Abstract
After termination, eukaryotic 80S ribosomes remain associated with mRNA, P-site deacylated tRNA and release factor eRF1, and must be recycled by dissociating these ligands and separating ribosomes into subunits. Although recycling of eukaryotic post-termination complexes (post-TCs) can be mediated by initiation factors eIF3, eIF1 and eIF1A (Pisarev et al., 2007), this energy-free mechanism can function only in a narrow range of low Mg2+ concentrations. Here we report that ABCE1, a conserved and essential member of the ATP-binding cassette (ABC) family of proteins, promotes eukaryotic ribosomal recycling over a wide range of Mg2+ concentrations. ABCE1 dissociates post-TCs into free 60S subunits and mRNA- and tRNA-bound 40S subunits. It can hydrolyze ATP, GTP, UTP and CTP. NTP hydrolysis by ABCE1 is stimulated by post-TCs and is required for its recycling activity. Importantly, ABCE1 dissociates only post-TCs obtained with eRF1/eRF3 (or eRF1 alone), but not post-TCs obtained with puromycin in eRF1's absence.
Keywords: ABCE1, recycling, ribosome, eRF1, eRF3
Introduction
Protein synthesis consists of four stages: initiation, elongation, termination and ribosomal recycling. During termination, when a stop codon enters the A-site, release factors (RFs/eRFs) mediate hydrolysis of the P-site peptidyl-tRNA. In bacteria, class-1 RF1 and RF2 promote hydrolysis of peptidyl-tRNA, whereas the class-2 RF3 mediates release of RF1/RF2 from post-termination ribosomes and dissociates itself after hydrolyzing GTP, yielding post-TCs that comprise 70S ribosomes, mRNA and P-site deacylated tRNA (Zavialov et al., 2001, Gao et al., 2007). Recycling of post-TCs requires elongation factor EF-G, the ribosome recycling factor RRF and initiation factor IF3 (reviewed in Petry et al., 2008). EF-G and RRF dissociate post-TCs into free 50S subunits and 30S subunits bound to mRNA and P-site deacylated tRNA in a GTP-dependent manner, after which IF3 induces tRNA release from 30S subunits, and mRNA then dissociates spontaneously. RRF binds to post-TCs in the ratcheted state, interacting with segments of 23S rRNA that are constituents of B2a and B3 inter-subunit bridges (Gao et al., 2005). It has been proposed that EF-G•GDP binds RRF-associated post-TCs and exchanges GDP for GTP; EF-G•GTP then induces rotation of RRF's head domain, which after GTP hydrolysis splits ribosomes into subunits (Gao et al., 2005).
The termination mechanism in eukaryotes differs significantly from that in bacteria: whereas RF3 mediates recycling of RF1/RF2 from post-TCs, eukaryotic class-2 eRF3 acts cooperatively with class-1 eRF1 to ensure rapid peptide release (Alkalaeva et al., 2006). In a current model, eRF1, eRF3 and GTP form a ternary complex, which induces conformational changes in eRF1 that likely increase its affinity to pre-termination complexes (pre-TCs) (Cheng et al., 2009 and references therein). Binding of eRF1/eRF3/GTP induces conformational changes in pre-TCs, which manifest as a 2nt forward shift of their toe-print (Alkalaeva et al., 2006). Subsequent GTP hydrolysis by eRF3 induces further conformational changes, likely in eRF1 (Cheng et al., 2009), required for positioning of eRF1's GGQ loop in the peptidyl transferase center and efficient peptide release (Alkalaeva et al., 2006; Fan-Minogue et al., 2008). In contrast to bacteria, at least one release factor, eRF1, remains bound to eukaryotic post-TCs, accounting for maintenance of the 2nt toe-print shift in post-TCs after peptide release (Pisarev et al., 2007). The ribosomal binding sites for eRF1/eRF3 and EF-G/RRF overlap, and since eRF1 remain associated with post-TCs, mechanisms of ribosomal recycling in the two kingdoms must differ. Consistently, eukaryotes do not encode a RRF homologue, and at a free (nucleotide-unbound) Mg2+ concentration of 1 mM, eukaryotic recycling can be mediated by initiation factors eIF3, eIF1, eIF1A and eIF3's loosely associated 3j subunit (Pisarev et al., 2007). eIF3, which binds to the solvent side of 40S subunits (Siridechadilok et al., 2005), promotes dissociation of post-TCs into 60S subunits and tRNA/mRNA-bound 40S subunits, and its activity is enhanced by eIF3j, eIF1 and eIF1A. eIF1 then induces release of P-site tRNA, followed by 3j-mediated dissociation of mRNA.
Although bacterial and proposed eukaryotic recycling mechanisms show some similarities, such as dissociation of post-TCs into free large subunits and tRNA/mRNA-bound small subunits, and subsequent promotion of release of deacylated tRNA by eIF1 and IF3, factors that bind to identical regions on small ribosomal subunits and perform equivalent roles during initiation (Lomakin et al., 2006 and references therein), the differences between these mechanisms outweigh their similarities. The principal difference is that in bacteria, recycling occurs from the intersubunit space and involves GTP hydrolysis, whereas in eukaryotes, in the current model it occurs without energy consumption and relies on eIF3 acting from the 40S subunit's solvent side. We have now determined that this “passive” mechanism of eukaryotic recycling functions only in a narrow range of low Mg2+ concentrations, but ribosomal recycling in a wide range of Mg2+ concentrations is promoted by ABCE1, an essential member of the ATP-binding cassette (ABC) family of proteins, which dissociates post-TCs into free 60S subunits and mRNA- and tRNA-bound 40S subunits.
Results
Initiation factors promote ribosomal recycling only at low Mg2+ concentrations
To investigate the ability of eIFs 3/1/1A to promote recycling at elevated Mg2+ concentrations, which reduce the flexibility of ribosomal subunits and stabilize their association (Shenvi et al., 2005), pre-TCs were assembled from 40S subunits, [32P]60S subunits, eIFs 2, 3, 1, 1A, 4A, 4B, 4F, 5 and 5B, elongation factors eEF1H and eEF2, and native aminoacylated Met-, Val-, His- and Leu-tRNA on MVHL-STOP mRNA encoding a MVHL tetrapeptide followed by a UAA stop codon (Fig. 1A), and purified by sucrose density gradient (SDG) centrifugation. We note that Mg2+ concentrations described throughout the text will refer to the free (nucleotide unbound) concentration. Whereas incubation of such pre-TCs with eRF1/eRF3 and eIFs 3/1/1A at 1 mM Mg2+ dissociates post-termination ribosomes into subunits (Pisarev et al., 2007; Fig. 1B), no dissociation occurred at 2.5 mM Mg2+ (Fig. 1B), even though peptide release is efficient even at 8 mM Mg2+ (Alkalaeva et al., 2006). Binding of eRF1/eRF3/GTP to pre-TCs at 1 mM Mg2+ shifts their toe-print 2nt forward from +16nt from the P-site CUU codon to +15nt from the A-site UAA stop codon (Alkalaeva et al., 2006), and this shift persists in post-TCs due to their continued association with eRF1 (or with eRF1/eRF3 if eRF3 does not dissociate after GTP hydrolysis) even after peptide release (Pisarev et al., 2007). However, the toe-print shift in post-TCs was less complete than in pre-TCs when peptide release was not allowed (e.g. in the presence of GMPPNP) suggesting that eRF(s) to some extent dissociate from post-TCs (Alkalaeva et al., 2006). We now found that at 2.5 mM Mg2+, the toe-print shift in post-TCs was preserved completely (Fig. 1C), indicating that eRF1 (or eRF1/eRF3) remained firmly bound to post-TCs. Consistently, whereas at 1 mM Mg2+, eRF1 dissociated from post-TCs after SDG centrifugation, it remained bound at 2.5 mM Mg2+ (Fig. 1D). Although no association of eRF3 with post-TCs was observed at any Mg2+ concentration (data not shown), we cannot exclude that eRF3 might have dissociated due to the stringency of centrifugation. Importantly, even though firm association of eRF1 (or eRF1/eRF3) with post-TCs at 2.5 mM Mg2+ likely contributes to their stability, it cannot account entirely for the inability of eIFs to promote recycling at this Mg2+ concentration, because post-TCs obtained by incubation with puromycin could also not be recycled (Fig. 1B).
Figure 1. Association of Post-TCs with eRF1 and their recycling by eIFs at Different Mg2+ Concentrations.
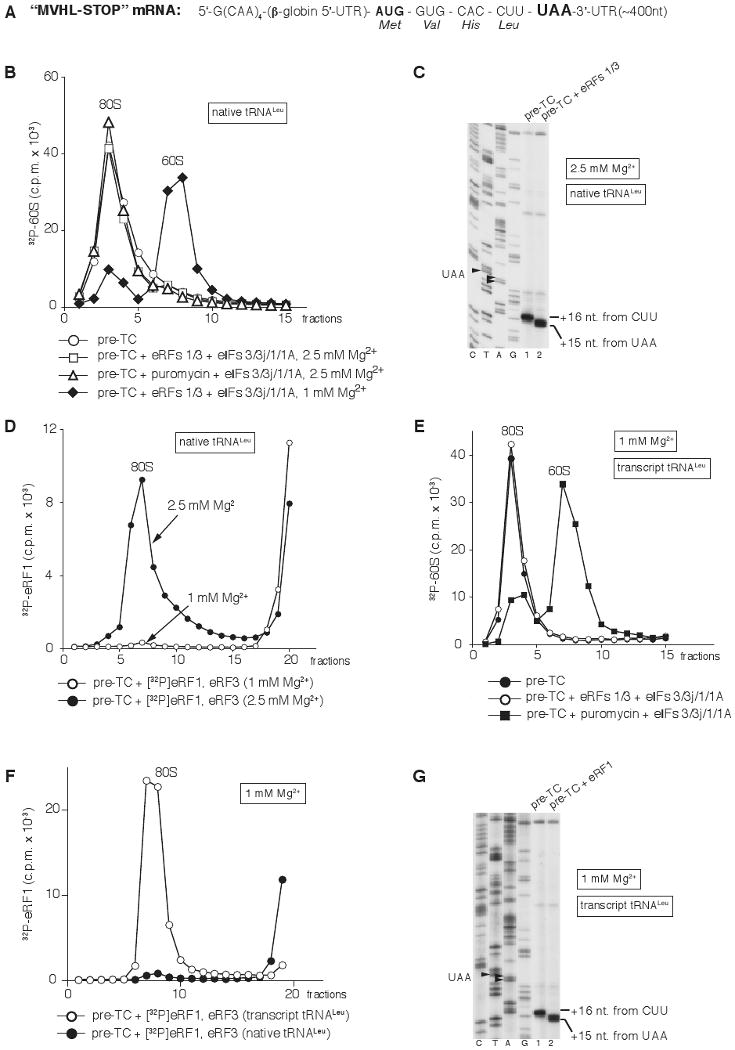
(A) Structure of MVHL-STOP mRNA.
(B) Dissociation of pre-TCs, assembled on MVHL-STOP mRNA with [32P]60S subunits and native Leu-tRNALeu after incubation with eRFs, eIFs, and puromycin at indicated Mg2+ concentrations, assayed by SDG centrifugation.
(C) Toeprinting analysis of ribosomal complexes obtained by incubating pre-TCs assembled on MVHL-STOP mRNA using native tRNALeu with eRFs at 2.5 mM Mg2+. Lanes C, T, A, and G depict cDNA sequences corresponding to MVHL-STOP mRNA.
(D) Association of [32P]eRF1 with post-TCs assembled on MVHL-STOP mRNA with native Leu-tRNALeu at indicated Mg2+ concentrations, assayed by SDG centrifugation.
(E) Dissociation of pre-TCs, assembled on MVHL-STOP mRNA with [32P]60S subunits and transcript Leu-tRNALeu after incubation with eRFs, eIFs, and puromycin at 1 mM Mg2+, assayed by SDG centrifugation.
(F) Association of [32P]eRF1 with post-TCs assembled on MVHL-STOP mRNA with native or transcript Leu-tRNALeu at 1 mM Mg2+, assayed by SDG centrifugation.
(G) Toeprinting analysis of ribosomal complexes obtained by incubating pre-TCs assembled on MVHL-STOP mRNA using transcript tRNALeu with eRF1 at 1 mM Mg2+. Lanes C, T, A, and G depict cDNA sequences corresponding to MVHL-STOP mRNA. Upper fractions were omitted for clarity from (B) and (D)–(F).
Interestingly, we also found that post-TCs assembled with P-site unmodified in vitro transcribed tRNALeu could not be recycled by eIFs even at 1 mM Mg2+ if peptide release was triggered by eRFs, but could if it was induced by puromycin (Fig. 1E). The inability of eIFs 3/1/1A to recycle post-TCs containing transcript tRNALeu that were obtained with eRFs could not be explained by incomplete peptide release, since it occurred as efficiently as on post-TCs with native tRNALeu (Supplemental Fig. S1), and was most likely due to increased retention of eRF1 (or eRF1/eRF3) on post-TCs after peptide release. Thus, in contrast to post-TCs with native tRNALeu, eRF1 remained on post-TCs with transcript tRNALeu after SDG centrifugation even at 1 mM Mg2+ (Fig. 1F). Moreover, in contrast to post-TCs with native tRNALeu, in which the 2nt toe-print shift required both eRFs (Alkalaeva et al., 2006), this shift could be induced by eRF1 alone in complexes with transcript tRNALeu (Fig. 1G). Nucleotide modifications stabilize tRNA conformation (Agris, 2008), and their absence in transcript tRNALeu likely altered its conformation and interactions with the ribosome or even eRF1 in a way that enhanced eRF1's association with post-TCs. We do not know which characteristics of transcript tRNALeu are responsible for strengthening the eRF1/post-TC interaction but note that post-TCs assembled with native tRNACys or tRNAGln behaved analogously to post-TCs assembled with native, rather than with transcript tRNALeu (data not shown).
Thus, elevation of the Mg2+ concentration or firm association of eRF1 (or eRF1/eRF3) with post-TCs prevent ribosomal recycling by eIFs 3/1/1A in the in vitro reconstituted system.
ABCE1 dissociates post-TCs into free 60S subunits and mRNA- and tRNA-bound 40S subunits over a wide range of Mg2+ concentrations
In RRL, however, recycling of pre-assembled post-TCs containing native or transcript tRNALeu occurred efficiently over a wide range of Mg2+ concentrations (Supplemental Fig. S2), indicating the existence of a different recycling mechanism. Purification from RRL of factor(s) that promoted recycling at elevated Mg2+ concentrations yielded an apparently homogenous ∼65 kDa protein (Fig. 2A) that was identified as ABCE1 (Supplementary Table S1), a member of the ABC family of proteins that are mostly involved in transport across membranes but also in DNA repair and translation (Rees et al., 2009). ABC proteins typically contain two nucleotide-binding domains (NBDs) arranged in a head-to-tail manner to form two composite active sites, and can generate a tweezer-like power stroke upon nucleotide-dependent conformational transitions between the stages of ATP binding and hydrolysis.
Figure 2. Dissociation of post-TCs by ABCE1.
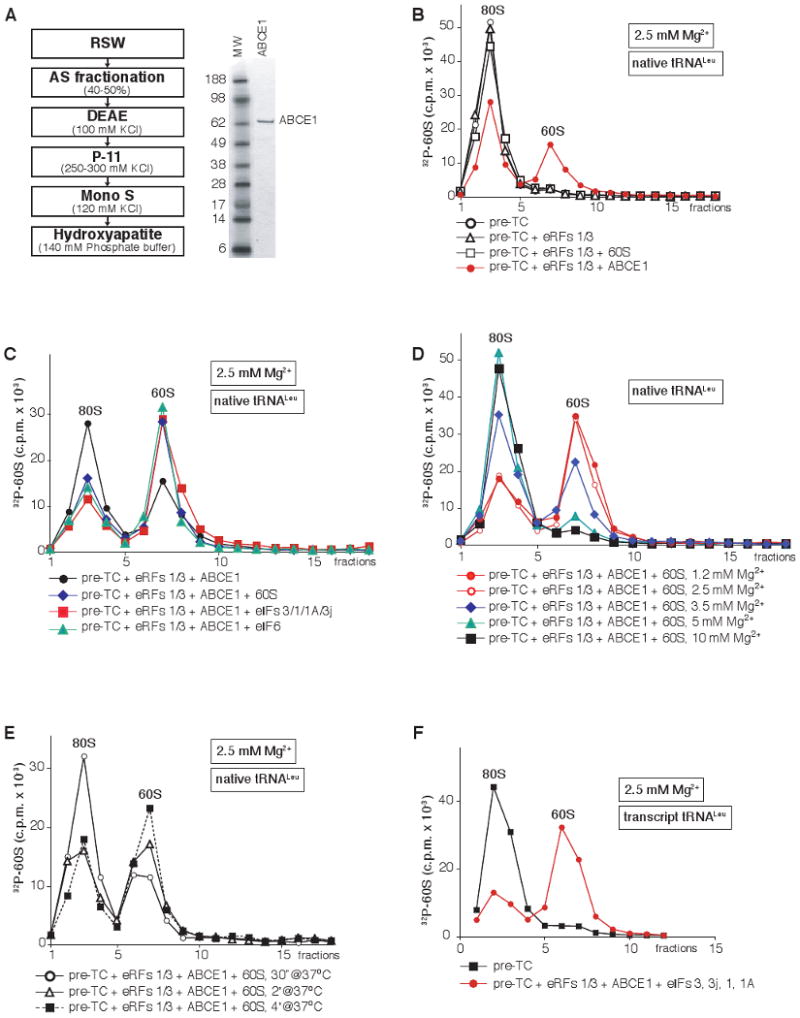
(A) Left panel - scheme for purification of ABCE1 from RRL, right panel - purified ABCE1 resolved by SDS-PAGE. (B-F) Dissociation of pre-TCs, assembled on MVHL-STOP mRNA with [32P]60S subunits and (B-E) native or (F) transcript Leu-tRNALeu, after incubation with different combinations of eRFs, ABCE1, eIFs and unlabeled 60S subunits at indicated Mg2+ concentrations, assayed by SDG centrifugation. Upper fractions were omitted for clarity.
Expression of ABCE1 in E. coli or S. cerevisiae did not yield active protein (data not shown), and earlier attempts to purify overexpressed ABCE1 from S. cerevisiae likely also resulted in its inactivation (Dong et al., 2004). For this reason, experiments described below were done using native ABCE1.
At 2.5 mM Mg2+, ABCE1 promoted efficient (∼40%) dissociation into subunits of post-TCs containing native tRNALeu (Fig. 2B). However, the level of dissociation by ABCE1 was substantially increased by inclusion in reaction mixtures of 60S subunits, eIF6 (a ribosome anti-association factor that binds 60S subunits; Si et al., 1997), or eIFs 3/1/1A (Fig. 2C) that can all prevent reassociation of recycled [32P]60S with 40S subunits, which indicated that dissociation was transient in nature. All further experiments on dissociation of post-TCs by ABCE1 were therefore done in the presence of eIF6 to trap recycled 60S subunits, or 60S subunits or eIFs to trap recycled 40S subunits, as appropriate.
Recycling by ABCE1 was equally efficient at 1.2 and 2.5 mM Mg2+, ∼30% less efficient at 3.5 mM Mg2+, marginal at 5 mM Mg2+, and did not occur at 10 mM Mg2+ (Fig. 2D). Importantly, the Mg2+-dependence of recycling observed in the reconstituted system in the presence of ABCE1 was similar to that in RRL (Supplemental Fig. S2), although we note that the concentration of free Mg2+ in RRL could not be calculated precisely. Recycling was rapid, and ∼30% of post-TCs were dissociated within 30 seconds of incubation (Fig. 2E). ABCE1 was also able to promote dissociation of post-TCs assembled with transcript tRNALeu (Fig. 2F).
After incubation of pre-TCs assembled using 32P-labeled Leu-tRNALeu or MVHL-STOP mRNA with eRF1/eRF3, ABCE1 and eIF6, both tRNA and mRNA were bound to recycled 40S subunits (Fig. 3A, black squares; Fig. 3B, red circles), indicating that ABCE1 dissociates post-TCs into free 60S subunits and mRNA/tRNA-bound 40S subunits. Consistently, in toe-printing experiments, incubation of post-TCs with ABCE1 reversed the 2nt toe-print shift back to +16nt from the P-site CUU codon indicating dissociation of post-TCs into free 60S subunits and 40S subunits associated with mRNA and P-site deacylated tRNA, which were responsible for the +16nt toe-print (Fig. 3E, lanes 2, 3). Inclusion of eIFs 3/3j/1A did not influence association of tRNA with recycled 40S subunits (Fig. 3A, black triangles), whereas ∼30% less tRNA was bound to 40S subunits in the presence of eIF1 (Fig. 3A, open red circles), and only ∼10% tRNA remained bound in the presence of eIFs 1/1A (Fig. 3A, filled red circles), indicating that eIF1 is the key factor in promoting release of tRNA from recycled 40S subunits, and that eIF1A strongly enhances its activity. Efficient eIF1/eIF1A-mediated dissociation of tRNA was accompanied by near-complete mRNA release (Fig. 3C). Inclusion in the reaction mixture of eIF3 in addition to eIFs 1/1A resulted in less complete dissociation of mRNA (Fig. 3D, blue triangles), consistent with the reported stabilization of 40S/mRNA association by eIF3 (Kolupaeva et al., 2005), and efficient dissociation of mRNA from recycled 40S subunits in the presence of eIF3 required eIF3j (Fig. 3D, red circles). As in the case of tRNA, association of mRNA with 40S subunits was not influenced by eIFs 3/3j/1A in the absence of eIF1 (Fig. 3D). However, in toe-printing experiments, predominant synthesis of full-length cDNA indicative of complete recycling was observed only in the presence of all four initiation factors (Fig. 3E, lane 4), but not eIF1 and eIF1A alone (Fig. 3E, lane 6). This indicates that in the absence of eIF3, eIFs 1/1A only weaken association of P-site deacylated tRNA with recycled 40S subunits, which leads to tRNA and subsequent mRNA dissociation during centrifugation, whereas release of tRNA and mRNA from recycled 40S subunits without SDG centrifugation also requires eIF3 and eIF3j. Thus, the mechanism of tRNA and mRNA release from 40S subunits during recycling at high Mg2+ concentrations in the presence of ABCE1 paralleled that observed at low Mg2+ concentrations in its absence (Pisarev et al., 2007).
Figure 3. Dissociation of tRNA and mRNA from recycled 40S subunits.
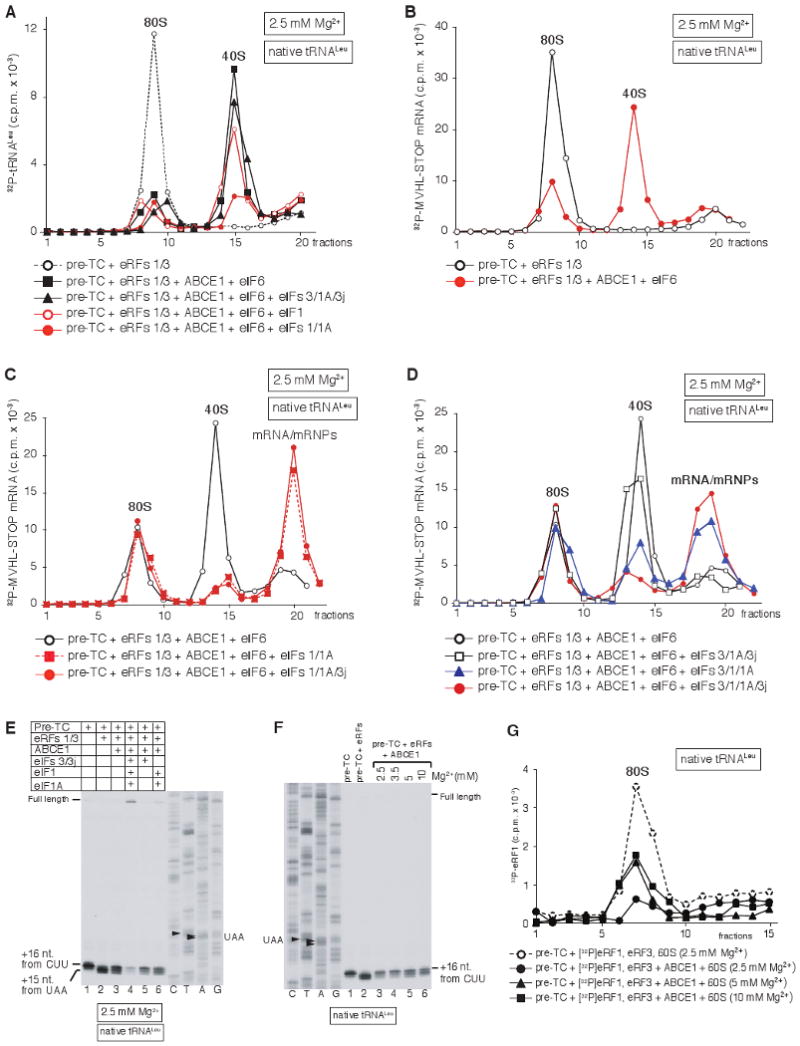
(A-D) Ribosomal association of (A) [32P]tRNALeu and (B-D) [32P]MVHL-STOP mRNA after incubating pre-TCs with eRFs, ABCE1 and eIFs at 2.5 mM Mg2+, as indicated, assayed by SDG centrifugation. (E, F) Toe-print analysis of ribosomal complexes, obtained by incubating pre-TCs, assembled on MVHL-STOP mRNA using native Leu-tRNALeu, with combinations of eRFs, ABCE1 and eIFs at indicated Mg2+ concentrations. Lanes C, T, A, G depict cDNA sequences corresponding to MVHL-STOP mRNA. The positions of full-length cDNA and of toe-prints corresponding to ribosomal complexes are indicated. (G) Influence of ABCE1 on association of [32P]eRF1 with post-TCs at different Mg2+ concentrations, assayed by SDG centrifugation. Upper fractions were omitted for clarity from panels A, G.
Interestingly, incubation of post-TCs with ABCE1 at 5 or 10 mM Mg2+, when dissociation of post-TCs into subunits does not occur (Fig. 2D), also reversed the 2nt toe-print shift (Fig. 3F). Even though post-TCs were not dissociated at such non-permissive Mg2+ concentrations, association of eRF1 with them was reduced (Fig. 3G, triangles and squares), indicating that ABCE1 may cause conformational changes in post-TCs that weaken the eRF1/post-TC interaction.
To exclude the possibility that ABCE1's recycling activity was either an artifact of the highly fractionated reconstituted system, or due to trace amounts of co-purifying protein(s), recycling of preassembled post-TCs was investigated in a HeLa cell extract, in which ABCE1's level was reduced by RNA interference. Transfection of HeLa cells with ABCE1 siRNA resulted in ∼80% depletion of ABCE1 mRNA (data not shown) and in 80-90% reduction in protein level (Fig. 4A), which was accompanied by a 4-fold reduction in translation of a capped polyadenylated reporter mRNA in the corresponding silenced HeLa cell extract comparing to mock depleted (GFP) extract (Fig. 4B). To estimate the effect of ABCE1-depletion on recycling, pre-TCs were assembled on MVHL-STOP mRNA, purified by SDG centrifugation, treated with eRFs 1/3 to induce peptide release, and then incubated with control or ABCE1-silenced HeLa cell extracts. Ribosomal association of MVHL mRNA was then assayed by toe-printing. Incubation with the control extract led to 3-5-fold more efficient dissociation of post-TCs and a concomitant increase in appearance of full-length cDNA, than incubation with ABCE1-silenced extract over a range of extract concentrations (Fig. 4C), indicating that recycling was impaired in the latter. Addition of purified ABCE1 to the ABCE1-silenced extract restored its recycling activity (Fig. 4C).
Figure 4. Ribosomal recycling is impaired in ABCE1-silenced HeLa cell extract: Dissociation of post-TCs by ABCE1 requires eRF1.
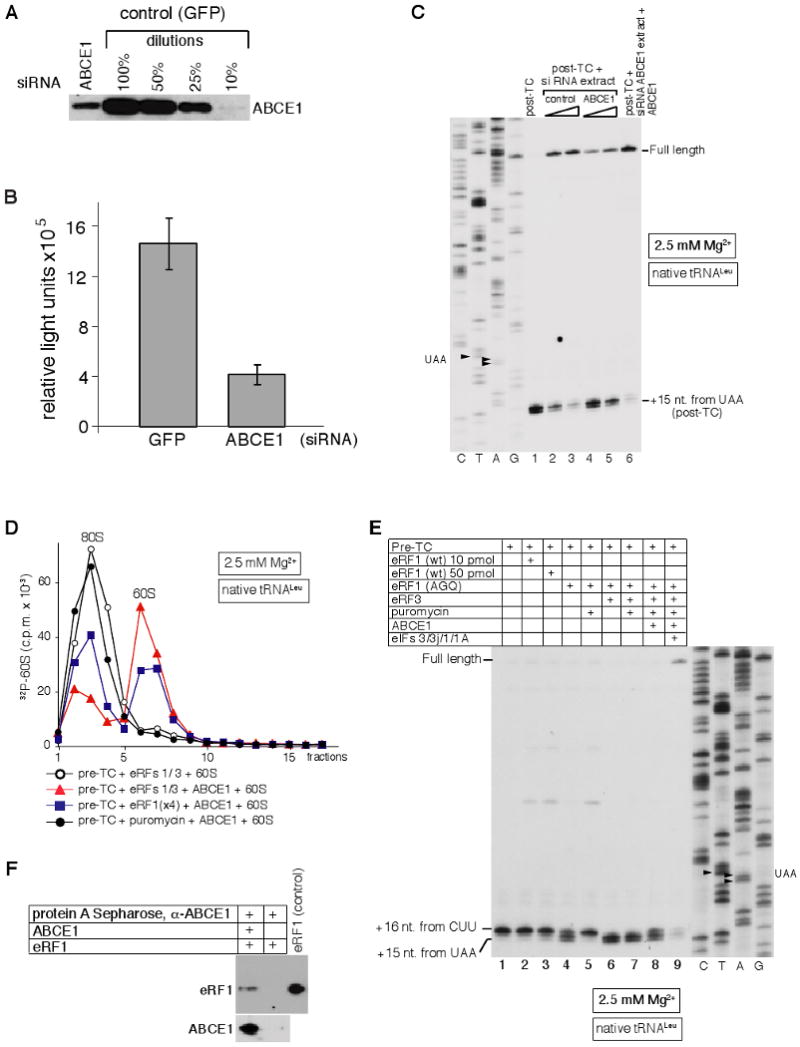
(A) Expression of ABCE1 in HeLa cells transfected with control (GFP) or ABCE1 siRNAs, assayed by western blotting. (B) Influence of ABCE1 silencing on protein synthesis assayed by the translation efficiency of a capped, polyadenylated luciferase reporter mRNA in control (GFP) and ABCE1-silenced HeLa cell extracts. (C) Toe-print analysis of recycling of post-TCs, assembled on the MVHL-STOP mRNA using the in vitro reconstituted translation system, in control or ABCE1-silenced HeLa cell extracts. Lanes C, T, A, G depict cDNA sequences corresponding to MVHL-STOP mRNA. The positions of full-length cDNA and of toe-prints corresponding to post-TCs are indicated. (D) Dissociation by ABCE1 of post-TCs, obtained by incubation of pre-TCs assembled on MVHL-STOP mRNA using [32P]60S subunits with eRF1/eRF3, eRF1 alone, or puromycin, assayed by SDG centrifugation. (E) Toe-print analysis of ribosomal complexes, obtained by incubating pre-TCs, assembled on MVHL-STOP mRNA using native Leu-tRNALeu, with combinations of eRFs, puromycin, ABCE1 and eIFs at 2.5 mM Mg2+. Lanes C, T, A, G depict cDNA sequences corresponding to MVHL-STOP mRNA. The positions of full-length cDNA and of toe-prints corresponding to ribosomal complexes are indicated. (F) Interaction between ABCE1 and eRF1 assayed by co-immunoprecipitation. Anti-ABCE1 antibodies with/without ABCE1 were bound to protein A sepharose, which was then incubated with eRF1. Proteins bound to the matrix were further analyzed by western blotting using anti-eRF1 (upper panel) and anti-ABCE1 (lower panel) antibodies.
Dissociation of post-TCs by ABCE1 requires eRF1
Importantly, ABCE1 dissociated efficiently only those post-TCs, in which peptide release had been triggered by both eRF1 and eRF3 (Fig. 4D, red triangles), whereas dissociation of post-TCs obtained with eRF1 alone was less efficient (blue squares), and occurred only at high concentrations of eRF1 that exceeded those required for peptide release. Moreover, post-TCs obtained with puromycin could not be dissociated by ABCE1 (black circles), indicating that eRF1 was essential for recycling of post-TCs by ABCE1.
The difference in the efficiency of recycling of post-TCs obtained with eRF1/eRF3 and with eRF1 alone must reflect differences in their composition and/or conformation. We noticed that when peptide release was induced by eRF1 alone, the 2nt toe-print shift of post-TCs occurred very inefficiently, and only at a very high eRF1 concentration (Fig. 4E, lanes 2, 3). However, in contrast to wt eRF1, the eRF1(AGQ) mutant with a substitution in the GGQ motif (which is inactive in peptide release) could efficiently induce the shift in eRF3's absence, but it disappeared upon puromycin-induced peptide release (Fig. 4E, lanes 4, 5). This suggested that wt eRF1 might also induce the shift during initial binding to pre-TCs, and that it reverts after peptide release. On the other hand, addition of puromycin to pre-TCs incubated with eRF1(AGQ)/eRF3 did not eliminate this shift (Fig. 4E, lanes 6, 7), and addition of ABCE1 and eIFs 3/3j/1/1A to such post-TCs obtained with eRF1(AGQ)/eRF3/puromycin promoted their recycling (Fig. 4E, lane 9). Since the 2nt toe-print shift most likely reflects association of eRF1 with ribosomal complexes, these observations suggest that eRF1 remains firmly bound to post-TCs only in the presence of eRF3. We also detected a direct physical interaction between eRF1 and ABCE1 using a pulldown assay (Fig. 4F), but the specificity of this interaction and its existence in the context of post-TCs remain to be investigated. Taken together, these data suggest that conformational changes induced in post-TCs by release factors or a physical interaction between eRF1 and ABCE1 are required for productive interaction of ABCE1 with post-TCs.
Ribosomal association of ABCE1
We next investigated the nucleotide-dependence of ribosomal binding of ABCE1 using SDG centrifugation followed by western blotting. In the presence of AMPPNP, ABCE1 bound stably to 40S subunits and to 43S complexes obtained with GMPPNP (Fig. 5A, lanes 1, 3), but not to 80S ribosomes (Fig. 5A, lane 7) or pre-TCs (Fig. 5C, lane 5), indicating that the ABCE1-binding site in both of them is occluded. However, in the presence of AMPPNP, ABCE1 efficiently associated with post-TCs containing transcript tRNALeu that were obtained with eRF1 alone (Fig. 5C, lane 1), and with pre-TCs bound to the eRF1(AGQ) mutant, which is inactive in peptide release (Fig. 5C, lane 7), indicating that AMPPNP-dependent binding of ABCE1 to 80S ribosomal complexes requires their association with eRF1, but does not depend on peptide release. The fact that ABCE1 did not associate with any ribosomal complexes in the presence of ATP or ADP (Figs. 5A-C) suggests that NTP hydrolysis by ABCE1 reduces its ribosomal affinity.
Figure 5. Nucleotide-dependence of ribosomal association of ABCE1.
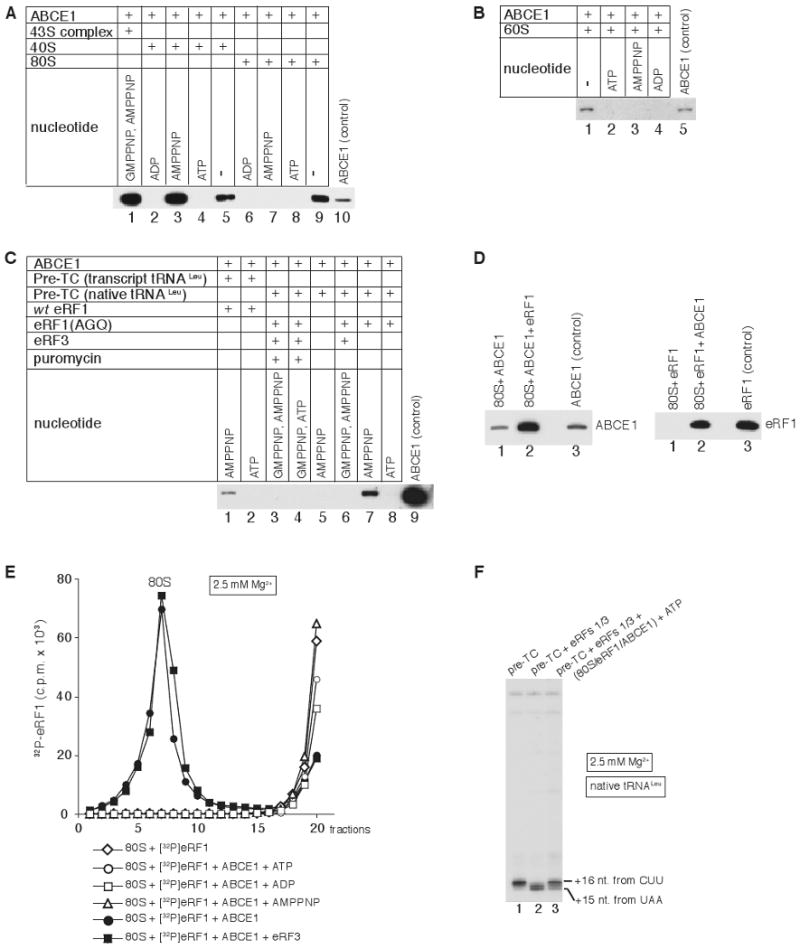
(A-C) Association of ABCE1 with (A) 40S subunits, 43S complexes and 80S ribosomes, (B) 60S subunits and (C) pre-TCs assembled with native or transcript Leu-tRNALeu and incubated with combinations of wt eRF1, eRF1(AGQ), eRF3 and puromycin, in the presence/absence of nucleotides, as indicated. Ribosomal peak fractions obtained by SDG centrifugation were analyzed by western blotting using anti-ABCE1 antibodies. (D) Association of ABCE1 and eRF1 with individual 80S ribosomes in the absence of nucleotides assayed by SDG centrifugation and western blotting of peak ribosomal fractions using anti-ABCE1 (left panel) and anti-eRF1 (right panel) antibodies. (E) Association of [32P]eRF1 with 80S ribosomes in the presence/absence of ABCE1 and nucleotides as indicated, assayed by SDG centrifugation. Upper fractions were omitted for clarity. (F) Toe-print analysis of ribosomal complexes obtained by incubating pre-TCs, assembled on MVHL-STOP mRNA using native tRNALeu, with eRF1/eRF3 and SDG-purified 80S/eRF1/ABCE1 complexes (panel E). Toe-prints corresponding to ribosomal complexes are indicated.
Importantly, addition of eRF3/GMPPNP to reaction mixtures containing pre-TCs and eRF1(AGQ) prevented association of ABCE1 with ribosomal complexes irrespective of whether or not peptide release was induced by puromycin (Fig. 5C, lanes 3, 6). Inhibition of ABCE1's ribosomal binding by eRF3/GMPPNP suggests a mechanism to prevent premature association of ABCE1 with termination complexes.
In the absence of nucleotides, ABCE1 alone associated weakly with 80S ribosomes, 40S and 60S subunits (Fig. 5A, lanes 5, 9; Fig. 5B, lane 1), whereas in the presence of eRF1, ABCE1 and eRF1 exhibited strong cooperative binding to 80S ribosomes, which was not influenced by eRF3 (Fig. 5D, lanes 1, 2 of both panels; Fig. 5E). eRF1's association with 80S ribosomes was not enhanced by ABCE1 in the presence of ATP, ADP or AMPPNP (Fig. 5E). In toe-printing experiments, addition of SDG-purified 80S/eRF1/ABCE1 complexes to post-TCs eliminated the 2nt toe-print shift (Fig. 5F), indicating that in the presence of ATP, ABCE1 can dissociate from 80S/eRF1/ABCE1 complexes and interact productively with post-TCs.
NTP hydrolysis by ABCE1 is required for its function in ribosomal recycling
ABCE1 had very low intrinsic NTPase activity and lacked specificity, hydrolyzing ATP, GTP, CTP and UTP (Fig. 6A, lanes 3, 7, 11, 15). ABCE1's NTPase activity was stimulated 2-3-fold by 80S ribosomes (Fig. 6A, lanes 4, 8, 12, 16). The low stimulation of ABCE1's NTPase activity by 80S ribosomes is consistent with the lack of stable interaction between ABCE1 and free 80S ribosomes (Fig. 5A). However, ABCE1's NTPase activity was very strongly stimulated in the presence of pre-TCs and eRF1(AGQ) or wt eRF1 (shown for GTP hydrolysis in Fig. 6B, filled circles and triangles). Stimulation by pre-TCs and wt eRF1 was reduced by eIF6, which most likely prevented reassociation of recycled ribosomal subunits thus allowing fewer rounds of NTP hydrolysis per ribosomal complex (Fig. 6B, open diamonds). Stimulation of NTP hydrolysis by pre-TCs alone was low (Fig. 6B, open squares) and similar to that observed for individual 80S ribosomes (Fig. 6A). Stimulation of ABCE1's NTPase activity by different ribosomal complexes therefore correlated with their ability to bind ABCE1.
Figure 6. NTPase activity of ABCE1 is required for its function in ribosomal recycling.
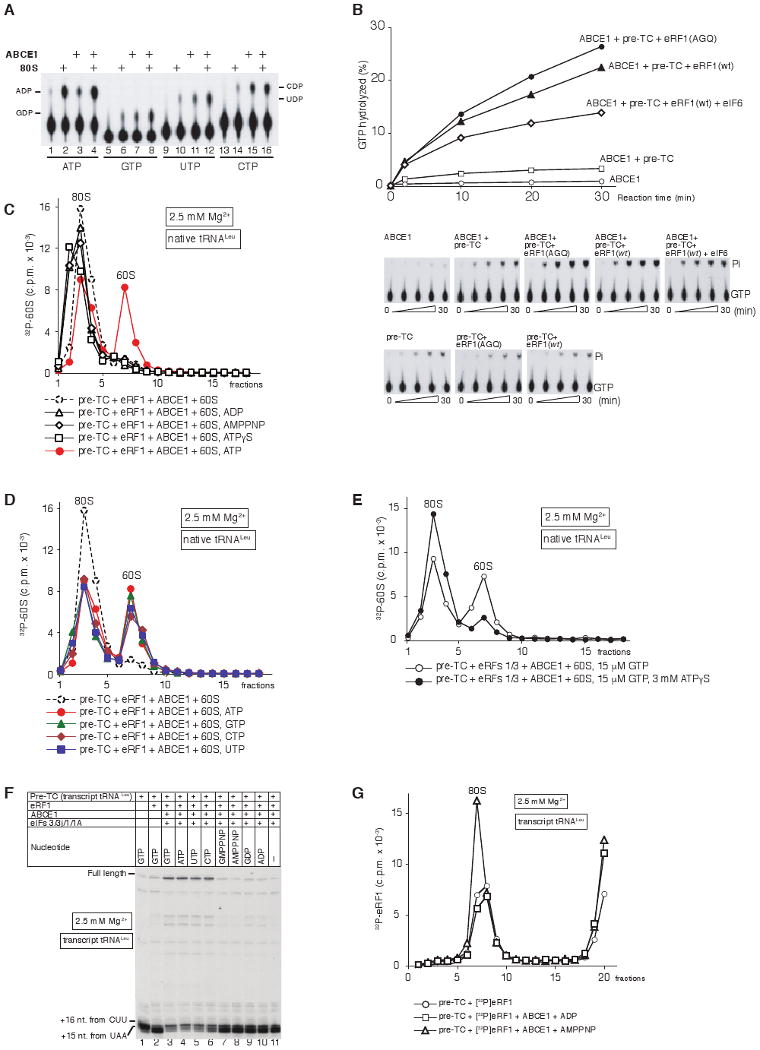
(A) Thin-layer chromatography analysis of ABCE1's NTPase activity in the presence/absence of 80S ribosomes. The positions of [α32P]-NDPs are indicated. (B) Time courses of GTP hydrolysis by ABCE1 in the presence/absence of pre-TC, pre-TC/eRF1(AGQ), pre-TC/eRF1(wt), or pre-TC/eRF1(wt)/eIF6, as indicated. 15 μl reaction mixtures containing 0.5 pmol ABCE1, 0.33 μM [γ-32P]GTP and combinations of 0.5 pmol pre-TC, 10 pmol eRF1(AGQ), 10 pmol eRF1(wt) and 10 pmol eIF6, were incubated at 37°C. Aliquots were removed after 2-30 minutes. GTP hydrolysis in the presence of both ABCE1 and pre-TCs (upper panels) was corrected to take into account the intrinsic GTPase activity of pre-TCs (lower panels). (C-E) Dissociation by ABCE1 of post-TCs, obtained by incubation of pre-TCs assembled on MVHL-STOP mRNA using native Leu-tRNALeu and [32P]60S subunits with (C, D) eRF1, or (E) eRF1/eRF3, depending on the presence/absence of nucleotides as indicated, assayed by SDG centrifugation. (F) Toe-print analysis of ribosomal complexes obtained by incubating pre-TCs assembled on MVHL-STOP mRNA using transcript tRNALeu with eRF1, ABCE1, eIFs and nucleotides, as indicated. The positions of full-length cDNA and of toe-prints corresponding to ribosomal complexes are indicated. (G) Association of [32P]eRF1 with post-TCs, assembled on MVHL-STOP mRNA with transcript Leu-tRNALeu, depending on the presence/absence of ABCE1 and nucleotides, assayed by SDG centrifugation. Upper fractions were omitted for clarity in panels C-E, G.
Since eRF3 must hydrolyze GTP to allow peptide release, and ABCE1 lacks nucleotide specificity and can also hydrolyze GTP, post-TCs obtained with eRF1/eRF3 could not be tested for stimulation of ABCE1's NTPase activity. For the same reason these complexes were not suitable for direct investigation of the role of ABCE1's NTPase activity in recycling, which was therefore instead first tested using post-TCs obtained with a high concentration of eRF1. Recycling was observed only in the presence of ATP, but not in the absence of nucleotides or in the presence of ADP, AMPPNP, or ATPγS (Fig. 6C). All four NTPs promoted similarly efficient recycling (Fig. 6D). Importantly, addition of excess of the slowly-hydrolysable ATP analogue ATPγS to post-TCs obtained with eRF1/eRF3/GTP strongly inhibited their recycling by ABCE1 (Fig. 6E). In similar experiments done using post-TCs obtained by incubating pre-TCs containing transcript tRNALeu with eRF1, ABCE1 promoted efficient recycling only in the presence of GTP, ATP, UTP or CTP (Fig. 6F, lanes 3-6), but not in the absence of nucleotides or in the presence of GMPPNP, AMPPNP, GDP or ADP, assayed using toe-printing (Fig. 6F, lanes 7-11). Interestingly, in the presence of AMPPNP, ABCE1 even stabilized the eRF1/post-TC interaction (Fig. 6G). Taken together, these results indicate that NTP hydrolysis by ABCE1 is required for its activity in recycling.
Discussion
The mechanism of ribosomal recycling is a long-standing unresolved question of eukaryotic translation. Although eukaryotic recycling can be mediated by eIF3, eIF1 and eIF1A (Pisarev et al., 2007), this energy-free mechanism can function only at low (∼1 mM) Mg2+ concentrations. We now report that recycling over a wide range of Mg2+ concentrations requires ABCE1, which promotes dissociation of post-termination complexes into free 60S subunits and tRNA- and mRNA-bound 40S subunits (Fig. 7). Release of tRNA and mRNA from recycled 40S subunits is mediated by eIFs 3, 1 and 1A. ABCE1 is most likely involved in recycling at all Mg2+ concentrations, accelerating the process at lower and becoming essential at higher concentrations. ABCE1 is the fourth ABC family member that is involved in eukaryotic translation. The others are: (i) ABC50, which stimulates formation of eIF2/GTP/Met-tRNAMeti complexes (Paytubi et al., 2009), (ii) GCN20, which functions with GCN1 in activating the GCN2 eIF2α kinase (Marton et al., 1997), and (iii) fungus-specific elongation factor eEF3, which facilitates release of deacylated tRNA from the E-site after translocation (Andersen et al., 2006).
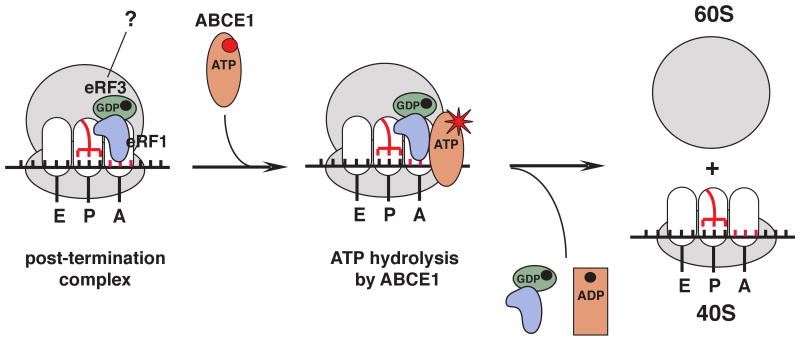
Figure 7. Model of ribosomal recycling by ABCE1.
ABCE1 binds to post-TCs containing eRF1 (or eRF1/eRF3, if eRF3 remains associated with ribosomal complexes), and after hydrolyzing ATP promotes their dissociation into 60S subunits and tRNA- and mRNA-bound 40S subunits.
ABCE1 is evolutionarily highly conserved, and is present in Archaea and all eukaryotes (Dean and Annilo, 2005). Although it was originally identified as an RNase L inhibitor (Bisbal et al., 1995), this could not be ABCE1's main function because RNAse L occurs only in mammals. Consistently, recent studies implicated it in more fundamental processes of translation (Andersen and Leevers, 2007; Dong et al., 2004; Chen et al., 2006) and ribosome biogenesis (Kispal et al., 2005; Yarunin et al., 2005). Depletion of ABCE1 in vivo in yeast, mammalian and Drosophila cells resulted in a decrease in polysomes and accumulation of mRNA-free 80S monomers (Andersen and Leevers, 2007; Dong et al., 2004; Chen et al., 2006), consistent with a defect in initiation. Strong reductions in 40S-associated eIF2 and eIF1 in ABCE1-depleted yeast cells led to the suggestion that it promotes 43S complex assembly (Dong et al., 2004), and since ABCE1 is much less abundant than eIF2 or eIF3 (Ghaemmaghami et al., 2003), it was proposed to act catalytically in this process (Dong et al., 2004). In contrast to these data, 90% depletion of ABCE1 in Drosophila cells did not influence levels of 40S-bound eIF2 and eIF3, but markedly reduced polysomes, suggesting that ABCE1 is involved in a step downstream of 43S complex formation (Andersen and Leevers, 2007). Consistently, we found that silencing of ABCE1 in HeLa cells impaired ribosomal recycling. Although a role for ABCE1 in initiation cannot be excluded by these data, the ‘initiation’ defect observed in cells with depleted ABCE1 might also be a consequence of ABCE1's function in recycling. Thus, impaired recycling, on one hand, would lead to disruption of the hypothetical ‘closed loop’ mechanism in which initiation is enhanced by preferential shunting of recycled 40S subunits to the 5′-end of the same mRNA due to its circularization through the cap-eIF4E/eIF4G/PABP-polyA/eRF3 chain of interactions (Uchida et al., 2002), whereas stalled elongation complexes that would presumably accumulate on mRNAs might, on the other hand, be targeted for degradation. Interestingly, some mutations in release factors also lead to changes in polysome profiles that are characteristic of initiation defects (e.g. Smirnov et al., 1976).
ABC proteins typically contain twin ABC-type NBDs, which can convert chemical energy into mechanical work (Rees et al., 2009). NBDs contain catalytic domains arranged in a head to tail orientation, which creates two composite nucleotide-binding sites. ATP binds at the interface of NBDs, with the ATP γ-phosphate located between the P-loop of the Walker A motif of one NBD and the signature motif of the other. The basis for the activity of ABC proteins is their ability to undergo cyclical conformational changes determined by changes in the relative positions of the NBDs, which depend on the nucleotide-bound state of the protein: the ATP-bound state is characterized by a closed, ‘dimerized’ conformation of the ABC cassettes with an extensive interface between NBD domains, whereas in the ADP-bound and nucleotide-free states, the separation between NBD domains is much greater. Thus, nucleotide-dependent conformational transitions between the stages of ATP binding and hydrolysis induce a tweezer-like power stroke between NBD domains that could cause conformational changes in associated domains and/or macromolecules.
ABCE1 comprises an N-terminal domain harboring two [4Fe-4S] clusters that is structurally related to bacterial-type ferredoxins, followed by two NBDs arranged by a hinge domain (Karcher et al., 2008). Mutation of conserved amino acids involved in ATP binding and hydrolysis, and of structurally important residues in the FeS domain are lethal (Dong et al., 2004; Kispal et al., 2005; Karcher et al., 2005; Barthelme et al., 2007) indicating that both NBDs and the FeS domain are essential for ABCE1's function. The fact that ABCE1's [4Fe-4S] clusters are stable down to redox potentials up to -560 mV argues against their electron transfer function (Barthelme et al., 2007), whereas the presence of a conserved positively charged patch on the FeS domain suggests that it might be involved in ABCE1's interaction with the ribosome (Karcher et al., 2008). The FeS domain also directly contacts NBD1's ATP-binding site, and might therefore transmit ATP-dependent structural changes in the NBDs to associated ligands, e.g. the ribosome.
Dissociation of post-TCs into ribosomal subunits by ABCE1 requires NTP hydrolysis and occurs only if peptide release is induced by eRF1/eRF3 or by eRF1 alone, but not by puromycin. Moreover, whereas dissociation of post-TCs obtained with eRF1/eRF3 is efficient, dissociation of post-TCs obtained with eRF1 is not and requires high concentrations of eRF1. Post-TCs obtained with eRF1/eRF3 remain bound to eRF1 (or possibly eRF1/eRF3) and are characterized by a 2nt. shift of their toe-print that likely results from conformational changes involving rotation of the head relative to the body of the 40S subunit that affects the extent to which reverse transcriptase penetrates into ribosomal complexes. In contrast, when peptide release is induced by eRF1 alone, eRF1 does not remain bound to post-TCs, and a weak 2nt toe-print shift is apparent only at high concentrations of eRF1. Thus ABCE1's ability to dissociate post-TCs correlates with their binding to eRF1, indicating that conformational changes induced in post-TCs by eRF1 and/or its physical presence on post-TCs determine their productive interaction with ABCE1. This, in turn, poses the question of why eRF1 remains firmly bound post-TCs only in the presence of eRF3. One possibility is that eRF3 remains associated with ribosomal complexes throughout the termination process and stabilizes the eRF1/post-TC interaction after peptide release. Alternatively, the conformations of eRF1, in which it induces peptide release with and without eRF3, may differ. eRF1 might establish additional ribosomal contacts in eRF3's presence that would allow it to remain bound to post-TCs even if eRF3 were to dissociate after GTP hydrolysis or peptide release. Thus, in the first scenario, eRF3 would have to remain associated with post-TCs after peptide release, whereas in the second, it would be allowed to dissociate. In either case, continuous or initial association of eRF3 with eRF1 and ribosomal complexes strengthens eRF1's interaction with post-TCs.
Consistent with previous reports (Andersen and Leevers, 2007; Dong et al., 2004; Kispal et al., 2005; Yarunin et al., 2005), we observed that in the AMPPNP-bound form, ABCE1 efficiently associated with 40S subunits and 43S complexes, but not with 80S ribosomes or pre-TCs, which suggests that ABCE1's binding site is occluded in both. However, it could stably bind to post- and even to pre-TCs that contained eRF1 and exhibit the 2nt toe-print shift: to post-TCs containing transcript tRNALeu and obtained with eRF1(wt), and to eRF1(AGQ)-bound pre-TCs. A potential direct contact between ABCE1 and eRF1 might contribute to ABCE1's binding to 80S ribosomal complexes, but it seems more likely that ABCE1's binding to ribosomal complexes is promoted by the putative conformational changes induced in them by eRF1, which unmask ABCE1's binding site. Interestingly, ribosomal binding of eEF3 also depends on the conformational state of 80S ribosomes: in its ATP-bound form, eEF3 binds most stably to ribosomes in the post-translocation state, whereas binding to empty ribosomes is weaker, and a rotated conformation of the head in pre-translocated ribosomes is not permissive for eEF3-binding (Andersen et al., 2006). Significantly, in its GTP-bound form, eRF3 prevents association of ABCE1 with eRF1-bound ribosomal complexes. Thus, before GTP hydrolysis, eRF3 either sterically blocks association of ABCE1 with ribosomal complexes or induces conformational changes that are unfavorable for its binding. To allow ATP-dependent binding of ABCE1 to post-TCs, either eRF3/GDP dissociates from ribosomal complexes, or in contrast to eRF3/GTP-bound ribosomal complexes, eRF3/GDP-bound post-TCs can productively interact with ABCE1.
Binding to post-TCs stimulates ABCE1's NTPase activity, which results in dissociation of post-TCs into 60S subunits and tRNA- and mRNA-bound 40S subunits. These data are consistent with a model in which mechanical work, into which ABCE1 converts the chemical energy of NTP hydrolysis, is used to dissociate post-TCs into subunits. The presence of eRF1 on post-TCs is required for ABCE1's binding, but eRF1's role in ribosomal recycling is likely more than just to create a binding site for ABCE1, and may involve transmission and even amplification of the impact of conformational changes in ABCE1 that occur upon NTP hydrolysis. Since no stable binding of ABCE1 with ribosomal subunits was observed in the presence of ADP, after dissociation of post-TCs into subunits, ABCE1 is most likely released into solution. Clarification of the molecular mechanism of ribosomal recycling by ABCE1 will require structural studies of post-TCs to determine the conformation and position of eRF1, and to establish the ribosomal location of ABCE1.
The mechanism of eukaryotic ribosomal recycling is therefore very different from that in bacteria. The differences are in part determined by differences in the preceeding stage of translation termination, after which eukaryotic post-TCs are stably bound at least to eRF1, in contrast to bacterial post-TCs, which do not remain associated with release factors. The fact that eRF1 is required for the next stage of ribosomal recycling accounts for our prior finding that eRF1 remains bound to post-TCs (Pisarev et al., 2007). Since ABCE1 also occurs in Archaea, but not in bacteria (Dean and Annilo, 2005), and as archaeal aRF1 is homologous to eukaryotic eRF1 (Atkinson et al., 2008), it is tempting to suggest that the mechanism of recycling in Archaea is similar to that in eukaryotes.
Experimental Procedures
Plasmid construction, ABCE1 silencing in HeLa cells, preparation of HeLa cell extract and the pull-down assay are described in Supplementary data, which also contain detailed protocols for all experimental procedures.
Purification of factors and ribosomal subunits
Native 40S and 60S subunits, eIF2, eIF3, eIF4F, eIF5B, eIF6, eEF1H and eEF2, and recombinant eIF1, eIF1A, eIF4A, eIF4B, eIF5, eIF3j, wt eRF1, eRF1(AGQ) and eRF3 were purified as described (Alkalaeva et al., 2006; Si et al., 1997). [32P]eRF1 and [32P]60S subunits were prepared as described (Pisarev et al., 2007).
ABCE1 purification
ABCE1 was purified from RRL on the basis of activity in promoting recycling at 2.5 mM Mg2+ of post-TCs formed on MVHL-STOP mRNA. Purification involved preparation of ribosomal salt wash, fractionation by ammonium sulphate precipitation, chromatography on DEAE cellulose and phosphocellulose, and FPLC on MonoS and hydroxyapatite columns.
Assembly of ribosomal complexes
Pre-TCs were assembled on MVHL-STOP mRNA with native or transcript aa-tRNAs and purified by SDG centrifugation (Alkalaeva et al., 2006).
Dissociation of post-TCs into subunits
Pre-TCs containing [32P]60S subunits were incubated with combinations of eRF1, eRF3, 1mM puromycin, ABCE1, eIF6, eIF3, eIF1, eIF1A, eIF3j and 60S subunits for 10 minutes at 37°C in buffer A (20 mM Tris, pH 7.5, 100 mM KCl, 0.25 mM spermidine, 2 mM DTT) supplemented with nucleotides and corresponding amounts of MgCl2 to achieve indicated Mg2+ concentrations, and centrifuged through 10-30% SDGs.
mRNA release
Pre-TCs containing [32P]MVHL-STOP mRNA were incubated with combinations of wt eRF1, eRF3, ABCE1, eIF6, eIF3, eIF1, eIF1A and eIF3j for 10 minutes at 37°C in buffer A supplemented with 0.2 mM ATP, 0.2 mM GTP and corresponding amounts of MgCl2 to achieve indicated Mg2+ concentrations, and centrifuged through 10-30% SDGs.
tRNA release
Pre-TCs containing native Leu-[5′-32P]tRNALeu were incubated with combinations of wt eRF1, eRF3, ABCE1, eIF6, eIF3, eIF1, eIF1A and eIF3j for 10 minutes at 37°C in buffer A supplemented with with 0.2 mM ATP, 0.2 mM GTP and corresponding amounts of MgCl2 to achieve indicated Mg2+ concentrations, and centrifuged through 10-30% SDGs.
Toe-printing analysis
Pre-TCs were incubated with combinations of wt eRF1, eRF1(AGQ), eRF3, 1mM puromycin, ABCE1, eIF3, eIF1, eIF1A, eIF3j and SDG-purified 80S/eRF1/ABCE1 complexes for 10 minutes at 37°C in buffer A supplemented with ATP, GTP, UTP, CTP, ADP, GDP, GMPPNP or AMPPNP and corresponding amounts of MgCl2 to achieve indicated Mg2+ concentrations. After incubation, the Mg2+ concentration was elevated to 20 mM to prevent further recycling. Ribosomal complexes were analyzed by primer extension.
Analysis of eRF1's association with post-TCs
For experiments shown in Figs. 1D, 1F, 3G and 6G, pre-TCs were incubated with [32P]eRF1 and combinations of eRF3, ABCE1 and 60S subunits for 10 minutes at 37°C in buffer A supplemented with GTP, ATP, ADP or AMPPNP and corresponding amounts of MgCl2 to achieve indicated Mg2+ concentrations. For experiments shown in Fig. 5E, 80S ribosomes were incubated with [32P]eRF1 in the absence/presence of ABCE1 and eRF3 for 10 minutes at 37°C in buffer A supplemented with 1 mM ATP, ADP or AMPPNP and corresponding amounts of MgCl2 to achieve indicated Mg2+ concentrations. Reaction mixtures were centrifuged through 10-30% SDGs.
Analysis of ribosomal binding of ABCE1
For experiments shown in Figs. 5A-B, ABCE1 was incubated with 80S ribosomes, 40S subunits, 60S subunits or 43S complexes in the presence/absence of various nucleotides (1 mM) for 10 minutes at 37°C in buffer A + 4 mM MgCl2. For experiments shown in Fig. 5C, ABCE1 was incubated with pre-TCs and combinations of wt eRF1, eRF1(AGQ), eRF3, 1 mM puromycin and nucleotides (1 mM) for 10 minutes at 37°C in buffer A + 4 mM MgCl2. For experiments shown in Fig. 5D, 80S ribosomes were incubated with ABCE1 and eRF1 individually or in combination for 10 minutes at 37°C in buffer A + 2.5 mM MgCl2. After incubation, reaction mixtures were centrifuged through 10-30% SDGs, and ribosomal complexes were analyzed by western blotting.
NTPase assay
For experiments shown in Figure 6A, ABCE1 was incubated with/without 80S ribosomes in buffer A supplemented with 2.5 mM MgCl2 and 0.33 μM [α-32P]ATP, [α-32P]GTP, [α-32P]UTP or [α-32P]CTP for 30 minutes at 37°C. For experiments shown in Figure 6B, ABCE1 was incubated with combinations of pre-TCs, eRF1 wt, eRF1(AGQ) mutant and eIF6 in buffer A supplemented with 0.33 μM [γ-32P]GTP and 2.5 mM MgCl2 for 2-30 minutes at 37°C. Reaction mixtures were analyzed by chromatography on polyethyleneimine cellulose.
Supplementary Material
Acknowledgments
We thank A. Komar for expression of ABCE1 in S. cerevisiae and for helpful discussion, and L.Yu. Frolova for eRF expression vectors. This work was supported by NIH Grant GM80623 to TVP, by Deutsche Forschungsgemeinschaft grant HE 1442/12-1 to MWH and by an EMBO long-term Fellowship and a Marie Curie International Incoming Fellowship from the EU under the seventh Framework to AMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- Andersen DS, Leevers SJ. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. J Biol Chem. 2007;282:14752–14760. doi: 10.1074/jbc.M701361200. [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Scheele U, Dinkelaker S, Janoschka A, Macmillan F, Albers SV, Driessen AJ, Stagni MS, Bill E, Meyer-Klaucke W, et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J Biol Chem. 2007;282:14598–14607. doi: 10.1074/jbc.M700825200. [DOI] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Karcher A, Büttner K, Märtens B, Jansen RP, Hopfner KP. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure. 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janáky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch AG. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paytubi S, Wang X, Lam YW, Izquierdo L, Hunter MJ, Jan E, Hundal HS, Proud CG. ABC50 promotes translation initiation in mammalian cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.031625. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin Struct Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenvi CL, Dong KC, Friedman EM, Hanson JA, Cate JH. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations-implications for the study of ribosome dynamics. RNA. 2005;11:1898–1908. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Chaudhuri J, Chevesich J, Maitra U. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc Natl Acad Sci. 1997;94:14285–14290. doi: 10.1073/pnas.94.26.14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Smirnov VN, Surguchov AP, Fominykch ES, Lizlova LV, Saprygina TV, Inge-Vechtomov SG. Recessive nonsense-suppression in yeast: further characterization of a defect in translation. FEBS Lett. 1976;66:12–15. doi: 10.1016/0014-5793(76)80573-x. [DOI] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J Biol Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.