Abstract
Fancc suppresses cross-linker–induced genotoxicity, modulates growth-inhibitory cytokine responses, and modulates endotoxin responses. Although loss of the latter function is known to account for endotoxin-induced marrow failure in murine Fancc (mFancc)–deficient mice, some argue that cytokine and endotoxin hypersensitivities devolve simply from genomic instability. Seeking to resolve this question, we planned to ectopically express instructive human FANCC (hFANCC) mutants in murine Fancc-deficient hematopoietic stem cells. To first assure that hFANCC cDNA was competent in murine cells, we compared hFANCC and mFancc in complementation assays for cross-linking agent hypersensitivity and endotoxin hypersensitivity. We found that mFancc complemented murine Fancc-deficient cells in both assays, but that hFANCC fully suppressed only endotoxin hypersensitivity, not cross-linking agent hypersensitivity. These results support the notions that Fancc is multifunctional and that structural prerequisites for its genoprotective functions differ from those required to constrain endotoxin responses known to lead to marrow failure in Fancc-deficient mice.
Introduction
Fanconi anemia (FA) is characterized by bone marrow failure (BMF), leukemia, and epithelial malignancies and is caused by inactivation of one of 14 different genes, including FANCC.1,2 Although FA proteins clearly prevent cross-linking-agent-induced genotoxicity, there is substantial evidence some of these proteins are multifunctional.3,4 For example, 2 human FANCC (hFANCC) point mutants have been described that fully complement cross-linking agent hypersensitivity (CAH) in hFANCC-deficient cells without altering other functions of hFANCC.3 What is less certain is the relevance of each disparate hFANCC function to hematopoietic stem cell (HSC) homeostasis.
Fancc−/− mice5,6 display CAH7 and endotoxin (lipopolysaccharide [LPS]) hypersensitivity (EH),8 and both cross-linking agents and LPS induce BMF at doses that are tolerated in wild-type mice. HSCs from Fancc−/− mice display reduced multilineage-repopulating ability in vivo,9,10 a defect that can be restored by ectopic hFANCC expression.11 Likewise, hFANCC protects against endotoxin-induced BMF.8 Although it has been assumed by some in this field that the sole function of FA proteins is genome stabilization and that the FA hematopoietic defect is simply a downstream consequence of genomic instability, no group has tested this notion directly in vivo. We therefore planned to do so by transducing Fancc-deficient HSCs with hFANCC separation-of-function mutants.3 Preparing for this study, we had to assure that hFANCC and murine Fancc (mFancc) were equivalent in a murine context, so we compared the relative efficacies of hFANCC and mFancc to complement CAH in Fancc-deficient cells. Surprisingly, we found that hFANCC, unlike mFancc, does not fully correct CAH, but that both hFANCC and mFancc correct EH.
Methods
Cell culture and reagents
Primary (GFAC2 and 5) and SV40-transformed (MEF 11.1 and 61) embryonic fibroblasts (MEFs) were derived from wild-type and Fancc-deficient mice, respectively.12 SV40-transformed-tongue-epithelial cells (6640SV) were isolated from Fancc-deficient mice by trypsinization. Progenitors and primary (NFF-6, 11Lu) and transformed (NFF6-U195, PD331T) fibroblasts were isolated and grown as previously described.11,13 Mitomycin C (MMC) and breakage assays were performed as previously described.14 Institutional Animal Care and Use Committee approval was given for isolating primary tissue after death and deriving cell lines.
hFANCC/mFancc cDNA cloning and transduction
hFANCC cDNA was isolated previously.15 Cloning of mFancc, virion production,16,17 and transduction methods are described in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Immunoblot assays
Whole-cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis14 and incubated with rabbit-polyclonal antibodies: anti-hFANCC (1:1000; Oregon Health & Science University FA Antibody Project) or anti-mFancd2 (1:200; kindly provided by Dr Alan D'Andrea, Harvard). Densitometry was measured using Gel Documentation System (Bio-Rad) and analyzed with Quantity One software.
IL-6 measurements
Cells were plated at 25 000 cells/well in 24-well dishes and treated plus or minus LPS (Sigma-Aldrich). After 24 hours, interleukin-6 (IL-6) was measured from supernatants with Mouse IL-6 Quantikine ELISA Kit (R&D Systems).
Results and discussion
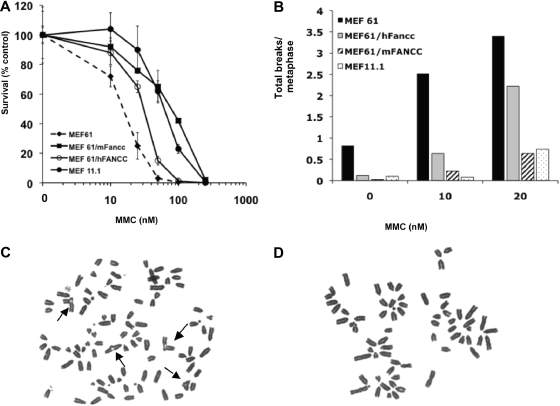
We ectopically expressed hFANCC and mFancc cDNA in Fancc-deficient MEFs (MEF61) and compared their capacity to complement CAH. We found that mFancc fully complemented the CAH to wild-type levels, but hFANCC had a marginal effect on MMC hypersensitivity (Figure 1A). Specifically MEF61 and hFANCC-transduced MEF61 (MEF61/hFANCC) cells showed similar EC50 (half-maximal effective concentration) values (≅ 20 and 30nM, respectively), but mFancc-transduced MEF61 cells (MEF61/mFancc) were fully complemented (EC50 ≅ 100nM) to wild-type levels (MEF11.1; EC50 ≅ 70nM). Chromosomal breakage (Figure 1B) mirrored the results discussed earlier in this paragraph. Specifically, MEF61/hFANCC cells displayed 2- to 3-fold more breaks than MEF61/mFancc or MEF11.1 and higher levels of quadriradial forms than MEF61/mFancc cells (Figure 1C and Figure 1D, respectively). Interestingly, the reverse was not true. We confirm, as reported previously,18 that mFancc fully complements CAH in hFANCC-deficient cells (supplemental Figure 1).
Figure 1.
Human FANCC does not fully correct Fancc-deficient MEFs. (A) Survivals of transformed Fancc-deficient MEFS (MEF61) transduced with human FANCC (MEF61/hFANCC) or murine Fancc (MEF61/mFancc) that were treated with MMC for 5 days (mean ± SD from one representative experiment of 3 performed in triplicate). MEF61/hFANCC cells were significantly more sensitive than MEF61/mFancc or wild-type (MEF11.1) cells (P < .001; analysis of variance [ANOVA]). MEF61/hFANCC cells were significantly more resistant to MMC than MEF61 cells (P < .001; ANOVA). (B) The average number of breaks per metaphase of the displayed MEFs treated with indicated concentrations of MMC for 24 hours; 50 metaphases were scored per cell line. Quadriradial forms were counted as 2 breaks per cell. (C-D) Metaphase spreads of MEF61/hFANCC (C) or MEF61/mFancc (D) cells treated with 20 ng/mL MMC for 24 hours. Arrows indicate radials.
Although our study does show marginal complementation by hFANCC, our results do not support the conclusions of Gush et al19 who reported that hFANCC is fully proficient in protecting Fancc-deficient cells from MMC-induced damage. We attribute the differences to the facts that in their study only one MMC dose was measured in vitro, low doses of MMC were used for the in vivo study, no chromosomal breakage was reported, and a direct mFancc comparison was not done.
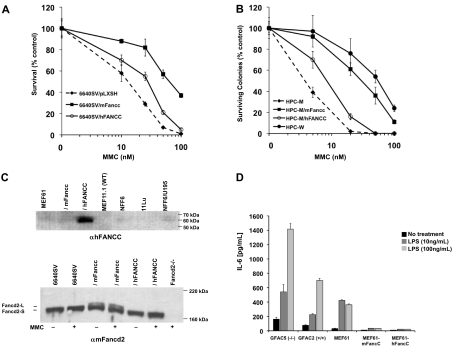
Failure of hFANCC to correct CAH was not cell type–specific. We measured CAH of Fancc-deficient tongue epithelial cells (6640SV; Figure 2A) and hematopoietic progenitors (HPC-M; Figure 2B) transduced with hFANCC or mFancc. 6640SV/hFANCC cells (EC50 ≅ 30nM) were significantly more sensitive to MMC than 6640SV/mFancc (EC50 ≅ 80nM). Likewise, HPC-M/hFANCC cells (EC50 ≅ 10nM) were more sensitive than HPC-M/mFancc (EC50 ≅ 30nM) and wild-type (HPC-W; EC50 ≅ 60nM).
Figure 2.
Human FANCC fails to complement DNA repair defects in Fancc-deficient cells but is sufficient for suppression of cytokine production. (A) Survivals of transformed Fancc-deficient tongue epithelial cells (6640SV/pLXSH) transduced with hFANCC (6640SV/hFANCC) or mFancc (6640SV/mFancc) and treated with MMC for 5 days (mean ± SD from one similar experiment of 3 performed in triplicate). 6640SV/hFANCC cells were significantly more sensitive to MMC than 6640SV/mFancc cells (P < .001; ANOVA). 6640SV/hFANCC cells were significantly more resistant to MMC than 6640SV cells (P < .001; ANOVA). (B) Colony assays of Fancc−/− hematopoietic progenitor cells (HPC-M) transduced with hFANCC (HPC-M/hFANCC) or mFancc (HPC-M/mFancc) and treated with MMC for 7 days (mean ± SD from one similar experiment of 2 performed in triplicate). HPC-M/hFANCC cells were 3-fold more sensitive to MMC than HPC-M/mFancc cells (P < .001; ANOVA), 6-fold more sensitive than wild-type cells (HPC-W; P < .001; ANOVA), and 3-fold more resistant to MMC than untransduced cells (HPC-M; P < .001; ANOVA). Transduction efficiencies of hFANCC/mFancc were measured by polymerase chain reaction with primers directed against green fluorescence protein and were similar (74% vs 79% for hFANCC and mFancc, respectively). (C) Immunoblots of fibroblast (top) or tongue epithelial (bottom) cell lines treated ± 150nM MMC for 24 hours. Lysates were blotted with either anti-FANCC antibody (top) or anti-Fancd2 antibody (bottom). (Top) Lanes 5 to 7 (NFF6, 11Lu, and NFF6/U195) represent normal human fibroblasts. The expression of hFANCC in transduced-murine fibroblasts (MEF61/hFANCC; lane 3) was more than 8-fold higher (by comparative densitometric analysis of lanes 3 and 7) than the level of expression required for a normal MMC response. WT indicates wild-type. (Bottom) Fancd2-L indicates mono-ubiquitinated Fancd2; and Fancd2-S, nonubiquitinated Fancd2. Fancd2−/− cells were isolated from Fancd2-deficient mice. (D) Primary (GFAC5 and 2) and transformed MEFs (MEF61) were treated with indicated concentrations of LPS for 24 hours. IL-6 secretion was measured in the supernatants by ELISA (mean ± SD from one similar experiment of 3 performed in triplicate). Fancc-deficient MEFs produced significantly higher levels of IL-6 than wild-type or transduced cells (P < .001 for both cell types).
Our results did not stem from low hFANCC expression in Fancc-deficient cells. We quantified both hFANCC mRNA and hFANCC protein levels. Although transduced hFANCC mRNA was approximately 3-fold lower than mFancc levels in MEF61 and 6640SV cells, hFANCC transcripts were approximately 67-fold higher than levels of endogenous mFancc in wild-type cells (supplemental Figure 2 and data not shown). Full-length hFANCC protein was also expressed at high levels in MEF61/hFANCC cells (Figure 2C top, lane 3) and by densitometry was more than 8-fold higher than endogenous hFANCC levels in normal human fibroblasts (lanes 5-7). Thus, hFANCC protein is expressed in murine cells at levels that would be more than sufficient to confer a normal MMC response.
Mono-ubiquitination of Fancd2 (mediated by Fancc) is required for cross-linking agent resistance.4 As expected, mFancc-transduced cells showed high levels of both nonubiquitinated and mono-ubiquitinated Fancd2 (Figure 2C bottom; Fancd2-S and -L, respectively; lanes 3 and 4). However, Fancd2-L was undetectable in hFANCC-transduced epithelial cells (lanes 5 and 6). Similar results were found using MEFs (supplemental Figure 3). Therefore, the lack of CAH complementation by hFANCC in mFancc-deficient cells is associated with its inability to facilitate mFancd2 ubiquitination. Although hFANCC was incapable of correcting the MMC phenotype, we did find that EH was completely corrected. Because endotoxin-exposed Fancc-deficient mice are known to produce extraordinarily high IL-6 levels,8 we confirmed that Fancc-deficient MEFs secreted more IL-6 after LPS stimulation than did wild-type cells (Figure 2D) and next showed that MEFs transduced with either hFANCC or mFancc were fully complemented.
hFANCC and mFancc share 79% amino acid similarity and 66% identity (supplemental Figure 4), which is similar to other FA core-complex proteins but substantially less than other DNA-repair proteins.18,20 Because hFANCC retains the ability to completely suppress EH in murine cells, our results suggest that requisite domains for this function remain intact in hFANCC, but domains required for optimal DNA repair in the context of a murine cell do not.
In light of the fact that that levels of hFANCC/mFancc RNA and hFANCC/mFancc protein in transduced MEFs and epithelial cells are so much higher than endogenous levels and that transduction efficiencies in hematopoietic cells are similar (74% and 79%, respectively), we conclude that hFANCC is hypomorphic in murine cells because the DNA damage defect persists and therefore argue that EH does not devolve simply from genomic instability. That hFANCC ameliorates the repopulating defect of Fancc-deficient HSCs to levels approximating wild-type cells,11,17 despite significantly reduced DNA-repair capacity (Figure 2B), indicates that noncanonical functions of FANCC/Fancc may be critical for supporting optimal hematopoiesis. To formally distinguish relative contributions of CAH and EH to BMF in mice, identification of separation-of-function mutants will be required. In effect, hFANCC cDNA meets this standard in murine cells and provides added opportunities to identify unique functional domains. Although such experiments will be informative, we also define 2 caveats for future research with murine FA models. First, it will be essential to confirm that any human FA cDNA is equivalent to murine cDNA in comprehensive functional assays. Second, to dissect Fancc's structure-function relationships, separation-of-function mutations must be developed using mFancc because hFANCC is hypomorphic. It will also be of interest to create murine homologs of known human FA mutations because, in light of our results, some of them might not inactivate mFancc function.
Supplementary Material
Acknowledgments
The authors thank Sunchin Kim, who provided technical assistance.
This work was supported by Fanconi Anemia Research Fund (L.E.H.), the Knight Cancer Institute at Oregon Health & Science University, Veterans Association Merit Review (G.C.B.), National Institutes of Health/National Heart, Lung, and Blood Institute (5P01-HL48546; G.C.B., S.B.O.), and National Institutes of Health/National Cancer Institute (1-R01-CA138237-01; D.W.C., G.C.B.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.E.H., G.C.B., and D.W.C. designed the study; L.E.H. and G.C.B. wrote the manuscript; L.E.H., W.W.K., J.E.Y., R.K.R., T.K., and Z.S. performed key experiments; S.B.O. conducted breakage and helped analyze data; and D.W.C. conducted hematopoietic progenitor assays and helped analyze data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laura E. Hays, 3710 SW US Veterans Hospital Rd, Portland Veterans Administration Medical Center, R&D-2, Portland, OR 97239; e-mail: haysl@ohsu.edu.
References
- 1.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668(1):11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 201;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 3.Pang Q, Christianson TA, Keeble W, et al. The Fanconi anemia complementation group C gene product: structural evidence of multifunctionality. Blood. 2001;98(5):1392–1401. doi: 10.1182/blood.v98.5.1392. [DOI] [PubMed] [Google Scholar]
- 4.Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43(3):147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Tomkins DJ, Auerbach W, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12(4):448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 6.Whitney MA, Royle G, Low MJ, et al. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88(1):49–58. [PubMed] [Google Scholar]
- 7.Carreau M, Gan OI, Liu L, et al. Bone marrow failure in the Fanconi anemia group C mouse model after DNA damage. Blood. 1998;91(8):2737–2744. [PubMed] [Google Scholar]
- 8.Sejas DP, Rani R, Qiu Y, et al. Inflammatory reactive oxygen species-mediated hemopoietic suppression in Fancc-deficient mice. J Immunol. 2007;178(8):5277–5287. doi: 10.4049/jimmunol.178.8.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haneline LS, Gobbett TA, Ramani R, et al. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94(1):1–8. [PubMed] [Google Scholar]
- 10.Habi O, Delisle MC, Messier N, Carreau M. Lack of self-renewal capacity in Fancc−/− stem cells after ex vivo expansion. Stem Cells. 2005;23(8):1135–1141. doi: 10.1634/stemcells.2004-0356. [DOI] [PubMed] [Google Scholar]
- 11.Haneline LS, Li X, Ciccone SL, et al. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc−/− hematopoietic stem cells and decreases the risk of clonal evolution. Blood. 2003;101(4):1299–1307. doi: 10.1182/blood-2002-08-2404. [DOI] [PubMed] [Google Scholar]
- 12.Fagerlie SR, Diaz J, Christianson TA, et al. Functional correction of FA-C cells with FANCC suppresses the expression of interferon gamma-inducible genes. Blood. 2001;97(10):3017–3024. doi: 10.1182/blood.v97.10.3017. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich MC, Dooley DC, Freed AC, et al. Constitutive expression of steel factor gene by human stromal cells. Blood. 1993;82(3):771–783. [PubMed] [Google Scholar]
- 14.Pejovic T, Yates JE, Liu HY, et al. Cytogenetic instability in ovarian epithelial cells from women at risk of ovarian cancer. Cancer Res. 2006;66(18):9017–9025. doi: 10.1158/0008-5472.CAN-06-0222. [DOI] [PubMed] [Google Scholar]
- 15.Rathbun RK, Faulkner GR, Ostroski MH, et al. Inactivation of the Fanconi anemia group C gene augments interferon-gamma-induced apoptotic responses in hematopoietic cells. Blood. 1997;90(3):974–985. [PubMed] [Google Scholar]
- 16.Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7(9):980–990. [PMC free article] [PubMed] [Google Scholar]
- 17.Si Y, Pulliam AC, Linka Y, et al. Overnight transduction with foamyviral vectors restores the long-term repopulating activity of Fancc−/− stem cells. Blood. 2008;112(12):4458–4465. doi: 10.1182/blood-2007-07-102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wevrick R, Clarke CA, Buchwald M. Cloning and analysis of the murine Fanconi anemia group C cDNA. Hum Mol Genet. 1993;2(6):655–662. doi: 10.1093/hmg/2.6.655. [DOI] [PubMed] [Google Scholar]
- 19.Gush KA, Fu KL, Grompe M, Walsh CE. Phenotypic correction of Fanconi anemia group C knockout mice. Blood. 2000;95(2):700–704. [PubMed] [Google Scholar]
- 20.Ahmad SI, Kirk SH. Molecular Mechanisms of Fanconi Anemia. New York, NY: Springer; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.