Abstract
The perceived taste intensities of quinine HCl, caffeine, sucrose octaacetate (SOA) and propylthiouracil (PROP) solutions were examined in 1457 twins and their siblings. Previous heritability modeling of these bitter stimuli indicated a common genetic factor for quinine, caffeine and SOA (22–28%), as well as separate specific genetic factors for PROP (72%) and quinine (15%). To identify the genes involved, we performed a genome-wide association study with the same sample as the modeling analysis, genotyped for approximately 610 000 single-nucleotide polymorphisms (SNPs). For caffeine and SOA, no SNP association reached a genome-wide statistical criterion. For PROP, the peak association was within TAS2R38 (rs713598, A49P, P = 1.6 × 10−104), which accounted for 45.9% of the trait variance. For quinine, the peak association was centered in a region that contains bitter receptor as well as salivary protein genes and explained 5.8% of the trait variance (TAS2R19, rs10772420, R299C, P = 1.8 × 10−15). We confirmed this association in a replication sample of twins of similar ancestry (P = 0.00001). The specific genetic factor for the perceived intensity of PROP was identified as the gene previously implicated in this trait (TAS2R38). For quinine, one or more bitter receptor or salivary proline-rich protein genes on chromosome 12 have alleles which affect its perception but tight linkage among very similar genes precludes the identification of a single causal genetic variant.
INTRODUCTION
In 1934, Professor R.A. Fisher began a series of studies in the Galton Laboratory at Oxford aimed at understanding the inheritance pattern of bitter taste perception for the then-new compound phenylthiocarbamide (PTC). These studies were interrupted by the start of World War II but the record cards were preserved and later analyzed by Dr D.S. Falconer, who published a popular paper about individual differences (1). What has been overlooked, then and now, is that people differ markedly in their ability to taste many other bitter compounds besides PTC and related structures. In Dr Fisher's study, the ability to perceive the bitterness of quinine was also tested and found to be markedly different among subjects and yet these results were overshadowed by his interest in the newer compound. In fact, sensory differences for many bitter compounds, i.e. PROP (propylthiouracil, a close chemical relative of PTC), caffeine, sucrose octaacetate (SOA) and quinine are heritable (2–4). For PROP and PTC, alleles of one of the 25 bitter receptors (TAS2R38) explain most of the genetic variation (5–7) although other modifier loci may exist (8–10). We have shown that, for caffeine, SOA and quinine, a common genetic factor accounts for 22–28% of the phenotypic variance, and a quinine-specific factor accounts for 15% of variation (4). However, unlike PROP and PTC, the genes responsible for variation in the perceived bitterness of these compounds have not been identified. To find these genes, we conducted genome-wide analyses in twins and their siblings (discovery sample) (11) and tested the associations in a second sample of twins (replication sample). We included PROP as a bitter taste stimulus to serve as a positive control because its genetic architecture is well understood.
RESULTS
Table 1 lists the sample sizes, age, sex and other details for the discovery and replication subject populations.
Table 1.
Discovery and replication sample characteristics
| Characteristic | Discovery | Replication |
|---|---|---|
| n | 1457 | 73 |
| Males/females | 671/786 | 16/57 |
| Age (mean + standard deviation) | 18 + 2 | 42 + 17 |
| Age range (years) | 11–25 | 21–82 |
| Race/ethnicity | Caucasian | Caucasian |
| Number of twins (MZ/DZ) | 405/847 | 52/21 |
| Number of siblings | 205 | 0 |
| Country ascertained | Australia | USA |
MZ, monozygotic; DZ, dizygotic.
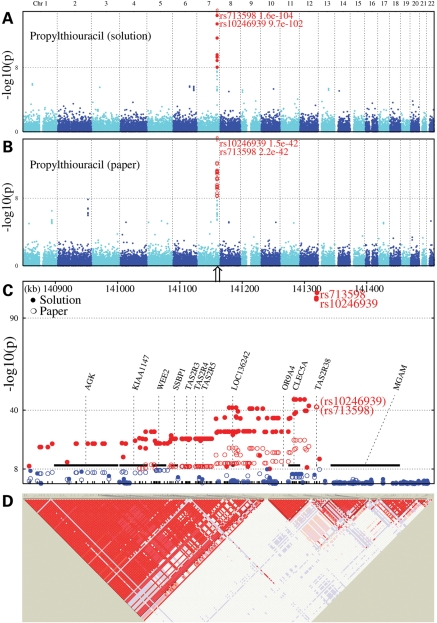
For caffeine and SOA, no single-nucleotide polymorphisms (SNPs) reached our genome-wide significance threshold of 1.136 × 10−8 (see reference 12 for rationale for threshold; Fig. 1). Previous research has demonstrated that a person's thresholds or perceived intensity ratings for caffeine, SOA and quinine are correlated (4,13,14), suggesting that these individual differences arise from a common mechanism. However, we found no overlapping genetic associations for all three taste stimuli. For the PROP solution, 157 SNPs (30 of which were genotyped) reached this criterion P-value, with TAS2R38 on chromosome 7 at the epicenter. The strongest signals in this region were observed at rs713598 (P = 1.6 × 10−104) and rs10246939 (P = 1.1 × 10−101). This region accounted for a maximum trait variance of 45.9%. When this association was accounted for (i.e. after conditioning on the genotyped peak SNP, rs10246939), there was little evidence for a nearby secondary peak (chromosome 7: rs13238628, P = 2.1 × 10−6.). The two highest secondary peaks identified after conditioning on the peak SNP may be due to chance (chromosome 2: rs4141835, P = 6.8 × 10−7; chromosome 7: rs4727180; P = 2.1 × 10−6). The results for PROP-saturated paper overlapped with those for the solution, and all SNPs were within or near TAS2R38 (rs1726866; P = 2.10 × 10−42). A region near a previously identified modifier locus (chromosome 5; TAS2R1) approached but did not meet the statistical criterion for genome-wide significance (rs6867567; P = 2.20 × 10−7). For other details, see Supplementary Material, Table S1.
Figure 1.
Genome-wide association for PROP perception (A and B), followed by regional association between 7q34 variants and PROP perception (C) and LD among markers for the 7q34 region (D). (A) and (B) The observed –log 10 P-values by position (Mbp) for bitterness of PROP tasted in solution (A) and tasted within a saturated paper strip (B). The horizontal dotted gray lines show the genome-wide association significance level corrected for the 4.4 independent traits analyzed (calculated using http://gump.qimr.edu.au/general/daleN/matSpD/). The regional association plot between 7q34 variants and PROP perception (C) indicates the location of known genes. Bitterness of PROP tasted in solution is indicated using closed circles and tasted on a saturated paper strip is shown using open circles. (D) Heat-map of the LD in the 7q34 region. The second LD block captures the high LD within the region of peak association. Bitter receptor gene nomenclature has recently changed and gene alias are available online (http://www.ensembl.org/index.html).
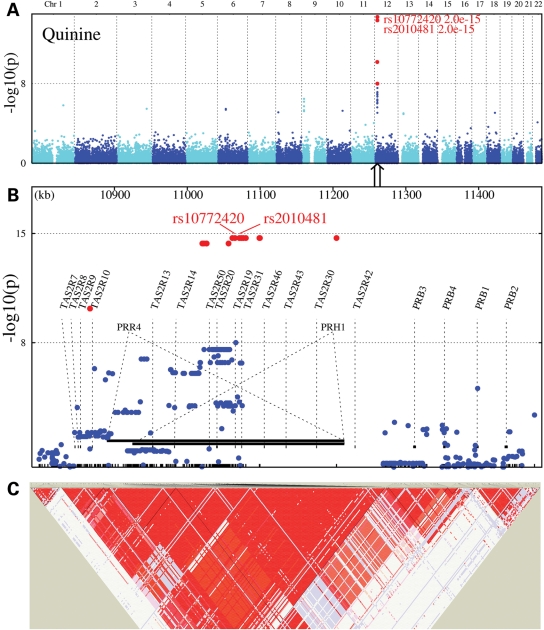
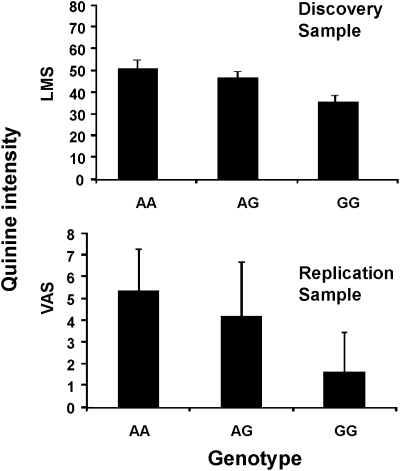
For quinine, 19 SNPs (12 of which were genotyped) from chromosome 12, near and within a cluster of bitter receptor and two salivary protein genes, were associated with bitterness perception. Eleven of the associated markers were within the introns of two proline-rich protein genes, and the other SNP coded for an arginine-to-cysteine substitution at amino acid 299 (R299C) in the bitter receptor TAS2R19 (rs10772420, R299C, P = 1.8 × 10−15; formerly known as TAS2R48) (Fig. 2). This region accounted for a maximum trait variance of 5.77%. After conditioning on the peak SNP (rs10772420), no secondary peaks reached the statistical criterion for genome-wide significance. We tested this association for quinine in an independent group of twins from the general population, collected as a part of a larger study on the genetics of taste perception conducted at a festival held in Twinsburg, OH, USA. One SNP (rs10772420) was genotyped, chosen from a subset of markers from the discovery sample because it was tightly associated with the ratings of quinine intensity and because it created an amino acid change in a bitter receptor (TaqMan, Applied Biosystems). The results indicated that the A allele of the genetic variant TAS2R19 R299C (rs10772420) was associated with more intense quinine perception [F(2,66) = 13.8, P = 0.00001] with the same direction of effect and the same allele associated with increased quinine sensitivity in both the discovery and replication samples (Fig. 3).
Figure 2.
Genome-wide association for quinine perception (A), followed by regional association between 12p13.2 variants and quinine perception (B) and LD among markers for the 12p13.2 region (C). See Figure 1 for details.
Figure 3.
Average bitter taste intensity of quinine as rated by people from the discovery (top) and replication (bottom) samples grouped by their rs10772420 genotype. LMS, labeled magnitude scale; VAS, visual analogue scale. Values are means and confidence intervals corrected for family relationship (upper panel) or means and standard deviations (lower panel). The TAS2R19 A allele corresponds to the cysteine amino acid at position 299 and is associated with more intense perception of quinine in both samples.
DISCUSSION
For quinine, the SNPs identified in the genome-wide association study implicate two orally expressed gene families: proline-rich proteins that are secreted in saliva (15) and bitter receptors that are found in taste receptor cells (16,17). These two types of genes have many family members and many alleles (18,19). Alleles from these gene clusters are in strong linkage disequilibrium (LD), thus the particular gene responsible for this taste trait cannot be readily identified by genetic association alone. Cell-based expression assays are useful for identifying receptor alleles and their functional responses and have been used to unequivocally identify alleles of a bitter receptor gene associated with taste insensitivity for other ligands (6). However, the assays conducted to date with quinine suggest that many bitter receptor genes from the cluster studied here respond to quinine (TAS2R7, 10, 14, 46 and 43) (7) although not all receptors could be tested due to technical limitations, e.g. TAS2R19. Also the specificity of these quinine responses cannot be adequately assessed at this time due to the ability of quinine to activate cells indirectly (20). Thus far, specific alleles have not been evaluated for their response to quinine. Furthermore, candidate bitter receptors may heterodimerize within native taste cells, adding to the complexity of identifying specific alleles (21). Although the identification of particular genes is difficult, this region is also supported by comparative studies. Quinine sensitivity in mice maps to the homologous region identified here (22–25).
For PROP, the region known to be associated with its perception was also supported in this analysis, i.e. TAS2R38 (6), accounting for almost half of the trait variance. Alleles near a bitter receptor on chromosome 5 previously associated with PROP perception also tended to be associated in this study (8,16).
Bitter receptors have been strongly selected during human evolution (26–29). Although the reason is not understood, flexibility within a population's ability to tolerate or avoid bitter foods might be useful in some environments and circumstances (e.g. to discover which plants are poisonous or, conversely, to tolerate nutritious plants that contain toxins) (30). However, the focus on oral toxin detection may be too narrow. Recently, the scope of the TAS2Rs and their role in detecting chemicals have been expanded to include the gut (31–34) and airways (35,36). Of particular note are two studies showing that bitter receptors respond to the chemicals secreted by bacteria and thus help the body fight infection, and their alleles have also been recently implicated in human diabetes (37). Thus, TAS2R selection could also be driven by chemicals in the nose, lungs, pancreas or gastrointestinal tract.
MATERIALS AND METHODS
Subjects
For the discovery sample, participants were a subset of adolescent and young adult twins and their singleton siblings (11) who have participated in previous studies of the genetics of skin moles and cognition. The sample for which taste sensitivity results were available consisted of females and males and included monozygotic (MZ) and dizygotic (DZ) twin pairs and their siblings. For the replication sample, experimenters recruited and tested participants at an annual convention of twins, Twins Days Festival, in Twinsburg, OH, USA. Testing occurred at the 2009 festival in August (Table 1). The discovery study was performed with the approval of the Queensland Institute of Medical Research (QIMR) Human Research Ethics Committee, and the replication study was performed with the approval of the Institutional Review Board at the University of Pennsylvania. Informed consent was obtained from all participants.
Phenotype definition
For the discovery sample, we measured bitter taste intensity for these four compounds by asking participants to rate two 1 ml samples of each solution on a paper-labeled magnitude scale marked with ‘no sensation’, ‘barely detectable’, ‘weak’, ‘moderate’, ‘strong’, ‘very strong’ and ‘strongest imaginable’, placed semi-logarithmically at 0, 2, 7, 20, 40, 61 and 114 mm, respectively (38). For the discovery sample, the procedures have been described elsewhere (4). They were asked to mark the location on the scale with a pen that best reflected their sensory experience, including regions between labels. The distance from ‘no sensation’ to their mark was measured in millimeters and used as the dependent variable in statistical analyses. The bitter solutions (0.05 m caffeine, 2.0 × 10−4m SOA, 1.81 × 10−4m quinine HCl, 6.0 × 10−4m PROP) were tested in the following order: SOA, caffeine, quinine, PROP; PROP, quinine, caffeine, SOA. After participants completed the tests with bitter solutions dissolved in water, they used the same scale to rate the taste intensity of a paper strip containing ∼1.2 mg of PROP that would dissolve in saliva upon being placed into the mouth. For the replication sample, participants rated their perceived intensity of quinine HCl (7.5 × 10−5m) on a 7.5 cm visual analogue scale, anchored on the left with ‘not at all bitter’ and on the right with ‘extremely bitter’.
Sample preparation and genotyping
DNA was derived from blood (discovery sample) or from cheek swabs (replication sample). For the discovery sample, genotyping was performed with the Illumina 610-Quad BeadChip, with 529 721 SNPs passing quality control, as outlined previously (39). For the replication sample, the genotyping procedure has been described previously (40).
Statistical analysis
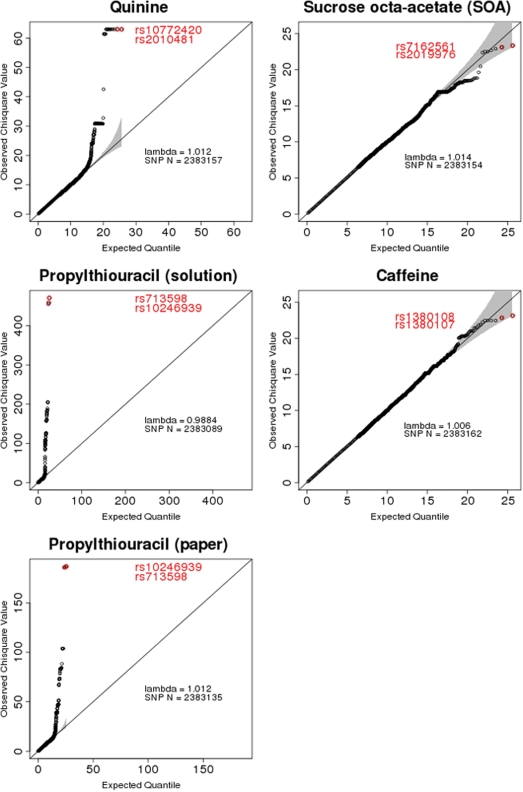
For the discovery sample, to gain the maximum amount of potential information for the association study, genomic coverage was extended to 2.3 m SNPs by imputation using the phased data from the HapMap samples of Caucasian European ancestry (Build 36, Release 22) and MACH 1.0 Markov chain-based haplotyper (41). Quality control filters were applied to the assayed genotypes to restrict the imputation to samples and SNPs with high data quality [i.e. imputation score <0.3 (indicating low imputation confidence; ∼3%), a minor allele frequency <0.01 or a Hardy–Weinberg equilibrium score of P < 10−6 (∼5%)]. Individual SNPs were tested for association with the family-based SCORE test implemented in the software Merlin (42), which accounts for the relatedness of individuals, including MZ twins, after excluding phenotypic outliers, adjusting for age and sex and normalizing each trait (4). The genomic inflation factor (λ) ranged between 0.9982 and 1.0014 (Fig. 4), indicating that potential technical or population stratification artifacts had a negligible impact on the results. For the replication sample, genotype, age, sex, and age by sex interaction were used as fixed factors in a general linear model with the rating of quinine perception as the dependent variable (STATISTICA v 8.0, StatSoft, St Louis, MO, USA). For this sample, only one MZ twin per pair was chosen randomly for inclusion in the analysis.
Figure 4.
The Q–Q plots for each of the five substances analyzed. The 95% confidence interval is shown in gray. Values lifting above the quinine plot were from a single location on chromosome 12 within and near the TAS2R19 gene; most of these for PROP (in solution and PROP papers) centered on chromosome 7 within and near the TAS2R38 gene. The excess of SNPs with small P-values is low, and all λ values are near 1.0.
In addition, from the GWAS, we extracted approximately 9000 SNPs, each 1 Mbp apart, and conducted family-based linkage analysis for (i) the intensity of quinine in solution, (ii) the intensity of PROP in a solution and (iii) the intensity of PROP in saturated filter paper (Supplementary Material, Fig. S1). No linkage was observed for quinine at the region of chromosome 12 [logarithm of odds, (LOD) = 0.73], but this was to be expected given the lack of statistical power (i.e. with 764 quasi-independent sib pairs and QTL additive variance of 23%, we had 20% power at P = 0.001). For PROP, we observed a strong linkage signal near the region of the TAS2R38 gene (LOD = 5.51) which diminishes for intensity ratings of PROP paper (LOD = 1.17). Whereas the QTL additive variance was 51% for the intensity rating of PROP in solution, it accounted for only 24% of the variance in the intensity rating of PROP paper.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Health and Medical Research Council (241944, 219178, 389875 to N.G.M.), the Australian Research Council (DP0212016, DP0664638 to N.G.M. and M.J.W.) and the National Institutes of Health (DC004698 to D.R.R., DC02995 to P.A.S.B.). S.E.M. is supported by the National Health and Medical Research Council Fellowship Scheme. Funding to pay the Open Access Charge was provided by institutional funds from the Monell Chemical Senses Center.
Supplementary Material
ACKNOWLEDGEMENTS
From the Monell Chemical Senses Center, we thank Kirsten J. Mascioli, Anilet Tharp and Christopher Tharp for making the taste tests, Alfred Goossens for his expertise in chemistry and Laura Alarcon, Ryan McDermott and Brian R Gantick for their assistance with data collection. From the Queensland Institute of Medical Research, we thank Ann Eldridge and Marlene Grace for phenotype collection, Alison Mackenzie, Romana Leisser and Kim Eldridge for project co-ordination and data entry, Jonathan Hansen for the cleaning and compiling of phenotypes, David Smyth and Daniel Park for computer support, Lisa Bowdler and Sara Smith for DNA processing and preparation, Scott Gordon for the cleaning and compiling of genotyping data and Dale Nyholt for his expertise in genomics and management of the genotype data. We thank Alexander A. Bachmanov, Gary K. Beauchamp, Michael G. Tordoff and Antti Knaapila for their comments on the manuscript before publication. Sandy Miller and the organizers of Twinsdays in Twinsburg OH assisted in data collection. Most of all, we thank the twins and their families for their participation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Falconer D.S. Sensory thresholds for solutions of phenyl-thio-carbamide. Ann. Eugen. 1947;13:211–222. [PubMed] [Google Scholar]
- 2.Fischer R., Griffin F. Quinine dimorphism: a cardinal determinant of taste sensitivity. Nature. 1963;200:343–347. doi: 10.1038/200343a0. doi:10.1038/200343a0. [DOI] [PubMed] [Google Scholar]
- 3.Smith S.E., Davies P.D. Quinine taste thresholds: a family study and a twin study. Ann. Hum. Genet. 1973;37:227–232. doi: 10.1111/j.1469-1809.1973.tb01830.x. doi:10.1111/j.1469-1809.1973.tb01830.x. [DOI] [PubMed] [Google Scholar]
- 4.Hansen J.L., Reed D.R., Wright M.J., Martin N.G., Breslin P.A. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCl, and caffeine. Chem. Senses. 2006;31:403–413. doi: 10.1093/chemse/bjj044. doi:10.1093/chemse/bjj044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim U.K., Jorgenson E., Coon H., Leppert M., Risch N., Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. doi:10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 6.Bufe B., Breslin P.A., Kuhn C., Reed D.R., Tharp C.D., Slack J.P., Kim U.K., Drayna D., Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. doi:10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 8.Reed D.R., Nanthakumar E., North M., Bell C., Bartoshuk L.M., Price R.A. Localization of a gene for bitter-taste perception to human chromosome 5p15. Am. J. Hum. Genet. 1999;64:1478–1480. doi: 10.1086/302367. doi:10.1086/302367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drayna D., Coon H., Kim U.K., Elsner T., Cromer K., Otterud B., Baird L., Peiffer A.P., Leppert M. Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum. Genet. 2003;112:567–572. doi: 10.1007/s00439-003-0911-y. [DOI] [PubMed] [Google Scholar]
- 10.Prodi D.A., Drayna D., Forabosco P., Palmas M.A., Maestrale G.B., Piras D., Pirastu M., Angius A. Bitter taste study in a Sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chem. Senses. 2004;29:697–702. doi: 10.1093/chemse/bjh074. doi:10.1093/chemse/bjh074. [DOI] [PubMed] [Google Scholar]
- 11.Wright M., Martin N. Brisbane Adolescent Twin Study: outline of study methods and research projects. Aust. J. Psychol. 2004;56:65–78. doi:10.1080/00049530410001734865. [Google Scholar]
- 12.Anonymous. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. doi:10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 13.Delwiche J.F., Buletic Z., Breslin P.A. Covariation in individuals' sensitivities to bitter compounds: evidence supporting multiple receptor/transduction mechanisms. Percept. Psychophys. 2001;63:761–776. doi: 10.3758/bf03194436. [DOI] [PubMed] [Google Scholar]
- 14.Yokomukai Y., Cowart B.J., Beauchamp G.K. Individual differences in sensitivity to bitter-tasting substances. Chem. Senses. 1993;18:669–681. doi:10.1093/chemse/18.6.669. [Google Scholar]
- 15.Spielman A.I. Interaction of saliva and taste. J. Dent. Res. 1990;69:838–843. doi: 10.1177/00220345900690030101. [DOI] [PubMed] [Google Scholar]
- 16.Adler E., Hoon M.A., Mueller K.L., Chandrashekar J., Ryba N.J.P., Zuker C.S. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. doi:10.1016/S0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 17.Chandrashekar J., Mueller K.L., Hoon M.A., Adler E., Feng L., Guo W., Zuker C.S., Ryba N. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. doi:10.1016/S0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 18.Kim U., Wooding S., Ricci D., Jorde L.B., Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum. Mutat. 2005;26:199–204. doi: 10.1002/humu.20203. doi:10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- 19.Azen E.A., Lush I.E., Taylor B.A. Close linkage of mouse genes for salivary proline-rich proteins (PRPs) and taste. Trends Genet. 1986;2:199–200. doi:10.1016/0168-9525(86)90228-3. [Google Scholar]
- 20.Greger R., Bleich M., Schlatter E. Ion channels in the thick ascending limb of Henle's loop. Renal Physiol. Biochem. 1990;13:37–50. doi: 10.1159/000173346. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn C., Bufe B., Batram C., Meyerhof W. Oligomerization of TAS2R bitter taste receptors. Chem. Senses. 2010;35:395–406. doi: 10.1093/chemse/bjq027. [DOI] [PubMed] [Google Scholar]
- 22.Lush I.E. The genetics of tasting in mice. III. Quinine. Genet. Res. 1984;44:151–160. doi: 10.1017/s0016672300026355. doi:10.1017/S0016672300026355. [DOI] [PubMed] [Google Scholar]
- 23.Boughter J.D., Harder D.B., Capeless C.G., Whitney G. Polygenic determination of quinine aversion among mice. Chem. Senses. 1992;17:427–434. doi:10.1093/chemse/17.4.427. [Google Scholar]
- 24.Blizard D.A., Kotlus B., Frank M.E. Quantitative trait loci associated with short-term intake of sucrose, saccharin and quinine solutions in laboratory mice. Chem. Senses. 1999;24:373–385. doi: 10.1093/chemse/24.4.373. doi:10.1093/chemse/24.4.373. [DOI] [PubMed] [Google Scholar]
- 25.Harder D.B., Whitney G. A common polygenic basis for quinine and PROP avoidance in mice. Chem. Senses. 1998;23:327–332. doi: 10.1093/chemse/23.3.327. doi:10.1093/chemse/23.3.327. [DOI] [PubMed] [Google Scholar]
- 26.Shi P., Zhang J., Yang H., Zhang Y.P. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol. Biol. Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. doi:10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- 27.Soranzo N., Bufe B., Sabeti P.C., Wilson J.F., Weale M.E., Marguerie R., Meyerhof W., Goldstein D.B. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr. Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. doi:10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 28.Wooding S., Kim U.K., Bamshad M.J., Larsen J., Jorde L.B., Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am. J. Hum. Genet. 2004;74:637–646. doi: 10.1086/383092. doi:10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lalueza-Fox C., Gigli E., de la Rasilla M., Fortea J., Rosas A. Bitter taste perception in Neanderthals through the analysis of the TAS2R38 gene. Biol. Lett. 2009;23:809–811. doi: 10.1098/rsbl.2009.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene L.S. Physical growth and development, neurological maturation, and behavioral functioning in two Ecuadorian Andean communities in which goiter is endemic. II. PTC taste sensitivity and neurological maturation. Am. J. Phys. Anthropol. 1974;41:139–151. doi: 10.1002/ajpa.1330410118. doi:10.1002/ajpa.1330410118. [DOI] [PubMed] [Google Scholar]
- 31.Kaji I., Karaki S.I., Fukami Y., Terasaki M., Kuwahara A. Secretory effects of a luminal bitter tastant and expressions of bitter taste receptors, T2Rs, in the human and rat large intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G971–G981. doi: 10.1152/ajpgi.90514.2008. [DOI] [PubMed] [Google Scholar]
- 32.Chen M.C., Wu S.V., Reeve J.R., Jr., Rozengurt E. Bitter stimuli induce Ca2+ signaling and CCK release in enteroendocrine STC-1 cells: role of L-type voltage-sensitive Ca2+ channels. Am. J. Physiol. Cell Physiol. 2006;291:C726–C739. doi: 10.1152/ajpcell.00003.2006. doi:10.1152/ajpcell.00003.2006. [DOI] [PubMed] [Google Scholar]
- 33.Rozengurt E. Taste receptors in the gastrointestinal tract. I. Bitter taste receptors and alpha-gustducin in the mammalian gut. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;291:G171–G177. doi: 10.1152/ajpgi.00073.2006. doi:10.1152/ajpgi.00073.2006. [DOI] [PubMed] [Google Scholar]
- 34.Jeon T.I., Zhu B., Larson J.L., Osborne T.F. SREBP-2 regulates gut peptide secretion through intestinal bitter taste receptor signaling in mice. J. Clin. Invest. 2008;118:3693–3700. doi: 10.1172/JCI36461. doi:10.1172/JCI36461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah A.S., Ben-Shahar Y., Moninger T.O., Kline J.N., Welsh M.J. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. doi:10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tizzano M., Gulbransen B.D., Vandenbeuch A., Clapp T.R., Herman J.P., Sibhatu H.M., Churchill M.E., Silver W.L., Kinnamon S.C., Finger T.E. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc. Natl Acad. Sci. USA. 107:3210–3215. doi: 10.1073/pnas.0911934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dotson C.D., Zhang L., Xu H., Shin Y.K., Vigues S., Ott S.H., Elson A.E., Choi H.J., Shaw H., Egan J.M., et al. Bitter taste receptors influence glucose homeostasis. PLoS ONE. 2008;3:e3974. doi: 10.1371/journal.pone.0003974. doi:10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green B., Shaffer G., Gilmore M. Derivation and evaluation of a semantic scale of oral sensation magnitude with apparent ratio qualities. Chem. Senses. 1993;18:683–702. doi:10.1093/chemse/18.6.683. [Google Scholar]
- 39.Medland S.E., Nyholt D.R., Painter J.N., McEvoy B.P., McRae A.F., Zhu G., Gordon S.D., Ferreira M.A., Wright M.J., Henders A.K., et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. doi:10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mennella J.A., Pepino M.Y., Reed D.R. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics. 2005;115:e216–e222. doi: 10.1542/peds.2004-1582. doi:10.1542/peds.2004-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Abecasis G. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 42.Chen W.M., Abecasis G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. doi:10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.