Abstract
Imatinib mesylate (IM) induces clinical remissions in chronic-phase chronic myeloid leukemia (CML) patients but IM resistance remains a problem. We recently identified several features of CML CD34+ stem/progenitor cells expected to confer resistance to BCR-ABL-targeted therapeutics. From a study of 25 initially chronic-phase patients, we now demonstrate that some, but not all, of these parameters correlate with subsequent clinical response to IM therapy. CD34+ cells from the 14 IM nonresponders demonstrated greater resistance to IM than the 11 IM responders in colony-forming cell assays in vitro (P < .001) and direct sequencing of cloned transcripts from CD34+ cells further revealed a higher incidence of BCR-ABL kinase domain mutations in the IM nonresponders (10%-40% vs 0%-20% in IM responders, P < .003). In contrast, CD34+ cells from IM nonresponders and IM responders were not distinguished by differences in BCR-ABL or transporter gene expression. Interestingly, one BCR-ABL mutation (V304D), predicted to destabilize the interaction between p210BCR-ABL and IM, was detectable in 14 of 20 patients. T315I mutant CD34+ cells found before IM treatment in 2 of 20 patients examined were preferentially amplified after IM treatment. Thus, 2 properties of pretreatment CML stem/progenitor cells correlate with subsequent response to IM therapy. Prospective assessment of these properties may allow improved patient management.
Introduction
Chronic myeloid leukemia (CML) is a clonal, multistep, multilineage myeloproliferative disorder. In patients with chronic-phase disease, the leukemic cells are characterized by the presence of a BCR-ABL fusion gene as the sole detectable chromosomal abnormality.1–3 The BCR-ABL encodes a chimeric oncoprotein (p210BCR-ABL) that displays constitutively elevated tyrosine kinase activity and drives the pathogenesis of the disease.4,5 The elevated tyrosine kinase activity of the BCR-ABL oncoprotein causes biochemical changes that affect the growth factor dependence, turnover, and genomic stability of primitive CD34+ leukemic cells in chronic-phase patients but has little impact on their ultimate differentiation.6–9 Recently, imatinib mesylate (IM), a small molecule inhibitor of the BCR-ABL tyrosine kinase,5 has shown great promise in suppressing the disease in chronic-phase CML patients.10,11 However, both initial and acquired resistance to IM has emerged as a significant clinical problem with mutations in the BCR-ABL kinase domain accounting for 60% to 90% of relapses.9,12–14 Dasatinib (DA) and nilotinib (NL) are more potent small molecule inhibitors of the BCR-ABL-encoded kinase than IM and are effective in some, but not all, patients with IM-resistant disease.15–17 Recent studies indicate that primitive CML cells are less responsive to IM and other tyrosine kinase inhibitors and are a reservoir for the emergence of IM-resistant subclones.18–22 We and others have further demonstrated that CML progenitor and/or stem cells possess multiple features that would be expected to promote both innate and acquired resistance to p210BCR-ABL-targeted therapeutics.8,19,20,23,24 These include elevated BCR-ABL expression and tyrosine kinase activity, increased expression of ABCB1/MDR and ABCG2 that efflux IM and other drugs used to treat acute leukemia,20,25 decreased expression of OCT1 that is responsible for IM uptake,26,27 and a high degree of instability of the BCR-ABL gene itself.23
In this study, we asked whether these properties of leukemic CD34+ cells obtained from chronic-phase CML patients before initiation of IM therapy would correlate with the subsequent response of the patient to IM therapy. We measured the sensitivity of their individual pretreatment clonogenic progenitors to IM exposure in vitro, the level of expression of BCR-ABL, ABCB1/MDR, and ABCG2 transcripts in their CD34+ cells, and the frequency of mutant BCR-ABL and ATPA1 transcripts in the same cells, and compared the results for clinically defined responders and nonresponders. The results confirm that CD34+ CML cells from individual chronic-phase patients harbor differences in all of these properties and that one of these correlates significantly with the patient's clinical IM response, suggesting that it might form the basis of a prospective test for optimizing CML patient management.
Methods
Patients
A total of 25 chronic-phase CML patients were studied, none of whom had been treated previously with any BCR-ABL-targeted drug. The clinical details of these patients and their subsequent classification as IM responders (n = 11) or nonresponders (n = 14) are given in Table 1. Responders are patients who achieved complete hematologic remission within 3 months, a major cytogenetic remission within 12 months, and a complete cytogenetic remission within 18 months.28 Conversely, nonresponders were patients who did not achieve these response criteria (primary resistance) or had evidence of loss of a complete hematologic remission or a complete cytogenetic remission (secondary resistance) or who developed blast crisis (BC) within 3 months of achieving a complete cytogenetic response (3 patients).
Table 1.
Characteristics of clinically classified IM responders and IM nonresponders
| Characteristic | IM responders | IM nonresponders | P* |
|---|---|---|---|
| Quantitative features | |||
| WBC at diagnosis | Range: [25.5, 450]; mean, 149; CI: [62.9, 235] | Range: [32.2, 536] mean, 204; CI: [101, 252] | .42 |
| Age, y | Range: [23,76]; mean, 52.64; CI: [49.47, 55.80] | Range: [29,77]; mean, 55.71; CI: [53.86, 57.57] | .60 |
| Mutant BCR-ABL transcripts, percentage | Range: [0, 20]; mean, 10.1; CI: [8.3, 12.2] | Range: [9, 40]; mean, 24.5; CI: [21.5, 26.5] | < .003† |
| NA | 1 (9.1%) | 4 (28.6%) | |
| No. of colonies as percentage of untreated cells CFC IM 1μM | Range: [14.2, 53]; mean, 35.6; CI: [32.7, 38.4] | Range: [40.6, 92] mean, 60.1; CI: [57.6, 62.6] | < .001 |
| CFC IM 5μM | Range: [7.7, 42.6]; mean, 22.7; CI: [20.5, 24.9] | Range: [34, 64]; mean, 48.66; CI: [47.4, 49.9] | < .001 |
| CFC IM 10μM | Range: [6.6, 42]; mean, 22.3; CI: [20, 24.6] | Range: [20.8, 61.3]; mean, 44.0; CI: [41.8, 46.1] | < .001 |
| Time from diagnosis to IM treatment, d | Range: [7, 335]; mean, 60.36; CI: [-122, 242] | Range: [7, 377]; mean, 107; CI: [-133, 347] | .29 |
| Qualitative features | |||
| Sokal risk score | .928‡ | ||
| Low | 1 (14.3%); CI: NA§ | 2 (9.1%); CI: [0.14, 1.22] | |
| Intermediate | 7 (42.9%); CI: [0.84, 0.94] | 6 (63.6%); CI: [0.89, 0.96] | |
| High | 3 (35.7%); CI: [0.58, 2.9] | 5 (27.3%); CI: [1.56, 2.55] | |
| NA | 0 | 1 (7.14%) | |
| Sex | > .311‖ | ||
| Male | 6 (54.6%) | 8 (57.1%) | |
| Female | 5 (45. 5%) | 6 (42.9%) | |
| Prior interferon | > .158‖ | ||
| Yes | 0 | 3 (21.4%) | |
| No | 11 (100%) | 11 (78.6%) | |
| Prior hydroxyurea | .072‖ | ||
| Yes | 7 (63.6%) | 14 (100%) | |
| No | 4 (36.4%) | 0 | |
| Daily IM dose | .161‡ | ||
| 400 mg | 9 (81.8%) | 8 (57.4%) | |
| 400-600 mg | 2 (18.2%) | 4 (28.6%) | |
| 400-800 mg | 0 | 2 (14.3%) | |
| Primary/secondary resistance | < .001 | ||
| NA¶ | 11 (100%) | 0 | |
| Primary resistance | 0 | 9 (64.3%) | |
| Secondary resistance | 0 | 5 (35.7%) |
NA indicates not available.
P values were calculated using the Welch 2-sample t test at the 5% significant level under the null hypothesis H0: mean (IM responders) = mean (IM nonresponders).
P value of mutant BCR-ABL transcript frequency was calculated after all the missing values were taken out of the sample.
Kruskal-Wallis test was used to calculate P values of ordered categorical variable for Sokal risk score and daily IM dose.
CI for low IM responders in the low Sokal risk group is not available because there is only one patient in this category.
Fisher exact test was used to calculate P values of nonordered categorical variable for sex, prior interferon, and prior hydroxyurea.
Primary/secondary resistance is not available for all IM responders.
Cells
Heparin-anticoagulated peripheral blood cells, apheresis cells, or bone marrow (BM) cells were obtained from patients before initiation of IM therapy. Low-density cells and, in some cases, CD34+ cell–enriched populations were isolated before cryopreservation.29 Cells were also obtained from BM cells harvested from healthy adult donors for use in allogeneic hematopoietic stem cell transplantations. In all cases, informed consent was obtained in accordance with the Declaration of Helsinki, and the procedures used were approved by the Research Ethics Board of the University of British Columbia. CD34+ cells (> 85% pure) were isolated immunomagnetically by positive selection of thawed cells using the EasySep CD34 selection kit (StemCell Technologies), and the purity of these was verified by restaining the isolated cells with a fluorescein isothiocyanate-labeled anti-CD34 antibody (StemCell Technologies) and analysis on a fluorescence-activated cell sorter.
In vitro colony assays
Assays for colony-forming cell (CFC) activity were performed by plating the CD34+ cells in methylcellulose medium (H4330, StemCell Technologies) supplemented with 20 ng/mL interleukin-3 (IL-3, from Novartis), 20 ng/mL IL-6 (Cangene), 20 ng/mL granulocyte colony-stimulating factor (StemCell Technologies), and 20 ng/mL granulocyte-macrophage colony-stimulating factor (Novartis) in the presence or absence of IM (0, 1, 5, or 10μM, Novartis). Steel factor was not added to avoid inhibitory effects of IM on KIT-dependent colony formation because IM also targets this receptor for steel factor.5 Colonies derived from burst-forming units–erythroid (BFU-E), multilineage granulopoietic, erythroid, macrophage, and megakaryocytic colony-forming units (CFU-GEMM), and granulocyte-macrophage colony-forming units (CFU-GM) were scored in situ after 14 days of incubation using an inverted microscope.
Suspension cultures
CD34+ cells were suspended in Iscove medium supplemented with bovine serum albumin, insulin, transferrin, and low-density lipoproteins (StemCell Technologies) and 10−4M 2-mercaptoethanol (Sigma-Aldrich). A total of 5 × 104 cells was then added to each well of a 24-well plate with or without IM in the presence of the following purified recombinant human growth factors: 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL granulocyte colony-stimulating factor, and 100 ng/mL Fms-like tyrosine kinase 3-ligand (Immunex). Viable cell numbers were determined after 3 weeks of incubation by hemacytometer counts of trypan blue-excluding cells.
Quantitative real time PCR analyses
Total RNA was extracted using the Absolutely RNA Microprep kit (Stratagene) or the PicoPure RNA isolation Kit (Arcturus). RNA (0.1-0.25 μg) was reverse transcribed with random hexamers in a 20-μL reaction volume using SuperScript III reverse transcriptase according to the manufacturer's instructions (Invitrogen). Reverse-transcribed polymerase chain reaction (RT-PCR) was then performed as previously described23 in a 25-μL reaction mixture consisting of 12.5 μL of 2 × POWER SYBR Green PCR Master Mix (Applied Biosystems), 1 μL of 20μM specific primers, 1 to 2 μL cDNA, and water as needed. Specific forward and reverse primers to produce approximately 100-bp amplicons for optimal amplification of reverse-transcribed cDNA for BCR-ABL were as follows: 5′-CATTCCGCTGACCATCAATAAG-3′ and 5′-GATGCTACTGGCCGCTGAAG-3′, for BCR were 5′-CCAACCTGCTCACCTTCCTTT-3′ and 5′-GTTGTGCAGGGACATCTTATTG-3′, for GAPDH were 5′-CCCATCACCATCTTCCAGGAG-3′ and 5′-CTTCTCCATGGTGGTGAAGACG-3′, for OCT1 were 5′-CGCCCAACTACATGTCCATGC-3′ and 5′-CCGAGCCAACAAATTCTGTGATT-3′, for ABCB1 were 5′-CAAATGCAAGAGGAGCAGCTTA-3′ and 5′-CCACTCTTCGAATAGCTGTCAA-3′, and for ABCG2 were 5′-GCTTTCTACCTTGTCGAAAACC-3′ and 5′-GAGACCAGGTTTCATGATCCC-3′. Optimal reaction conditions for the amplification of these genes were 40 cycles of 2-step PCR (95°C for 15 seconds followed by 60°C for 1 minute with a single fluorescence measurement) after initial denaturation (50°C for 2 minutes and 94°C for 10 minutes). RT-PCR data were also analyzed by melt curve analysis for verification of true target amplicons and absence of primer dimers. The 7500 Real Time PCR System Software (Applied Biosystems) was used for data analysis.
To compare BCR-ABL, BCR, and transporter gene transcript levels, the threshold cycle (Ct) values of all samples were first normalized to the Ct value of the housekeeping gene GAPDH in the same sample. This normalized value was compared with the normalized value of the reference sample (eg, normal BM cells) using the following formula: relative expression of the gene in the sample of interest normalized to that of the reference sample = 2 − ΔΔCt where ΔΔCt is ΔCt, sample −ΔCt, reference. ΔCt, sample is the Ct value for any sample normalized to GAPDH, and ΔCt, reference is the Ct value for the calibrator (eg, normal BM cells) also normalized to the Ct value for GAPDH. The efficiency of amplification of GAPDH, BCR-ABL, BCR, and transporter gene sequences was confirmed to be close to 100% from the observed linear relationship obtained between the value for ΔCt and the template cDNA dilution.
Mutational analysis
cDNA was obtained as described in “Quantitative real time PCR analyses,” and the major tyrosine kinase domain of BCR-ABL was amplified using a 2-step procedure.23,30 First, a 1.3-kb BCR-ABL fusion fragment was generated by addition of 1 to 2 μL of the cDNA to 22.5 μL Accuprime Pfx SuperMix (Invitrogen), 1 μL 0.5M KCl, and 0.25 μL forward BCR-ATPB1 (5′-GAAGCTTCTCCCTGGCATCCGTG-3′) and reverse ABL-ATPB2 (5′-GCCAGGCTCTCGGGTGCAGTCC-3) primers at 20μM concentrations. The second nested PCR was performed by addition of 0.5 μL of the first PCR reaction product to 45 μL of Accuprime Pfx SuperMix, 4.5 μL of PCR water, and 0.5 μL of forward primer ABL-ATPB1 (5′-GCGCAACAAGCCCACTGTCATTGG-3′) and reverse primer ABL-ATPB2, each at a concentration of 20μM. The 0.6-kb product derived from the major ABL kinase domain (residues 220-420) was run on a TAE gel, purified, and cloned into the pCR2.1-TOPO vector (Invitrogen). Ten to 30 bacterial colonies were produced on LB media, and plasmid DNA was then isolated from the bacteria using standard methods. Sequencing was performed using a 3730XL DNA Analyzer System (Applied Biosystems). A cloned 0.6-kb fragment derived from the wild-type major ABL kinase domain in extracts obtained from normal CD34+ BM cells was sequenced using the ABL-ATPB1 and ABL-ATPB2 primers.
Sequence analysis of the entire 3.7-kb sodium/potassium transporting ATPase subunit α-1 gene (ATPA1)31 was performed using the same procedure of PCR amplification, cloning, and plasmid extraction as described in the previous paragraph for BCR-ABL transcripts. A total of 1 to 2 μL of patient cDNA was amplified with 45 μL of Accuprimer Pfx SuperMix and 0.5 μL of 20μM forward and reverse primer pairs designed to generate 3 overlapping fragments each approximately 1.2 kb in length. Primers used to generate the N-terminus fragment were: ATPA1F1 (5′-TTGGACGTGATAAGTATGAGC-3′) and ATPA1R1 (5′-CCATGCGTTTGGCAGTAAGTG-3′), for the middle fragment were: ATPA1F2 (5′-GGTATCATCGTAGCCAATGTG-3′) and ATPA1R2 (5′-ACGATAGCACCCTGTCTTTGG-3′); and for the C-terminus fragment were: ATPA1F3 (5′-ACCACACTGAGATAGTGTTTGC-3′) and ATPA1R3 (5′-GGGCGTCGCCTGATGATGAG-3′).
For mutation frequency determinations, a mutant transcript was defined as one or more mutations per cloned fragment to provide an indication of the prevalence of mutant CD34+ cells present in the population analyzed. However, to ascertain the validity of the mutation(s) detected required that the same sequence be confirmed in both directions by sequencing each clone.
Molecular modeling and analysis
The following published x-ray structures were used as references for the modeling and analysis: 2HYY (human C-ABL bound to IM), 1IEP (mouse c-abl bound to IM), 2GQG (human C-ABL bound to DA), and 3CS9 (human C-ABL bound to NL).16,32,33 Molecular modeling of selected mutants was performed using the Swiss pdb Viewer.34 (Figure 4C was generated using the program PyMol.35)
Figure 4.
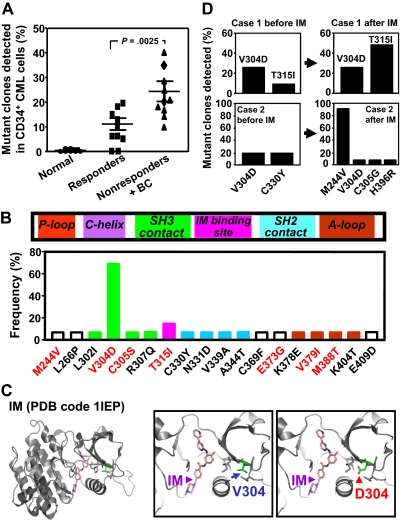
Comparison of the frequency and characterization of mutant BCR-ABL transcripts in CD34+ cells from IM responders and nonresponders. (A) BCR-ABL kinase domain transcript fragments in extracts of patients CD34+ cells were cloned and sequenced (10-30 clones per sample), and the frequency of mutant clones detected in each patients' sample was then calculated. Cross bars indicate the mean plus or minus SEM of data for each patient group. The difference between the frequencies of mutant transcripts detected in the IM nonresponders' cells (10 patients) and the 10 responders' cells (10 patients) is significant (P < .003). No mutations were found in CD34+ BM cells from 6 normal persons. (B) Locations of the single amino acid substitutions identified in relation to subdomains of the BCR-ABL kinase domain. The prevalence of each mutation among the 20 patients studied is shown as the proportion of patients in which each mutation was seen. Red letters indicate amino acid changes previously associated with IM resistance in patients. P-Loop indicates phosphate-binding loop; C-helix, catalytic domain; SH3 contact, Src homology 3 domain contact; IM binding site, imatinib binding site; SH2 domain, Src homology domain contact; and A-loop, activation loop. (C) Localization of the residue V304 based on the crystal structure of the c-ABL kinase domain in complex with IM (PDB code 1IEP).32 Substitution of V304 with a polar residue aspartic acid (V304D) would perturb the conformation and/or structure of the helix that forms part of the IM-binding groove. (D) Frequency of various mutant BCR-ABL transcripts in CD34+ CML cells before and after IM treatment from 2 IM nonresponders calculated as described for panel A.
Statistical analysis
Results are shown as the mean plus or minus SEM of values obtained in independent experiments. Differences between groups were assessed using the Welch 2-sample t test, Kruskal-Wallis test, and Fisher exact test. In the multivariable analysis, a logistic model was used to study the dependence between the clinical IM response and other independent variables listed in Table 1. In the preliminary model, the variables that were both mathematically solvable and whose P values were less or equal to 10% were analyzed (except BCR-ABL mutation because of some missing values, Table 1). Because prior interferon treatment, prior hydroxyurea treatment, and primary/secondary resistance could cause singular solutions in the preliminary model, they could not be included. Thus, the preliminary model was: logit ([IM response = 1]) = β0 + β1 CFC1 (1μM IM) + β2 CFC5 (5μM IM). A stepwise Akaike information criterion was used as a model selection criterion, and the result was verified by odds ratio.36 In this context, only inhibition of colony formation in the presence of 5μM IM was selected for use in the final multivariable model at a 5% level of significance (Table 2). The final model was thus as follows: logit [(IM response = 1]) = β0 + β1CFC5. Receiver operating characteristic (ROC) analysis37 was performed to test how well the final model predicted its covariates. The data were analyzed using the statistical software R (Version 2.8.1).38
Table 2.
Estimation of the coefficients of the logistic model used to analyze endpoints assessed for their association with a positive clinical IM response
| Variable | Coefficient estimate | SE | P |
|---|---|---|---|
| Intercept | 10.8 | 4.82 | < .026* |
| CFC (IM 1μM) | 0.004 | 0.079 | .962 |
| CFC (IM 5μM) | −0.305 | 0.147 | < .039* |
CFC refers to the extent of IM inhibition of colony formation by CD34+ CFCs.
The estimated coefficients and P values of the final model show that CFC (IM 5μM) has a significant impact on the IM response at the 5% level.
Results
IM responders and nonresponders do not have distinguishing clinical features
We studied 25 chronic-phase CML patients classified clinically as chronic-phase patients before IM treatment. Eleven of these subsequently proved to be IM responders and 14 to be IM nonresponders.28 The latter group included 9 patients with primary IM resistance and another 5 with secondary resistance, including 3 patients who developed BC within 3 months of an initial complete cytogenetic remission. Analysis of the pretreatment clinical data showed that the 14 nonresponders tended to have higher Sokal scores than the 11 responders, although these differences were not significant (P = .9, Table 1). It was also noted that none of the 5 patients with secondary IM resistance had a high Sokal risk score (> 1.2) compared with the patients with primary IM resistance (5 of 9 with a Sokal risk score > 1.2, Table 3). The average white blood cell count was also higher for the IM nonresponders than the responders, but this difference was not significant (P = .42, Table 1). All IM nonresponders had received hydroxyurea, and 3 of them had also received interferon treatment before initiation of IM treatment. Seven of 11 IM responders had received prior hydroxyurea, and none had received prior interferon (P = .07 and P = .16). These differences could cause singular solutions in the multivariable analysis and therefore could not be included (Table 2). The average time from diagnosis to beginning of IM treatment was longer for the IM nonresponders than the responders, but these differences were not significant (P = .29, Table 1).
Table 3.
Characteristics of clinically classified IM nonresponders with primary and secondary resistance
| Characteristics | Primary resistance | Secondary resistance | P* |
|---|---|---|---|
| Quantitative features | |||
| WBC at diagnosis | Range: [32.2, 536]; mean, 195; CI: [39.8, 350] | Range: [122, 480] mean, 221; CI: [36.7, 406] | .79 |
| Age, y | Range: [43, 77]; mean, 58; CI: [49.6, 66.4] | Range: [29,66]; mean, 51.6; CI: [34.2, 69.0] | .41 |
| Mutant BCR-ABL transcripts, percentage | Range: [18, 35]; mean, 25.8; CI: [13.5, 38.0] | Range: [9, 40]; mean, 22.6; CI: [8.48, 36.7] | .64† |
| NA | 4 (28.6%) | 0 | |
| No. of colonies as percentage of untreated cells CFC IM 1μM | Range: [41.1, 67]; mean, 54.1; CI: [47.1, 61.1] | Range: [40.6, 92] mean=70.9; CI: [44.3, 97.4] | .16 |
| CFC IM 5μM | Range: [36, 56.6]; mean, 47.7; CI: [43.0, 52.5] | Range: [34, 64]; mean, 50.3; CI: [36.0, 64.7] | .66 |
| CFC IM 10μM | Range: [29.1, 61.3]; mean, 44.1; CI: [34.9, 53.3] | Range: [20.8, 60.3]; mean, 43.8; CI: [21.0, 66.5] | .97 |
| Time from diagnosis to IM treatment, d | Range: [7, 302]; mean, 127.4; CI: [12.0, 178.5] | Range: [13, 377]; mean, 127; CI: [-66.7, 321] | .70 |
| Qualitative features | |||
| Sokal risk score | .434‡ | ||
| High | 5 (55.6%) | 0 | |
| Intermediate | 3 (33.3%) | 3 (60%) | |
| Low | 1 (11.1%) | 1 (20%) | |
| NA | 0 | 1 (20%) | |
| Sex | .21§ | ||
| Female | 5 (55.6%) | 1 (20%) | |
| Male | 4 (44.5%) | 4 (80%) | |
| Prior interferon | .247§ | ||
| No | 8 (88.9) | 3 (60%) | |
| Yes | 1 (11.1%) | 2 (40%) | |
| Prior hydroxyurea | 1§ | ||
| No | 9 (100%) | 5 (100%) | |
| Yes | 0 | 0 | |
| Daily IM dose | .177‡ | ||
| 400 mg | 6 (66.7%) | 2 (40%) | |
| 400-600 mg | 3 (33.3%) | 1 (20%) | |
| 400-800 mg | 0 | 2 (40%) |
P values were calculated using the Welch 2-sample t test at the 5% significant level under the null hypothesis H0: mean (primary resistance) = mean (secondary resistance).
P value of mutant BCR-ABL transcript frequency was calculated after all the missing values were taken out of the sample.
Kruskal-Wallis test was used to calculate P values of ordered categorical variable for Sokal risk score and daily IM dose.
Fisher exact test was used to calculate P values of nonordered categorical variable for sex, prior interferon, and prior hydroxyurea.
Exposure to IM in vitro differentially inhibits colony formation by leukemic CFCs from IM nonresponders and responders
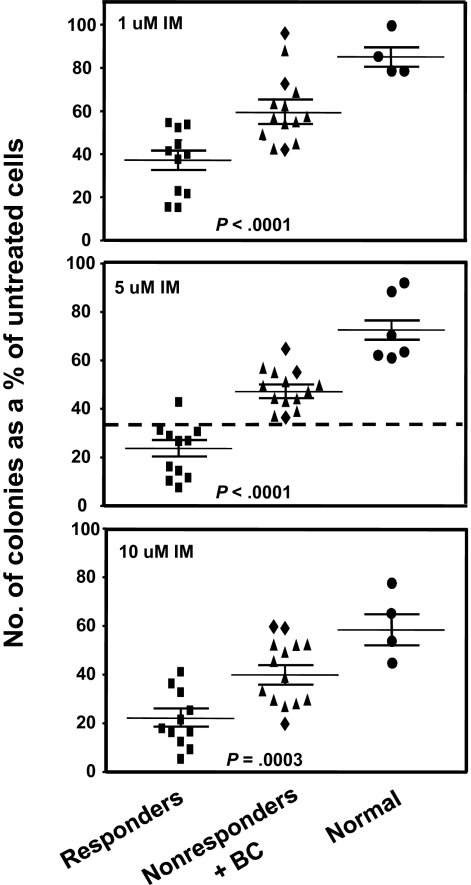
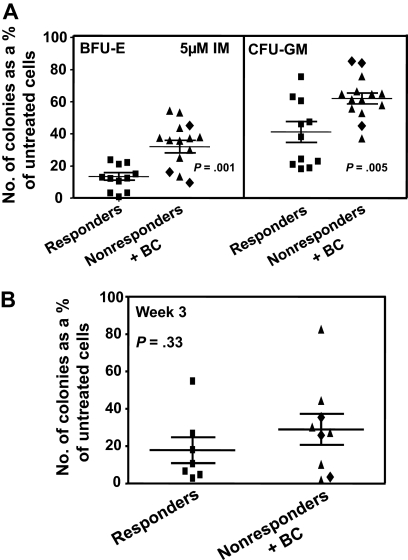
We assessed the IM sensitivity of the pretreatment leukemic CD34+ CFCs from all patients by performing standard in vitro colony assays in media supplemented with 0, 1, 5, or 10μM IM. IM inhibition of colony formation was consistently and significantly less (P < .001) for the 14 nonresponders, including the 3 patients who developed BC rapidly after an initial remission (Figure 1). Indeed, the CFCs from 2 of the latter showed the greatest resistance to IM in vitro, producing as many colonies in the presence of 10μM IM as CFCs from normal donors (Figure 1). The greater resistance of the CFCs from the IM nonresponder patients to IM in vitro was shared by both primitive erythroid (BFU-E) and granulopoietic progenitors (CFU-GM), although for both responder and nonresponder patients, the granulopoietic progenitors were generally more IM-resistant than the erythroid progenitors (Figure 2A).
Figure 1.
Comparison of the effect of different concentrations of IM on colony formation by CFCs from IM responders and nonresponders. CD34+ cells from 11 patients classified clinically as imatinib mesylate (IM) responders and 14 as nonresponders were assayed in duplicate 1-mL methylcellulose cultures containing 0, 1, 5, or 10μM IM. Two weeks later, the colonies were enumerated to determine the numbers derived from BFU-E, CFU-GEMM, or CFU-GM. The total number of colonies obtained in the IM-supplemented cultures was then calculated as a percentage of those obtained in the absence of IM. (♦) Results obtained from the 3 patients who rapidly developed BC; and cross bars, mean plus or minus SEM of data for each patient group. Differences between the results for the IM responders and nonresponders at all doses of IM tested are significant (P < .001).
Figure 2.
Comparison of the effect of IM in vitro on different subtypes of CD34+ cells from IM responders and nonresponders. (A) Comparison of the effect of 5μM IM on the ability of different subtypes of CFCs in unmanipulated CD34+ CML cells from IM responders and nonresponders to form colonies in vitro (same experiments and method of data calculation as shown in the middle panel of Figure 1). Differences between the results for BFU-E and CFU-GM from IM responders and nonresponders are significant (P ≤ .005). (B) The effect of 5μM IM on the patient-specific yield of CFCs in 3-week liquid suspension cultures initiated with the same cells as plated directly in methylcellulose in panel A. (♦) Results obtained from the 3 patients who rapidly developed BC. No significant differences were found between the effect of IM on CFC production in 3-week cultures initiated with CD34+ cells from the 2 patient groups (P = .33). In both panels, cross bars represent the mean plus or minus SEM of data for each patient group.
We also examined whether continuous exposure to 5μM IM in vitro would differentially affect the number of CFCs produced in 3-week liquid suspension cultures initiated with CD34+ cells from the both groups of patients. In contrast to the data for colony formation from CFCs, IM effects on the generation of CFCs were not significantly different (9 nonresponders and 7 responders analyzed, P = .33, Figure 2B).
CD34+ cells from IM nonresponders and responders are not distinguished by differences in their levels of BCR-ABL and transporter gene expression
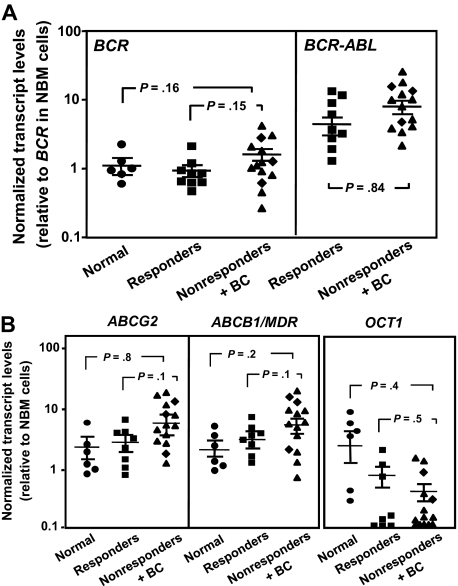
We also asked whether quantitative RT-PCR–determined levels of BCR-ABL, OCT1, ABCB1, and ABCG2 transcripts might be different in the leukemic CD34+ cells from IM nonresponders and responders before treatment because the expression of these genes in CD34+ progenitor and/or stem cells has been found to be altered in a fashion that would be expected to contribute to a more IM-resistant phenotype.20 However, none of these measurements identified significant differences between the responders and nonresponders studied (P > .05, Figure 3).
Figure 3.
Comparison of BCR-ABL and transport gene transcript levels in CD34+ cells from IM responders and nonresponders. (A) Comparison of BCR (left panel) and BCR-ABL (right panel) transcript levels in CD34+ CML cells from IM responders (n = 9) and nonresponders (n = 14) relative to BCR transcript levels in CD34+ cells isolated from normal persons (n = 6). (B) Comparison of ABCG2 (left panel), ABCB1/MDR (middle panel), and OCT1 (right panel) transcript levels relative to GAPDH transcripts in CD34+ CML cells from IM responders and nonresponders. (♦) Results obtained from the 3 patients who rapidly developed BC. Each data point represents the average of a triplicate quantitative RT-PCR measurement of each transcript normalized as described in “Methods.” Cross bars represent the mean ± SEM of data for each patient group. The differences between the results for IM responders and nonresponders are not significant (P > .05).
BCR-ABL transcripts in IM nonresponder CD34+ cells contain a higher frequency of kinase domain mutations than those from IM responders
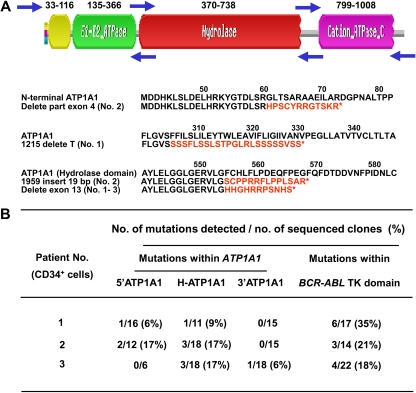
To investigate whether IM responders and nonresponders might have different abilities to regulate their genomic integrity, we first prepared cDNAs from RNA extracts of their CD34+ cells. We then cloned the major tyrosine kinase domain of the BCR-ABL transcripts present as well as overlapping fragments spanning the complete ATP1A1 transcript31 and amplified these in bacteria. A similar analysis of ATP1A1 transcripts was included to determine the genomic generality of any increased instability evident in the BCR-ABL gene of CML stem/progenitor cells, as previously suggested by studies of BCR-ABL-transduced murine cells.39 Finally, we sequenced the cloned fragments (10-30 clones per sample) to determine the numbers and types of mutations present in each. From this analysis, we found that the cloned BCR-ABL kinase domain fragments from 18 of 20 CML patients' CD34+ cells contained at least one mutation. The actual frequency of mutant transcripts per sample ranged from 6% to 40% (a mutant transcript being defined as one or more mutations per cloned fragment, Figure 4A). Mutant ATP1A1 transcripts were readily detectable in all 3 CML patients investigated (15%-34% of ATP1A1 transcripts found to contain one or more mutations, all IM nonresponders, Figure 5).
Figure 5.
Detection of mutant ATP1A1 transcripts in CD34+ cells from IM nonresponders. (A) Structure of the ATP1A1 gene and location of 3 forward and reverse primer pairs used to generate 3 overlapping fragments for PCR amplification of the entire ATP1A1 transcript cDNA (3.7 kb). Frameshift mutations (in red) that generate new amino acid sequences and premature stop codons (*) within the ATP1A1 gene were identified in CD34+ CML cells from all 3 IM nonresponders studied. (B) Specific ATP1A1 mutations identified in the pretreatment CD34+ cells from the 3 IM nonresponders studied. The mutations detected within the BCR-ABL tyrosine kinase domain from the same patients' cells are also indicated.
To evaluate the possibility of false-positive mutant transcript detection using the method adopted here, we used the same approach to examine the integrity of the c-ABL and ATP1A1 gene in CD34+ normal BM cells. This analysis failed to reveal any mutant c-ABL transcripts in 150 sequenced clones (from 6 normal donors) and only a single point mutation (L767P) in 80 ATP1A1 transcript clones (from 3 normal donors). These findings demonstrate the high fidelity of the method used here to quantify the higher levels of genomic change evident in CML CD34+ cells.
Interestingly, comparison of the mutant BCR-ABL transcript data from the IM responders and nonresponders showed that the frequencies of mutant transcripts were significantly higher in the IM nonresponders (10%-40%, vs 0%-20%, P < .003, 10 patients in each group studied).
Multivariable statistical analysis identifies a significant correlation between clinical IM response and the colony-forming response of the patient's pretreatment CD34+ cells
To evaluate the extent to which each of the various parameters studied was independently associated with subsequent clinical response to IM, a multivariable statistical model was used. The preliminary model was generated using the independent variables that were both mathematically solvable in the model and whose P values were less than or equal to 10% (the frequency of mutant BCR-ABL transcripts was omitted because of some missing values). This analysis revealed a significant correlation of the clinical IM response with the in vitro colony-forming response obtained when CD34+ stem/progenitor cells were cultured in 5μM IM (Table 2); this was also verified by odds ratio (odds = exp (−19.24) = 0 < 1). The finding of a significant correlation between these 2 parameters was confirmed by ROC analysis, which tests how well a final model predicts its covariates (the data not shown). Thus, these analyses indicate the clinically defined responders and nonresponders were significantly different from each other with respect to the inhibited growth response of their pretreatment CFC to 5μM IM.
Characterization of BCR-ABL and ATP1A1 mutations found in IM-naive CD34+ CML cells
In total, 18 different mutations within the major kinase domain of BCR-ABL (residues 220-420) were identified in the CD34+ cells from the 20 CML patients studied (2 of whom did not show a detectable level of mutant BCR-ABL+ cells). Two of the mutations identified (M244V and T315I) have been frequently associated with clinical IM resistance and one (T315I) with resistance to all available BCR-ABL inhibitors.40–42 Four amino acid substitutions (C305S, E373G, V379I, and M388T) were found at a low frequencies. Although most of the mutations were detected in only one sample, one (V304D) was present in 14 cases (Figure 4B). Other mutations at amino acid residue 304 (V304G or V304S) have been previously documented at a low frequency in clinical specimens from IM-resistant patients,40–42 and in CD34+ cells from IM-treated CML patients in complete cytogenetic remission (V304S).21 Molecular modeling analysis of the published crystal structures of the kinase domain of the BCR-ABL oncoprotein in complex with IM, DA, or NL reveals that valine 304 (V304) localizes to the hydrophobic interior of the kinase domain and packs against a helix that forms part of the IM binding groove.16,32,33 Substituting the nonpolar V304 with the polar residue, aspartic acid, would be predicted to dramatically perturb the conformation and/or structure of this helix and destabilize interactions with IM and NL (Figure 4C). A similar effect on DA binding would not be expected because DA binds both the active (open) and inactive (closed) conformations of the kinase domain and its binding does not involve the aforementioned helix.33 Several of the other mutations that we identified also map to the vicinity of or within the drug-binding cleft, suggesting they would also disrupt IM, DA, or NL binding (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
We were able to obtain post-IM treatment samples from 2 of the nonresponders and reanalyze the sequence of the kinase domain of the BCR-ABL transcripts in the CD34+ leukemic cells present at that later time. In the first patient, 10% of the pretreatment transcripts contained a T315I mutation and 25% contained a V304D mutation. After IM therapy, the frequency of T315I-mutant BCR-ABL transcripts increased 5-fold to 50% (Figure 4D), although the corresponding frequency of V304D mutant transcripts remained the same, at 25%. In the second patient, V304D mutant cells also persisted after IM therapy and additional mutations (M244V, C305G, and H396R) became evident at variable frequencies in the posttreatment cells (Figure 4D). Sequencing of cDNAs prepared from extracts of unpurified blood cells from both these patients also revealed the later prevalence of T315I mutant cells in patient 1 and M244V mutant cells in patient 2. However, the other mutations (V304D, C305G, H396R, and C330Y) detectable in their CD34+ cells at a relatively low frequency were not detectable in the total blood cell population.
The mutations identified in the ATP1A1 transcripts isolated from the CML CD34+ cells included point mutations, insertions, and deletions of parts of exon 4 and 13 (Figure 5). Notably, most of these mutations were found in the hydrolase domain of ATP1A1, and these were found in all 3 CML samples studied. The mutations included deletions of exon 13 within the hydrolase domain, resulting in premature termination codons and hence truncated proteins (Figure 5A).
Discussion
Despite the effectiveness of IM therapy in inducing high rates of initial hematologic and cytogenetic responses in chronic-phase CML patients, IM resistance continues to pose a major challenge for the management of some patients. The disease may be unresponsive to IM at the time of diagnosis, or resistance can become a problem a few months after an initial response. Occasionally, BC may occur unexpectedly after an initial response. For patients who have, or are prone, to develop more advanced-phase disease, the earliest possible use of alternative therapeutic options is critical. Predictive tests are urgently needed to identify patients who are unlikely to benefit from IM therapy. To date, none of the conventional clinical risk evaluation methods has proven useful for discriminating these patients. Optimal patient management therefore currently depends on repeated quantitative RT-PCR monitoring of peripheral blood BCR-ABL transcript levels bolstered by a search for mutant clones using sequencing when BCR-ABL transcript levels appear to be increasing despite continued IM treatment. Although these strategies can usually forecast IM-resistant relapses, the information may come too late for application of alternative therapies.
In chronic-phase CML, the stem/progenitor cells are intrinsically less sensitive to IM and other tyrosine kinase inhibitors than the more mature CML cells that compose the bulk (> 90%) of the leukemic clone. These rare cells are also a critical target population for generating genetically IM-resistant subclones.18–22 Therefore, it has been of interest to determine whether any distinguishing features of the leukemic (CD34+) stem/progenitor cells from CML patients might vary among patients in a fashion that correlates with the subsequent clinical response to IM therapy. Here we found that the growth-inhibiting activity of 5μM IM on leukemic CFCs in an in vitro colony assay was a significantly distinguishing difference between a small group of 25 patients independently defined as clinical IM responders and nonresponders. Interestingly, in the CD34+ pretreatment cells from 2 of 3 patients who developed BC rapidly after an initial clinical response to IM, the CFCs appeared completely resistant to IM in this assay and generated colonies at a level comparable with normal controls. Thus, it seems likely that biologically IM-resistant cells were already dominant in the stem/progenitor compartments of these 2 patients before the IM therapy was started, even though these resistant cells may not yet have become prevalent in the bulk population. Notably, no difference was identified between responders and nonresponders in terms of the effect of IM on the generation of CFCs in a 3-week liquid suspension culture system. Perhaps this is the result of the origin of these CFCs from more primitive CML cells that are innately more IM-resistant than the CFCs they generate.18–20 Although the numbers of responder and nonresponder cases studied here was quite small (11 and 14, respectively), the fact that their CFC responses to 5μM IM in vitro showed little overlap (Figure 1) suggests that pretreatment CFC sensitivity to IM in vitro may serve as a useful predictor of subsequent clinical response to IM.
Previous studies by our laboratory20 and others26,27,42,43 have demonstrated that primitive CML cells display a gene expression profile and associated activities that would be consistent with their relative IM resistance. This includes an exaggerated reduction in the levels of OCT1 transcripts in the CD34+CD38− stem cell–enriched population (< 5% of total CD34+ cells) compared with more mature CD34+CD38+ progenitor cells (∼ 95% of the total CD34+ population).20 Conversely, expression of both ABCB1 and ABCG2 is more highly elevated in the CD34+CD38− cells than in the CD34+CD38+ cells. However, in the present study, transcript levels of these transporter genes were not found to be significantly different in CD34+ CML cells from responders versus nonresponders. This is consistent with a recent study that shows OCT-1 mRNA expression is not associated with molecular response, although significant changes may be observed by measurement of OCT-1 protein activity.44 This suggests that transporter mRNA expression is not always indicative of protein function, a result that is not unexpected given the marked posttranscriptional regulation of these transporters.27,42,44 Thus, measurement of protein activity of these genes may be required for distinguishing differences between IM responders and nonresponders.
Most chronic-phase CML patients fail to achieve complete molecular remission, some experiencing an expansion of IM-resistant leukemic cells that have acquired mutations that confer additional IM-resistant features.13,45,46 The most common of these are point mutations in the kinase domain (accounting for 60%-90% of relapses).13,14,30,47,48 To date, mutations in more than 50 different amino acid positions within the BCR-ABL kinase domain have been reported in IM-resistant patients.40–42 We have recently demonstrated that the BCR-ABL fusion gene in very primitive (CD34+CD38−) CML cells is highly unstable.23 This is reflected in the unusually high frequency of BCR-ABL mutations that accumulate in this compartment, even in the absence of their selection by in vivo treatment with IM. Rapid acquisition of new mutations in the BCR-ABL gene can also be seen in the progeny of CML stem cells stimulated to proliferate and differentiate in vitro.23 Here we confirm this finding and further demonstrate that a similar level of genomic instability is not evident in the CD34+ BM cells from normal adults. In addition, we show that the genomic instability previously documented for the BCR-ABL gene in CD34+ CML cells is not unique to this locus. The demonstration of a similarly high frequency of mutations affecting the ATP1A1 gene in the CD34+ cells from 3 CML patients and not in normal CD34+ cells is strongly suggestive of a genome-wide instability. Taken together, these findings support the concept that primitive leukemic CML cells are intrinsically endowed with a tendency to continuously acquire new mutations at a greatly elevated rate independent of therapy. Some of the acquired secondary mutations would be expected to confer IM resistance; others could lead to disease progression. It is thus the nature and timing of these chance secondary events relative to the size of the primary clone at diagnosis and implementation of IM treatment that may explain the variable clinical responses of individual patients.23,30,48,49 The present study now shows, for the first time, that CML patients defined retrospectively as clinical IM nonresponders and responders display significant differences in the frequency of mutant BCR-ABL transcripts present in their pretreatment CD34+ cells.
A surprising discovery was the presence of BCR-ABL transcripts carrying the V304D mutation at readily detectable levels (> 5% of transcripts affected) in the CD34+ cells from most (70%) of the 20 patients studied. Mutations at the 304 site have been detected previously as V304G or V304S in clinical specimens from IM-resistant patients at a very low frequency,40–42 in CD34+ cells from IM-treated CML patients in complete cytogenetic remission (V304S),21 and in the progeny of cultured CML stem (lin−CD34+CD38−) cells (V304A).23 Here we found that the V304D mutation was also detectable in posttreatment CD34+ cells from 2 of 2 nonresponders examined. Interestingly, the V304D mutation has recently been described as a novel mutation emerging as a DA-resistant mutation.50 Molecular modeling analysis of the published crystal structures of the kinase domain of the BCR-ABL oncoprotein in complex with IM, DA, and NL indicates that this mutation is critical for IM and NL binding and would be predicted to dramatically perturb the local structure and destabilize interactions with IM and NL. However, a similar effect on DA binding would not be expected because DA binds both the active (open) and inactive (closed) conformations of the BCR-ABL kinase domain,16,32,33 although the V304D mutation has recently been reported in 4 of 15 patients resistant to DA.50 Because the V304D mutation was not frequently detected in unpurified cells from IM-resistant patients, it is unlikely that this mutation confers a growth advantage on resistant cells, although the modeling analysis suggests it affects IM binding. Whether this mutant clone plays a role in affecting intrinsic properties of primitive CML cells involved in drug resistance and/or disease progression or whether it simply acts as a surrogate marker of the increased genetic instability requires further investigation.
Interestingly, both a multivariable statistical model and ROC analysis of the various parameters studied demonstrated a significant correlation of IM response with the in vitro colony-forming response of CD34+ cells to 5μM IM (Table 2). Thus, CD34+ CML cells from individual chronic-phase patients harbor differences in certain biologic properties that may be predictive of how they will respond to IM therapy. The ability to develop rapid and robust tests to predict individual patients' responses to IM therapy could have a profound impact on CML patient management by providing a foundation for more effective treatment decisions.
Supplementary Material
Acknowledgments
The authors thank Dr M. Barnett, Dr D. Hogge, Dr J. Lavoie, Dr S. Nantel, Dr T. Nevill, Dr J. Shepherd, Dr K. Song, Dr H. Sutherland, and Dr C. Toze (all members of the Leukemia/BMT Program of British Columbia, Vancouver, BC) for providing patient samples; the Terry Fox Laboratory FACS Facility for assistance in cell sorting; the Stem Cell Assay Laboratory for cell processing and cryopreservation of normal and CML samples; Novartis (Basel, Switzerland) for providing IM and NL; Bristol-Myers Squibb for DA; and StemCell Technologies for culture reagents.
This work was supported by the Canadian Cancer Society, the Cancer Research Society, and the Leukemia & Lymphoma Society of Canada (X.J.), and the Canadian Cancer Society Research Institute (A.E., X.J., and C.E). X.J. and R.R.B. are Michael Smith Foundation for Health Research Scholars. Infrastructure support was provided by the British Columbia Cancer Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: X.J., D.F., C.S., A.E., and C.E. developed the concept for the study; X.J. designed and supervised the experiments; D.F., F.N., A.T., J.G., T.H., and H.J. provided clinical samples and clinical data; K.L., K.M.S., E.P., R.V., and P.L. performed the experiments and analyzed the data; M.Y. and R.R.B. performed statistical analysis; C.Y. and P.L. performed molecular modeling and analysis; X.J., A.R., and C.E. wrote the manuscript; and all authors commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiaoyan Jiang, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC V5Z 1L3, BC, Canada; e-mail: xjiang@bccrc.ca.
References
- 1.Goldman JM, Melo JV. Chronic myeloid leukemia: advances in biology and new approaches to treatment. N Engl J Med. 2003;349(15):1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 2.Jiang X. Cancer Nanotechnology. Stevenson Ranch, CA: American Scientific Publishers; 2007. Molecular and cellular mechanisms of deregulated hematopoietic stem cell functions in chronic myeloid leukemia. pp. 135–157. [Google Scholar]
- 3.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8(5):341–350. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 4.Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247(4946):1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 6.Holyoake TL, Jiang X, Drummond MW, Eaves AC, Eaves CJ. Elucidating critical mechanisms of deregulated stem cell turnover in the chronic phase of chronic myeloid leukemia. Leukemia. 2002;16(4):549–558. doi: 10.1038/sj.leu.2402444. [DOI] [PubMed] [Google Scholar]
- 7.Penserga ET, Skorski T. Fusion tyrosine kinases: a result and cause of genomic instability. Oncogene. 2007;26(1):11–20. doi: 10.1038/sj.onc.1209756. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Smith C, Eaves A, Eaves C. The challenges of targeting chronic myeloid leukemia stem cells. Clin Lymphoma Myeloma. 2007;7(suppl):S71–S80. doi: 10.3816/clm.2007.s.005. [DOI] [PubMed] [Google Scholar]
- 9.Valent P. Emerging stem cell concepts for imatinib-resistant chronic myeloid leukaemia: implications for the biology, management, and therapy of the disease. Br J Haematol. 2008;142(3):361–378. doi: 10.1111/j.1365-2141.2008.07197.x. [DOI] [PubMed] [Google Scholar]
- 10.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346(9):645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 11.Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 12.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 13.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105(7):2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 14.O'Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2006;16(1):92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305(5682):399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 16.Weisberg E, Manley PW, Breitenstein W, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7(2):129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354(24):2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 18.Graham SM, Jorgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 19.Copland M, Hamilton A, Elrick LJ, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML, but does not eliminate the quiescent fraction. Blood. 2006;107(11):4532–4539. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X, Zhao Y, Smith C, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21(5):926–935. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 21.Chu S, Xu H, Shah NP, et al. Detection of BCR-ABL kinase mutations in CD34+ cells from chronic myelogenous leukemia patients in complete cytogenetic remission on imatinib mesylate treatment. Blood. 2005;105(5):2093–2098. doi: 10.1182/blood-2004-03-1114. [DOI] [PubMed] [Google Scholar]
- 22.Konig H, Copland M, Chu S, Jove R, Holyoake TL, Bhatia R. Effects of dasatinib on SRC kinase activity and downstream intracellular signaling in primitive chronic myelogenous leukemia hematopoietic cells. Cancer Res. 2008;68(23):9624–9633. doi: 10.1158/0008-5472.CAN-08-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang X, Saw KM, Eaves A, Eaves C. Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst. 2007;99(9):680–693. doi: 10.1093/jnci/djk150. [DOI] [PubMed] [Google Scholar]
- 24.Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 25.Lepper ER, Nooter K, Verweij J, Acharya MR, Figg WD, Sparreboom A. Mechanisms of resistance to anticancer drugs: the role of the polymorphic ABC transporters ABCB1 and ABCG2. Pharmacogenomics. 2005;6(2):115–138. doi: 10.1517/14622416.6.2.115. [DOI] [PubMed] [Google Scholar]
- 26.White DL, Saunders VA, Dang P, et al. OCT-1 mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107); reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood. 2006;108(2):697–704. doi: 10.1182/blood-2005-11-4687. [DOI] [PubMed] [Google Scholar]
- 27.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: implications for drug resistance. Blood. 2004;104(12):3739–3745. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 28.Forrest DL, Jiang X, Eaves CJ, Smith CL. An approach to the management of chronic myeloid leukemia in British Columbia. Curr Oncol. 2008;15(2):48–55. doi: 10.3747/co.v15i2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holyoake TL, Jiang X, Jorgensen HG, et al. Primitive quiescent leukemic cells from patients with chronic myeloid leukemia spontaneously initiate factor-independent growth in vitro in association with up-regulation of expression of interleukin-3. Blood. 2001;97(3):720–728. doi: 10.1182/blood.v97.3.720. [DOI] [PubMed] [Google Scholar]
- 30.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2(2):117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami K, Ohta T, Nojima H, Nagano K. Primary structure of the alpha-subunit of human Na,K-ATPase deduced from cDNA sequence. J Biochem. 1986;100(2):389–397. doi: 10.1093/oxfordjournals.jbchem.a121726. [DOI] [PubMed] [Google Scholar]
- 32.Nagar B, Bornmann WG, Pellicena P, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res. 2002;62(15):4236–4243. [PubMed] [Google Scholar]
- 33.Tokarski JS, Newitt J, Chang CYJ, et al. The structure of dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66(11):5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 34.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMol Molecular Graphics System. South San Francisco, CA: DeLano Scientific; 2002. [Google Scholar]
- 36.Imaizumi M, Usa T, Tominaga T, et al. Radiation dose-response relationships for thyroid nodules and autoimmune thyroid diseases in Hiroshima and Nagasaki atomic bomb survivors 55-58 years after radiation exposure. JAMA. 2006;295(9):1011–1022. doi: 10.1001/jama.295.9.1011. [DOI] [PubMed] [Google Scholar]
- 37.Fawcett T. ROC Graphs: Notes and Practical Considerations for Researchers. Palo Alto, CA: HP Laboratories; 2004. pp. 1–38. [Tech Report HPL-2003-4] [Google Scholar]
- 38.R Development Core Team. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 39.Koptyra M, Falinski R, Nowicki MO, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108(1):319–327. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes T, Deininger M, Hochhaus A, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108(1):28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations and the unsettled problem of Bcr-AblT315I: looking into the future of controlling drug resistance in chronic myeloid leukemia. Clin Lymphoma Myeloma. 2007;7(suppl):S120–S130. doi: 10.3816/clm.2007.s.012. [DOI] [PubMed] [Google Scholar]
- 42.Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8(11):1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 43.Mahon FX, Belloc F, Lagarde V, et al. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood. 2003;101(6):2368–2373. doi: 10.1182/blood.V101.6.2368. [DOI] [PubMed] [Google Scholar]
- 44.White DL, Saunders VA, Dang P, et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood. 2007;110(12):4064–4072. doi: 10.1182/blood-2007-06-093617. [DOI] [PubMed] [Google Scholar]
- 45.Hughes TP, Kaeda J, Branford S, et al. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349(15):1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- 46.Branford S, Rudzki Z, Walsh S, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102(1):276–283. doi: 10.1182/blood-2002-09-2896. [DOI] [PubMed] [Google Scholar]
- 47.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112(6):831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 48.Roche-Lestienne C, Lai JL, Darre S, Facon T, Preudhomme C. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N Engl J Med. 2003;348(22):2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- 49.Sorel N, Bonnet ML, Guillier M, Guilhot F, Brizard A, Turhan AG. Evidence of ABL-kinase domain mutations in highly purified primitive stem cell populations of patients with chronic myelogenous leukemia. Biochem Biophys Res Commun. 2004;323(3):728–730. doi: 10.1016/j.bbrc.2004.08.169. [DOI] [PubMed] [Google Scholar]
- 50.Krijanovski Y, Donato N, Sun H, et al. Dasatinib resistance in patients with chronic myelogenous leukemia: identification of a novel bcr-abl kinase domain mutation. Clin Leukemia. 2008;2:267–271. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.