L-Cysteine | − | + |
| Phosphorous source | Pyrophosphate | + | +++ |
| Chemicals | Lincomycin, Cloxacillin, Enoxacin, Ceftriaxone, Ofloxacin, Spiramycin, Cefoxitin, Chloramphenicol, ,7-Dichloro-8- hydroxyquinoline, 5-Chloro-7-Iodo-8- Hydroxyquinoline, Dichlofluanid, Cefamandole, Methyltrioctylammonium Chloride, Harmane, Chlorhexidine, Disulphiram, Proflavine, Dodine, Oxytetracycline, Tolylfluanid |
− | + |
| Amoxicillin, Colistin, Nafcillin, Dodecyltrimethyl Ammonium Bromide, Cefuroxime, 9-Aminoacridine, Chelerythrine, Cefmetazole, Cetoperazone, Thiamphenicol, Pipemidic Acid, Antimony (III) chloride, Josamycin |
− | ++ | |
| Penicillin G | − | +++ | |
| Vancomycin, Hygromycin B | + | − | |
| Cephalothin, Oxolinic acid, Moxalactam, Acriflavine, Sodium Orthovanadate, Cefamandole, Iodonitro Tetrazolium Violet, Orphenadrine |
+ | +++ | |
| Nitrofurazone, Poly-L-lysine, Tinidazole, Ornidazole |
++ | − | |
| Apramycin | +++ | − | |
bacteria grew the same or less than the negative control which was a well without substrate
bacteria grew 1- to 2-fold more than the negative control
bacterial grew 2- to 3-fold more than the negative control
bacteria grew >3-fold more than the negative control
Unexpectedly there was little similarity between the effects measured in two E. coli O157:H7 strains after the loss of pO157 even though the two plasmids were 99% identical. Since genotypic diversities among E. coli O157:H7 isolates are well known, we performed the PM assays with three wild-type strains, 43894, Sakai, and WSU180 to determine phenotypic diversities among these strains. The three wild-type strains grew similarly in 840 of the 900 nutrient sources tested that included various carbon, nitrogen, sulfur, and phosphorus sources. The differences in utilization of nutrient sources are shown in Table 4. No pattern of differential nutrient source metabolisms among the wild-type strains was identified except for the utilization of dipeptide nitrogen sources. Compared to Sakai and WSU180, 43894 grew less well when dipeptide sources contained aromatic residues such as Trp, Tyr, or Phe. In contrast, 43894 grew better than Sakai or WSU180 when other dipeptides nitrogen sources were supplied. The three wild-type strains grew similarly in more than 1,000 other conditions such as osmotic pressure, pH, and chemical sensitivity (see Supplement Table 1). The differences in growth under various stress conditions among the wild-type E. coli O157:H7 strains are listed in Table 5. In general, 43894 was more resistant than Sakai or WSU180 to most stress conditions, however, Sakai grew better than the other two when furaltadone, 2-phenylphenol or tinidazol were applied, and WSU180 grew better than the other two strains when 1-chloro-2,4-dinitrobenzene, peperacillin, or sodium orthovanadate were added. The phenotypic diversity among wild-type E. coli O157:H7 strains can be one explanation for the different effects of pO157. 43894-Cu showed increased growth with Trp, Tyr, or Phe but Sakai-Cu not. The pO157 in Sakai may affect usage or transport of aromatic amino acids similarly in 43894, but phenotypic changes in Sakai-Cu are not as easily distinguished as in 43894-Cu because Sakai uses these aromatic amino acids more efficiently than 43894 to begin with. The same reasoning may explain the enhanced antibiotic resistance measured in Sakai-Cu but not 43894-Cu. Further studies to test this hypothesis are ongoing in our laboratory. Also, although total DNA concentration does not usually effect transcription and we did not test for it, another mechanism to explain phenotypic change may involve global influences of the pO157 on RNA polymerase distribution.
Table 4.
Metabolic capacities of wild-type E. coli O157:H7 strains
| Mode of Action | Substrate(s) | 43894 | Sakai | WSU180 |
|---|---|---|---|---|
| Carbon source | α-Keto-Butyric Acid, α-Hydroxy-Butyric Acid, L-Galactonic Acid-g-Lactone |
+ | +++ | ++ |
| Dihydroxy-Acetone | + | +++ | +++ | |
| Nitrogen source | D-Lysine, D-Galactosamine, Thymidine | + | − | − |
| L-Glutamic Acid | + | − | + | |
| Guanine | + | + | − | |
| L-Alanine | ++ | − | + | |
|
Phosphorus Source |
Tripolyphosphate | + | + | − |
| 2-Aminoethyl Phosphonic Acid | ++ | + | − | |
| Sulfur source | Lanthionine | + | − | ++ |
|
Nutritional supplement |
L-Isoleucine + L-Valine, L-Leucine, D;L-Diamino- Pimelic Acid, Nicotinic Acid, L-Glutamic Acid, Pyridoxamine |
− | + | − |
| Quinolinic Acid, Inosine + Thiamine, α-Keto-Butyric Acid, D;L-Carnitine, Choline |
+ | + | − | |
|
Dipeptide Nitrogen source |
Leu-Val | − | − | + |
| Tyr-His | − | + | − | |
| Trp-Lys, Trp-Phe, Trp-Trp | − | + | ++ | |
| Trp-Tyr | − | ++ | + | |
| Tyr-Tyr | − | ++ | ++ | |
| Ala-His, Ala-Lys, Arg-Asp, Arg-Leu, Arg-Ser, Arg- Val, His-Pro, Ile-Ala, Gly-Leu, Ile-Arg, Leu-Arg, Leu-Gly, Lys-Gly |
+ | − | − | |
| Ile-Gln, Gly-Arg, Ala-Tyr, Leu-Ala, Lys-Ser, Ala-lle | + | − | + | |
| Ile-Ser, Gly-Thr | + | + | − | |
| Ala-His, Pro-Lys, Val-Gln, Val-Ser | ++ | − | − | |
| His-Ala, Pro-Val | ++ | − | + | |
bacteria grew the same or less than the negative control which was a well without substrate
bacteria grew 1- to 2-fold more than the negative control
bacterial grew 2- to 3-fold more than the negative control
bacteria grew >3-fold more than the negative control
Table 5.
Osmotic, pH, and chemical sensitivities of wild-type E. coli O157:H7 strains
| Mode of Action | Substrate(s) | 43894 | Sakai | WSU180 |
|---|---|---|---|---|
|
Osmotic sensitivity |
4% Sodium Lactate | + | − | − |
|
pH, decarboxylase |
pH 4.5 + L-Norvaline | + | − | − |
| pH 4.5 + a- Amino-N-Butyric Acid | + | − | − | |
| pH 4.5 + 5-Hydroxy-L-Lysine | + | − | − | |
| pH 4.5 + Urea | + | − | − | |
| pH 4.5 + b-Hydroxy Glutamate | + | − | + | |
| pH 4.5 + D,L Diamino-Pimelic Acid | + | + | − | |
| pH, deaminase | pH 9.5 + Agmatine | + | − | + |
| pH 9.5 + L-Homoarginine | + | + | − | |
|
Chemical sensitivity |
Cefoxitin, Disulphiram, Methyltrioctylammonium Chloride, Polymyxin B |
− | − | + |
| Ciprofloxacin | − | + | − | |
| Chloroxylenol, Orphenadrine, Oxolinic acid | − | + | + | |
| 1-Chloro-2,4-Dinitrobenzene Piperacillin, Sodium Orthovanadate |
− | + | ++ | |
| Furaltadone | − | ++ | − | |
| 2- Phenylphenol | − | +++ | − | |
| 2,2`-Dipyridyl, 2-Nitroimidazole, 3, 4- Dimethoxybenzyl alcohol, 5,7-Dichloro-8- hydroxyquinoline, 5-Fluoro-5'-deoxyuridine, Captan, Capreomycin, Cefmetazole, Cupric chloride, D,L- Methionine, Hydroxamate, D,L-Serine, Hydroxamate, D-Cycloserine, Gallic Acid, Lithium Chloride, Neomycin, Phleomycin, Procaine, Sodium Nitrite, Tobramycin, Tolylfluanid, Umbelliferone |
+ | − | − | |
| 2,4-Dintrophenol, Blasticidin S, Colistin, Proflavine, Thioridazine, Thiosalicylate, Trifluoperazine |
+ | − | + | |
| 18-Crown-6-Ether, Cobalt chloride, L-Aspartic-b- Hydroxamate, Sanguinarine |
+ | + | − | |
| Tinidazole | + | ++ | − | |
| Amoxicillin, Chlorhexidine, Chromium Chloride, Guanidine hydrochloride, Hydroxylamine, Phenyl- Methyl-Sulfonyl-Fluoride (PMSF), Protamine Sulfate, Sisomicin |
++ | − | − | |
| Ethionamide | ++ | − | + | |
| Geneticin (G418) | ++ | − | + | |
| 5-Azacytidine, Semicarbazide hydrochloride, Troleandomycin |
++ | + | − | |
| Aluminum Sulfate, Dihydrostreptomycin | +++ | − | − | |
| Kanamycin | +++ | + | − | |
| Zinc chloride | +++ | + | + | |
bacteria grew the same or less than the negative control which was a well without substrate
bacteria grew 1- to 2-fold more than the negative control
bacterial grew 2- to 3-fold more than the negative control
bacteria grew >3-fold more than the negative control
The genomic diversity among E. coli O157:H7 isolates has been described and some differences have been linked to virulence (Ohnishi et al., 2002; Ogura et al., 2006; Zhang et al., 2006; Ogura et al., 2007; Manning et al., 2008; Wu et al., 2008). However, a systematic analysis of E. coli O157:H7 phenotypic diversity has not been done previously. An important aspect of E. coli O157:H7 fitness likely includes its ability to survive and persist in a variety of environments including animal hair coats, soils, farm water troughs, and acidic conditions (Yoon and Hovde, 2008). The ability of the bacteria to utilize various substrates likely plays a key role in survival and growth in different environmental niches. Several studies have discussed the role of pO157 in virulence and shown pO157 genetic diversity among strains using DNA microarray or PCR-based techniques (Ohnishi et al., 2002; Zhang et al., 2002; Wu et al., 2008). In this study, we showed the loss of pO157 had the common phenotypic effects of increased acid resistance and reduced colonization of cattle for Sakai and 43894 strains. However, using PM technology, we found a large number of phenotypic changes that accompanied the loss of pO157 that were not common between Sakai and 43894. These differences occurred despite the identical (except for one gene) DNA sequences of the pO157. Nonetheless, E. coli O157:H7 strains that have lost the pO157 would likely be less virulent. Adherence and colonization ability are important characteristics for pathogenic bacteria to cause disease. Supporting this notion, recent studies report that clinical genotypes of E. coli O157:H7 exhibit increased expression of pO157 genes and decreased expression of genes associated acid resistance compared to bovine-biased genotype (Vanaja et al., 2009). Further analyses that combine PM technology with DNA microarray, whole genome sequencing, and/or gene expression are needed to detail the phenotypes of E. coli O157:H7 and predict the role in virulence and survival on the farm.
Supplementary Material
Acknowledgments
We would like to thank Michael Ziman (Biolog, Hayward, CA, USA) for helpful discussions on the Phenotypic Microarrays results, Lonie Austin for animal care and handling, and Dr. C. Sasakawa for providing Sakai and Sakai-Cu. This work was supported, in part, by the Idaho Agriculture Experiment Station, the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 04-04562, Public Health Service grants U54-AI-57141 and P20-RR16454 from the National Institutes of Health, and by grants from the Idaho Beef Council.
References
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. In: Joklik WK, editor. Microbiology: A Centenary Perspective. Washington D.C., USA: ASM Press; 1999. pp. 40–45. [Google Scholar]
- Benjamin MM, Datta AR. Acid tolerance of enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Shaikh N, Holt NJ, Tarr PI, Konkel ME, Malik-Kale P, Walsh CW, Whittam TS, Bono JL. Greater Diversity of Shiga Toxin-Encoding Bacteriophage Insertion Sites among Escherichia coli O157: H7 Isolates from Cattle than in Those from Humans. Appl. Environ. Microbiol. 2007;73:671–679. doi: 10.1128/AEM.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype MicroArrays for High-Throughput Phenotypic Testing and Assay of Gene Function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157: H7. Nucl. Acids Res. 2002;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli A, Morabito S, Brugère H, Oswald E. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- Cheville AM, Arnold KW, Buchrieser C, Cheng CM, Kaspar CW. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157: H7. Appl. Environ. Microbiol. 1996;62:1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cray WC, Jr, Moon HW. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Grauke LJ, Kudva IT, Yoon JW, Hunt CW, Williams CJ, Hovde CJ. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 2002;68:2269–2277. doi: 10.1128/AEM.68.5.2269-2277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T. Complete Genome Sequence of Enterohemorrhagic Escherichia coli O157: H7 and Genomic Comparison with a Laboratory Strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE. Complete Genome Sequence of the Genetically Tractable Hydrogenotrophic Methanogen Methanococcus maripaludis. J. Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Nietfeldt J, Benson AK. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157: H7 strains in cattle. Proc. Natl. Acad. Sci. USA. 1999;96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Li J, Sheng H, Besser TE, Potter K, Hovde CJ. Escherichia coli O157: H7 Colonization at the Rectoanal Junction of Long-Duration Culture-Positive Cattle. Appl. Environ. Microbiol. 2007;73:1380–1382. doi: 10.1128/AEM.02242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Sheng H, Seo KS, Park YH, Hovde CJ. Characterization of an Escherichia coli O157: H7 Plasmid O157 Deletion Mutant and Its Survival and Persistence in Cattle. Appl. Environ. Microbiol. 2007;73:2037–2047. doi: 10.1128/AEM.02643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K. Complete Nucleotide Sequences of 93-kb and 3.3-kb Plasmids of an Enterohemorrhagic Escherichia coli O157: H7 Derived from Sakai Outbreak. DNA Res. 1998;5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE. Variation in virulence among clades of Escherichia coli O157: H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA. 2008;105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-Related Illness and Death in the United States. J. Environ. Health. 2000;62:9–18. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Mammel MK, LeClerc JE, Cebula TA. Altered Utilization of N-Acetyl-d-Galactosamine by Escherichia coli O157: H7 from the 2006 Spinach Outbreak. J. Bacteriol. 2008;190:1710–1717. doi: 10.1128/JB.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DG, Gally DL. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller AC, McEllistrem MC, Pacheco AGF, Boxrud DJ, Harrison LH. Multilocus Variable-Number Tandem Repeat Analysis Distinguishes Outbreak and Sporadic Escherichia coli O157: H7 Isolates. J. Clin. Microbiol. 2003;41:5389–5397. doi: 10.1128/JCM.41.12.5389-5397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller AC, McEllistrem MC, Stine OC, Morris JJG, Boxrud DJ, Dixon B, Harrison LH. Multilocus Sequence Typing Reveals a Lack of Diversity among Escherichia coli O157: H7 Isolates That Are Distinct by Pulsed-Field Gel Electrophoresis. J. Clin. Microbiol. 2003;41:675–679. doi: 10.1128/JCM.41.2.675-679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien AD, Kaper JB. Shiga Toxin-Producing Escherichia coli: Yesterday, Today, and Tomorrow. In: Kaper JB, O'Brien AD, editors. Escherichia coli O157: H7 and Other Shiga Toxin-Producing E. coli Strains. Washington D.C., USA: ASM Press; 1998. pp. 1–12. [Google Scholar]
- Ogura Y, Kurokawa K, Ooka T, Tashiro K, Tobe T, Ohnishi M, Nakayama K, Morimoto T, Terajima J, Watanabe H. Complexity of the Genomic Diversity in Enterohemorrhagic Escherichia coli O157 Revealed by the Combinational Use of the O157 Sakai OligoDNA Microarray and the Whole Genome PCR scanning. DNA Res. 2006;13:3–14. doi: 10.1093/dnares/dsi026. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Ooka T, Terajima J, Nougayrede JP, Kurokawa K, Tashiro K, Tobe T, Nakayama K, Kuhara S, Oswald E. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 2007;8:R138. doi: 10.1186/gb-2007-8-7-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi M, Terajima J, Kurokawa K, Nakayama K, Murata T, Tamura K, Ogura Y, Watanabe H, Hayashi T. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA. 2002;99:17043–17048. doi: 10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DH, Sheng HQ, Wynia SA, Hovde CJ. Rectoanal Mucosal Swab Culture Is More Sensitive Than Fecal Culture and Distinguishes Escherichia coli O157: H7-Colonized Cattle and Those Transiently Shedding the Same Organism. J. Clin. Microbiol. 2003;41:4924–4929. doi: 10.1128/JCM.41.11.4924-4929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Washington, D. C., USA: Cold Spring Harbor Laboratory Press; 2001. Purification of Closed Circular DNA by Equilibrium Centrifugation in CsCl-Ethidium Bromide Gradients: Discontinuous Gradients; pp. 1.69–1.71. [Google Scholar]
- Schmidt H, Kernbach C, Karch H. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157: H7. Microbiol. 1996;142:907–914. doi: 10.1099/00221287-142-4-907. [DOI] [PubMed] [Google Scholar]
- Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. Role of Escherichia coli O157: H7 Virulence Factors in Colonization at the Bovine Terminal Rectal Mucosa. Infect. Immun. 2006;74:4685–4693. doi: 10.1128/IAI.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina a OA, Krin E, Laurent-Winter C, Hommais F, Danchin A, Bertin b PN. Regulation of bacterial motility in response to low pH in Escherichia coli: the role of H-NS protein. Microbiol. 2002;148:1543–1551. doi: 10.1099/00221287-148-5-1543. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Tatsuno I, Horie M, Abe H, Miki T, Makino K, Shinagawa H, Taguchi H, Kamiya S, Hayashi T. toxB Gene on pO157 of Enterohemorrhagic Escherichia coli O157: H7 Is Required for Full Epithelial Cell Adherence Phenotype. Infect. Immun. 2001;69:6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja SK, Springman AC, Besser TE, Whittam TS, Manning SD. Differential Expression of Virulence and Stress Fitness Genes between Clinical and Bovine-biased Genotypes of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2009 doi: 10.1128/AEM.01666-09. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JS. The Ecology Cycle of Escherichia coli O157:H7. In: Stewart CS, Flint HJ, editors. Escherichia coli O157 in Farm Animals. New York, USA: CABI Publishing; 1999. pp. 195–224. [Google Scholar]
- Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Wu G, Carter B, Mafura M, Liebana E, Woodward MJ, Anjum MF. Genetic diversity among Escherichia coli O157:H7 isolates and identification of genes linked to human infections. Infect. Immun. 2008;76:845–856. doi: 10.1128/IAI.00956-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JW, Hovde CJ. All blood, No stool: enterohemorrhagic Escherichia coli O157: H7 infection. J. Vet. Sci. 2008;9:219–231. doi: 10.4142/jvs.2008.9.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WL, Kohler B, Oswald E, Beutin L, Karch H, Morabito S, Caprioli A, Suerbaum S, Schmidt H. Genetic Diversity of Intimin Genes of Attaching and Effacing Escherichia coli Strains. J. Clin. Microbiol. 2002;40:4486–4492. doi: 10.1128/JCM.40.12.4486-4492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lei XH, Bochner BR, Wanner BL. Phenotype MicroArray Analysis of Escherichia coli K-12 Mutants with Deletions of All Two-Component Systems. J. Bacteriol. 2003;185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.