Abstract
The SIRTUIN1 (SIRT1) deacetylase responds to changes in nutrient availability and regulates mammalian physiology and metabolism. Human and mouse SIRT1 are transcriptionally repressed by p53 via p53 response elements in their proximal promoters. Here, we identify a novel p53-binding sequence in the distal human SIRT1 promoter that is required for nutrient-sensitive SIRT1 transcription. In addition, we show that a common single-nucleotide (C/T) variation in this sequence affects nutrient deprivation-induced SIRT1 transcription, and calorie restriction-induced SIRT1 expression. The p53-binding sequence lies in a region of the SIRT1 promoter that also binds the transcriptional repressor Hypermethylated-In-Cancer-1 (HIC1). Nutrient deprivation increases occupancy by p53, while decreasing occupancy by HIC1, of this region of the promoter. HIC1 and p53 compete with each other for promoter occupancy. In comparison with the T variation, the C variation disrupts the mirror image symmetry of the p53-binding sequence, resulting in decreased binding to p53, decreased nutrient sensitivity of the promoter and impaired calorie restriction-stimulated tissue expression of SIRT1 and SIRT1 target genes AMPKα2 and PGC-1β. Thus, a common SNP in a novel p53-binding sequence in the human SIRT1 promoter affects nutrient-sensitive SIRT1 expression, and could have a significant impact on calorie restriction-induced, SIRT1-mediated, changes in human metabolism and physiology.
INTRODUCTION
A reduction in available nutrients extends lifespan of yeast through mechanisms mediated by the class III nicotinamide adenine dinucleotide-dependent histone deacetylase (HDAC) Sir2 (silencing information regulator 2) (1). SIRT1 is the closest mammalian ortholog of Sir2 and targets histones as well as many non-histone proteins. SIRT1 mediates many of the changes in mammalian metabolism and physiology that occur during times of limited nutrient availability, including increased physical activity (2), hepatic gluconeogenesis (3) and lipolysis in white adipose tissue (4). In addition to its role in nutrient-deprived states, SIRT1 also regulates mammalian metabolic pathways during times of ample nutrient availability. Basal SIRT1 expression controls muscle fatty acid oxidation (5), hepatic lipid metabolism (6), serum glucose levels (7), pancreatic insulin secretion with glucose challenge (8) and insulin sensitivity in peripheral tissue (9).
SIRT1 expression is upregulated in peripheral tissues in fasting animals (3,7,10,11), calorie-restricted humans (12) and in cells deprived of nutrients (11). Exercise training also increases SIRT1 activity in peripheral tissues (13). SIRT1 upregulation during the fasted state or with nutrient deprivation may occur at both the transcriptional and post-transcriptional levels (14). The transcription factor p53 plays an important role in regulating SIRT1 transcription (11). The mouse and human SIRT1 promoters have p53 consensus elements near their transcription start sites that mediate p53-induced repression of SIRT1 expression during nutrient abundance (11,15). Nutrient deprivation results in binding of the Forkhead transcription factor Foxo3a to p53 and relieves p53-mediated repression of SIRT1 transcription via these elements (11). A region further upstream in the human SIRT1 promoter also functions as a repressor element by binding to the repressor Hypermethylated-In-Cancer-1 (HIC1), leading to suppression of SIRT1 expression during times of nutrient abundance (16). Mimicking calorie restriction by inhibiting glycolysis relieves HIC1-mediated repression of SIRT1 transcription, stimulating SIRT1 expression. Thus, both HIC1 and p53 serve as transcriptional repressors of SIRT1 during nutrient abundance, and nutrient deprivation relieves this repression.
Some of the most significantly upregulated genes in calorie restricted non-aged mice are transcriptional targets of p53, including cyclin-dependent kinase inhibitor 1A (p21/WAF) and DNA-damage-inducible transcript 4 (Ddit4) (17), suggesting that p53 may transcriptionally upregulate a sub-set of its target genes during nutrient-deprived states. Although p53 serves in one capacity as a repressor of SIRT1 transcription during nutrient-replete conditions, the requirement for p53 in nutrient deprivation-induced SIRT1 expression does not rule out the possibility that p53 may also function in another capacity as an activator of SIRT1 transcription during nutrient-deprived conditions. This prompted us to seek novel nutrient-sensitive regulatory elements in the SIRT1 gene that could serve as binding sites for p53. Herein, we report the existence of a sequence in the human SIRT1 promoter that binds to p53 during nutrient stress, and promotes the transcription of SIRT1. Importantly, we demonstrate the existence of a common single-nucleotide variation in this sequence that affects p53 binding and nutrient stress-induced SIRT1 transcription.
RESULTS
An element in the human SIRT1 promoter is a binding site for p53
Our initial goal was to identify novel elements in the human SIRT1 promoter that regulate SIRT1 transcription in a nutrient-sensitive fashion. Recognizing the importance of p53 in regulating SIRT1 expression induced by nutrient withdrawal, we limited our search to putative p53-binding elements in the human SIRT1 gene. Examination of the human SIRT1 gene reveals a 20-nucleotide element 1099 bp upstream of the transcription start site that bears significant homology to the consensus p53-binding sequence (Fig. 1A). This element contains two p53 consensus half-sites, with the highly conserved palindromic C(A/T)∣(T/A)G nucleotides important for p53 binding (18,19), preserved in each half-site. Therefore, we first asked whether this 20 bp element binds to p53 in vitro. Electrophoretic mobility shift assays demonstrated that a synthetic oligonucleotide with a sequence corresponding to this 20 bp element in the human SIRT1 promoter (referred to as SIRT1 promoter oligo-T) binds to recombinant p53 (Fig. 1B and C). In addition, p53 in nuclear extracts binds to SIRT1 promoter oligo-T (Fig. 1D and E). The multiple bands with the nuclear extracts likely reflect the presence of nuclear proteins that bind to p53 (as evident by super-shifting or blocking of these bands in the presence of p53 antibody), which are absent in assays with recombinant p53. To demonstrate the importance of the conserved C(A/T)∣(T/A)G nucleotides in this sequence to p53 binding, we performed electrophoretic mobility shift assays with an oligonucleotide which is deleted in nucleotides CTAG in the first half-site of this p53-binding sequence (referred to as SIRT1 promoter oligo-CTAG-del). Compared with wild-type oligonucleotide (SIRT1 promoter oligo-T), there was decreased binding of recombinant p53 to the SIRT1 promoter oligo-CTAG-del (Fig. 1F). Moreover, in competition electrophoretic mobility shift assays, SIRT1 promoter oligo-CTAG-del was a weaker competitor than SIRT1 promoter oligo-T (Fig. 1G). Thus, this 20 bp element in the human SIRT1 promoter binds to p53 in vitro, and deletion of key nucleotides that are conserved in p53-binding sequences weakens this interaction.
Figure 1.
The C/T SNP in the human SIRT1 promoter is in a p53-binding sequence and affects p53 binding in vitro. (A) Location of the putative p53-binding sequence and the (C/T) variation in this sequence. The two HIC1-binding sites in relation to the p53 site are shown. A half-site from the consensus p53-binding sequence is displayed. Pu, purine; Py, pyrimidine. (B) Electrophoretic mobility shift assay (EMSA) showing binding of recombinant p53 to an oligonucleotide corresponding to the putative p53-binding site in the human SIRT1 promoter (SIRT1 promoter oligo-T), and to an oligonucleotide that exactly matches the consensus p53-binding site (P53 consensus oligo). (C) EMSA showing supershift with p53 antibody of recombinant p53 bound to SIRT1 promoter oligo-T. (D) EMSA showing supershift with p53 antibody of p53 expressed in HEK 293 cells bound to SIRT1 promoter oligo-T. (E) EMSA showing binding of p53 expressed in cells to SIRT1 promoter oligo-T (corresponding to the T variation in the p53-binding site), p53 consensus oligo and an oligonucleotide corresponding to the p53-binding site with the C variation (SIRT1 promoter oligo-C). Lanes are from the same gel. (F) EMSA showing binding of recombinant p53 to SIRT1 promoter oligo-T, SIRT1 promoter oligo-C and an oligonucleotide corresponding to the p53-binding site with the nucleotides CTAG deleted in the first half-site (SIRT1 promoter oligo CTAG-del). (G) EMSA showing competition for binding to recombinant p53 between SIRT1 promoter oligo-T, SIRT1 promoter oligo-C, SIRT1 promoter oligo-CTAG-del and p53 consensus oligo. Labeled SIRT1 promoter oligo-T was used in all competitions. All data shown are representative of three independent experiments.
A C/T SNP in the p53-binding site affects p53 binding in vitro
The International HapMap Project has identified the existence of a single-nucleotide C/T variation (rs3758391) in this newly identified p53-binding sequence. In human genome databases, this single-nucleotide polymorphism is reported to be common with an overall average heterozygosity frequency of 50% in the HapMap Project. In white Americans of Northern and Western European ancestry and African-Americans, the T allele is the minor allele with a frequency of 25–35%, whereas in a Japanese and Chinese cohort, the T allele is the major allele with a frequency of 80–90% (http://www.hapmap.org/). Strikingly, the C allele of this SNP disrupts the mirror-image symmetry in the conserved C(A/T)∣(T/A)G nucleotides of the second half-site of the p53 consensus binding sequence, while the T allele does not (Fig. 1A), raising the possibility that this variation may affect binding of the sequence to p53. To determine the significance of this single-nucleotide variation to p53 binding, we first compared binding of oligonucleotides with the C and T alleles to recombinant p53. Electrophoretic mobility shift assays showed that p53 bound with lesser affinity to the oligonucleotide with the C allele (referred to as SIRT1 promoter oligo-C) than to the oligonucleotide with the T allele (SIRT1 promoter oligo-T) (Fig. 1F). Similarly, p53 expressed in cells bound with lower affinity to SIRT1 promoter oligo-C than to SIRT1 promoter oligo-T (Fig. 1E). In electrophoretic mobility shift competition assays as well, the SIRT1 promoter oligo-C was a weaker competitor than SIRT1 promoter oligo-T for binding to recombinant p53 (Fig. 1G). Therefore, change of a single nucleotide from T to C in a conserved region of the p53-binding element, corresponding to the C/T SNP rs3758391 in humans, results in decreased in vitro binding to p53.
The C/T SNP affects SIRT1 promoter occupancy by p53
We then determined whether p53 binds to this sequence in the SIRT1 promoter in the genomic context. Occupancy by p53 of this region of the human SIRT1 promoter was examined in p53-expressing human embryonic kidney (HEK 293) by chromatin immunoprecipitation (ChIP) assays. Under resting nutrient-replete conditions, endogenous p53 occupied the genomic region in the human SIRT1 promoter encompassing this p53-binding element (Fig. 2A). Next, we asked whether the C/T variation affects occupancy by p53 of this region in the SIRT1 promoter. Several different human cell lines were genotyped to identify the two that differed with respect to the C/T variation. HEK 293 cells were homozygous (CC), whereas human cervical carcinoma (HeLa) cells were heterozygous (CT) for this variation (Supplementary Material, Fig. S1). Because HEK 293 cells have greater levels of endogenous p53 protein than HeLa cells, we expressed ectopic p53 in HeLa cells to achieve similar p53 expression in both cell lines. Comparison of ChIP assays in HEK 293 and HeLa cells showed that occupancy by p53 of this region of the SIRT1 promoter was greater in HeLa cells, despite lower expression of p53 in this cell line (Fig. 2B). Thus, there is greater occupancy by p53 of this region of the SIRT1 promoter in a cell line that has a T allele, compared with one that does not.
Figure 2.
The C/T SNP affects occupancy by p53 of the SIRT1 promoter. (A) ChIP assay for endogenous p53 in HEK 293 cells showing amplification of a 169 bp genomic region in the SIRT1 promoter encompassing the p53-binding site (arrow). (B) ChIP assay showing greater occupancy by p53 of the SIRT1 promoter in HeLa cells compared with HEK 293 cells. HeLa cells were transfected with p53 to achieve expression of the protein. N-IgG, non-immune IgG; p53, p53 antibody; input, non-immunoprecipitated HEK 293 cell chromatin. All data shown are representative of three independent experiments.
SIRT1 promoter activation by nutrient stress is mediated by the p53-binding element and is modulated by the C/T SNP in this element
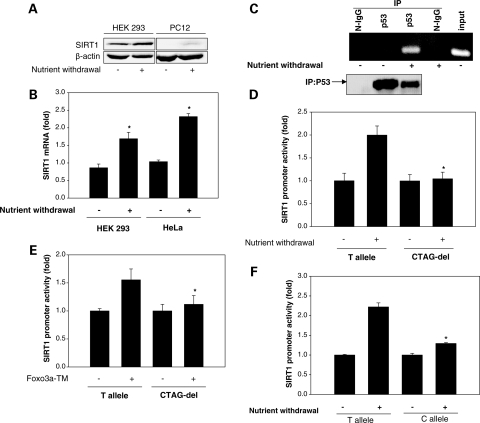
Next, we examined whether this p53-binding element plays a part in regulation of SIRT1 transcription induced by nutrient stress. Nutrient stress in cultured cells was induced by nutrient withdrawal to simulate calorie restriction. Nutrient withdrawal led to an increase in the SIRT1 protein and mRNA (Fig. 3A and B). This increase in SIRT1 transcription was accompanied by increased binding of p53 to the SIRT1 promoter (Fig. 3C). In parallel with the increase in SIRT1 expression, nutrient withdrawal stimulated activity of the human SIRT1 promoter, as assessed using a reporter construct of the human SIRT1 promoter that includes the p53-binding site (Fig. 3D). To determine whether this increase in SIRT1 promoter activity is mediated by the p53-binding element, we compared activation of the wild-type SIRT1 promoter (bearing the T allele) with that of an SIRT1 promoter deleted in nucleotides CTAG (CTAG-del), having shown that deletion of these nucleotides in the first p53 consensus half-site decreases in vitro binding to p53. In contrast to the wild-type promoter, nutrient withdrawal did not stimulate activity of the CTAG-del SIRT1 promoter (Fig. 3D). Thus, disruption of conserved features of the p53-binding site abrogates induction of SIRT1 promoter activity by nutrient withdrawal, underscoring the importance of this element to nutrient stress-stimulated SIRT1 transcription.
Figure 3.
The p53-binding sequence mediates, and the C/T variation in it affects, nutrient stress-stimulated SIRT1 promoter activity. (A) Immunoblot showing increase in SIRT1 protein with nutrient withdrawal. (B) Quantitative real-time PCR showing increase in SIRT1 mRNA with nutrient withdrawal. *P < 0.05 compared with no nutrient withdrawal. (C) ChIP showing increase in occupancy of the SIRT1 promoter by p53 with nutrient withdrawal. A 169 bp genomic region in the SIRT1 promoter encompassing the p53-binding site was amplified. Non-immunoprecipitated HEK 293 cell chromatin was used as input. (D) Promoter–reporter assay in HEK 293 cells showing nutrient withdrawal-stimulated change in activity of an SIRT1 promoter with the T allele in the p53-binding sequence (T allele), and an SIRT1 promoter with deletion of CTAG nucleotides in the first half-site of the p53-binding sequence (CTAG-del). *P < 0.05 compared with T allele. (E) Promoter–reporter assay in HEK 293 cells showing active Foxo3a (TM)-stimulated change in activity of the SIRT1 T allele promoter and the SIRT1 CTAG-del promoter. *P < 0.05 compared with T allele. (F) Promoter–reporter assay in HEK 293 cells showing nutrient withdrawal-stimulated change in activity of SIRT1 promoters with the C and T alleles in the p53-binding sequence. *P < 0.05 compared with T allele. All data shown are representative of three independent experiments.
Hypothesizing that this p53-binding site may confer responsiveness to p53 manipulation, we examined the effect of p53 up- and downregulation on SIRT1 promoter activity and expression. In the absence of nutrient stress, we did not find a robust effect of p53 manipulation on SIRT1 expression or promoter activity. Overexpression of p53 in HEK 293 cells, without nutrient stress, did not change SIRT1 expression (Supplementary Material, Fig. S2) or promoter activity (Supplementary Material, Fig. S3). Similarly, in nutrient-replete conditions, siRNA-mediated knockdown of p53 did not appreciably affect SIRT1 expression in HEK 293 cells (Supplementary Material, Fig. S4). Thus, during abundant availability of nutrients, SIRT1 transcription is not subject to regulation by p53 levels alone.
Nutrient stress-induced murine SIRT1 upregulation has been attributed, in part, to a forkhead-dependent mechanism, whereby, activation and binding of the Forkhead transcription factor Foxo3a to p53 with nutrient stress relieve p53-mediated repression of SIRT1 transcription (11). We therefore asked whether the p53-binding element plays a part in SIRT1 transcription induced by activation of forkhead signaling. Activity of the wild-type human SIRT1 promoter (bearing the T allele) was stimulated by expression of phosphorylation-deficient constitutively active Foxo3a-TM (Fig. 3E). However, activity of the CTAG-del SIRT1 promoter was not increased by expression of Foxo3a-TM (Fig. 3E). Therefore, mimicking nutrient stress by activating Forkhead signaling increases human SIRT1 transcription, and this increase is also mediated by the p53-binding element.
We then asked whether the single-nucleotide C/T variation in the p53-binding element affects SIRT1 promoter activation by nutrient stress. Consistent with decreased binding of p53 to an oligonucleotide and promoter with the C allele, nutrient withdrawal-induced activation was significantly lower with an SIRT1 promoter bearing the C allele when compared with a promoter with the T allele (Fig. 3F). Therefore, the single-nucleotide change from C to T in the p53-binding element results in a significant reduction in SIRT1 promoter activation by nutrient stress.
p53 and HIC1 reciprocally bind to the human SIRT1 promoter and compete with each other for promoter occupancy
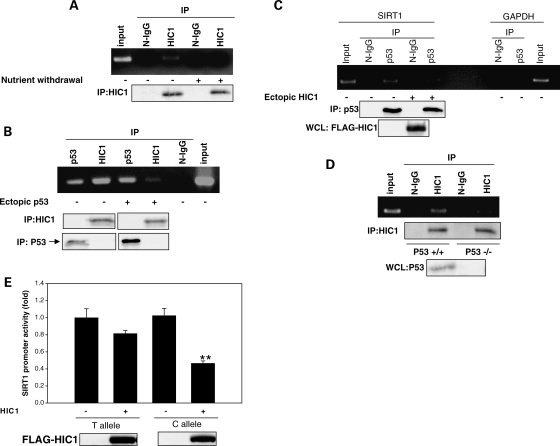
SIRT1 expression is downregulated by the transcriptional repressor HIC1 (20), and inhibition of glycolysis stimulates SIRT1 transcription, in part, by relieving HIC1-mediated repression of SIRT1 (16). There are two HIC1-binding sites in the human SIRT1 promoter, and these sites are in the same region of the promoter as the p53-binding element, lying −13 and +43 nucleotides from it (Fig. 1A). We were intrigued by the possibility that the proximity of the HIC1 sites and the p53-binding element on the promoter may have functional relevance to the regulation of SIRT1 transcription. To explore this further, we first examined change in binding of HIC1 to the SIRT1 promoter with nutrient stress. ChIP assays showed that nutrient withdrawal decreased occupancy by HIC1 of this region of the SIRT1 promoter (Fig. 4A). Next, we examined whether p53 and HIC1 compete with each other for occupancy of this region of the promoter. Overexpression of p53 decreased occupancy of the promoter by HIC1 (Fig. 4B). Conversely, overexpression of HIC1 decreased occupancy of the SIRT1 promoter by p53 (Fig. 4C). Moreover, occupancy of the promoter by HIC1 was greater in the p53-deficient human colon carcinoma cell line HCT 116 (p53−/−) compared with its isogenic control HCT 116 (p53+/+) cell line (Fig. 4D). These findings show that nutrient stress decreases binding of HIC1 to the same region of the promoter occupied by p53. Moreover, these data show that p53 and HIC1 antagonize each another for promoter occupancy.
Figure 4.
P53 and HIC1 compete for occupancy of the SIRT1 promoter. (A) ChIP in HEK 293 cells showing a decrease in occupancy of the SIRT1 promoter by HIC1 with nutrient withdrawal. (B). ChIP in HEK 293 cells showing a decrease in occupancy of the SIRT1 promoter by HIC1 with p53 overexpression. (C) ChIP in HEK 293 cells showing a decrease in occupancy of the SIRT1 promoter by p53 with HIC1 overexpression. (D) ChIP assay for endogenous HIC1 in p53−/− and p53+/+ HCT 116 cells. Non-immunoprecipitated HCT 116 P53+/+ chromatin was used as input. In all ChIP assays, a 179 bp genomic region in the SIRT1 promoter encompassing the two HIC1- and p53-binding sites was amplified. N-IgG, non-immune IgG; p53, p53 antibody; HIC1, HIC1 antibody. (E) Promoter–reporter assay in HEK 293 cells showing change in activity of the SIRT1 promoter with the T allele and the SIRT1 promoter with the C allele with expression of HIC1. **P<0.01 compared with T allele. All data shown are representative of three independent experiments.
Knowing that p53 and HIC1 compete with each other for occupancy of this region of the promoter, and the C/T SNP affects binding of p53 to this region, we reasoned that this genetic variation may also impact HIC1-induced repression of SIRT1 transcription. We therefore determined the effect of the rs3758391 SNP on HIC1-induced change in SIRT1 promoter activity. Overexpression of HIC1 repressed activity of the SIRT1 promoter bearing the C allele to a significantly greater degree than an SIRT1 promoter with the T allele (Fig. 4E). Thus, HIC1-stimulated repression of SIRT1 transcription is also affected by the C/T variation. This finding suggests that decreased binding of p53 to the promoter with the C allele renders it more susceptible to HIC1-induced repression.
The C/T SNP affects tissue SIRT1 upregulation in calorie-restricted humans
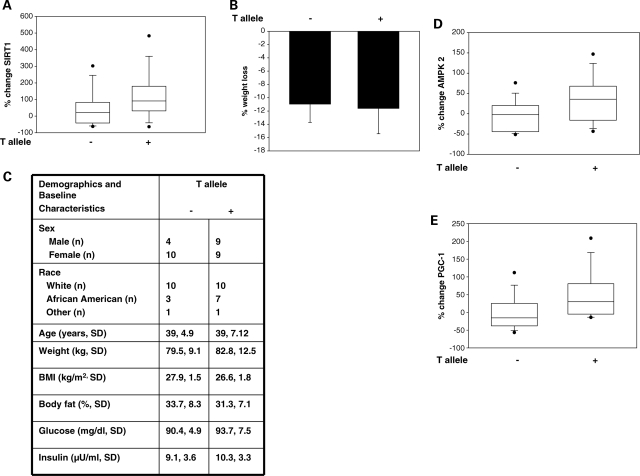
Long-term calorie restriction in humans decreases energy expenditure, improves whole-body metabolic efficiency and increases SIRT1 expression and mitochondrial biogenesis in skeletal muscle (12,21). Having shown the impact of the C/T variation on SIRT1 transcription stimulated by nutrient withdrawal in vitro, we next explored any association of this SNP with transcription of tissue SIRT1 triggered by calorie restriction in humans. Skeletal muscle SIRT1 expression was measured in a cohort of overweight individuals at baseline and after 3 and 6 months of a calorie-restricted diet (CR), or on a combined regimen of calorie restriction plus exercise (CREX). All subjects were genotyped for the rs3758391 SNP. Overall, both 3 and 6 months of CR or CREX led to a significant induction of skeletal muscle SIRT1 expression (Supplementary Material, Fig. S5). Genotype- and allele-specific differences in skeletal muscle SIRT1 expression at baseline and after 3 and 6 months of CR or CREX were determined. Six months of CR or CREX led to a larger increase in skeletal muscle SIRT1 in individuals who had at least one T allele (CT and TT genotypes) when compared with individuals who did not have a T allele (CC genotype) (Fig. 5A). However, the C/T SNP was not associated with baseline skeletal muscle SIRT1 expression (Supplementary Material, Fig. S6) or skeletal muscle SIRT1 induction after a period of 3 months of CR or CREX (Supplementary Material, Fig. S7). The presence of a T allele did not affect the magnitude of weight loss with CR or CREX (Fig. 5B). Furthermore, there were no differences in demographic, anthropometric and baseline clinical characteristics between those that had the T allele and those that did not (Fig. 5C). Thus, this SNP is not associated with a change in baseline SIRT1 expression but does affect upregulation of SIRT1 in response to long-term CR or CREX.
Figure 5.
The C/T SNP is associated with calorie restriction-induced tissue SIRT1, AMPKα2 and PGC-1β expression. (A) Presence of the T allele is associated with a larger increase in skeletal muscle SIRT1 mRNA after 6 months of CR or CREX; P = 0.035 (n = 32). (B) Presence of the T allele does not affect weight loss after 6 months of CR or CREX; P = NS (n = 32). Error bars are ±SD. (C) Demographics and baseline clinical characteristics of subjects on 6 months of CR or CREX with and without the T allele. Values are means ± SD. There was no statistical difference in any of the characteristics between those with and without the T allele. (D) Presence of the T allele is associated with a larger increase in skeletal muscle AMPKα2 mRNA after 6 months of CR or CREX; P = 0.013 (n = 32). (E) Presence of the T allele is associated with a larger increase in skeletal muscle PGC-1β mRNA after 6 months of CR or CREX; P = 0.016 (n = 32). Each box plot shows the distribution of the change in SIRT1 expression from 25 to 75th percentile, and the lines inside the boxes denote the medians. Whiskers denote the intervals between the 5 and 95th percentiles, with dots representing data points outside these percentiles.
AMP-regulated kinase (AMPK) and peroxisome proliferator-activated receptor gamma coactivator-1-β (PGC-1β) are two targets of SIRT1 (22,23) that regulate mitochondrial function and energy utilization. To determine whether the allele-specific difference in SIRT1 upregulation with CR or CREX translated into a difference in upregulation of SIRT1 target genes, we examined the change in skeletal muscle AMPKα2 and PGC-1β in humans on long-term CR or CREX. Similar to that observed with SIRT1, there was an allele-specific difference in upregulation of AMPKα2 and PGC-1β after 6 months of CR or CREX, with the presence of the T allele being associated with larger increases in skeletal muscle AMPKα2 (Fig. 5D) and PGC-1β (Fig. 5E). These findings suggest the rs3758391 SNP in SIRT1 also affects the expression of nutrient-sensitive target genes of SIRT1.
DISCUSSION
SIRT1 is an important mediator of metabolic adaptation in response to nutrient availability. Therefore, nutrient-sensitive mechanisms that regulate SIRT1 expression and activity would be expected to have a broad impact on biologic parameters. Understanding the regulation of human SIRT1 expression is particularly relevant given the intensive efforts to target SIRT1 to treat human diseases. In contrast to the known and rapidly growing list of targets of SIRT1, relatively little is known about how SIRT1 expression and function are controlled. Our work identifies a novel element in the human SIRT1 gene as important for nutrient-sensitive regulation of its expression. Moreover, the finding of a functional genetic variation in this element suggests a genotype-specific heterogeneity with respect to SIRT1-mediated phenotypes in response to changes in energy availability.
Nutrient withdrawal increases p53 occupancy of the SIRT1 promoter region encompassing the p53-binding element that we identified. Thus, this novel p53-binding element most likely serves as a positive regulatory element for nutrient stress-stimulated SIRT1 transcription, and therefore differs from the p53-binding sequence previously shown to repress SIRT1 transcription (11). The relative contributions of these two p53-binding sequences to SIRT1 transcription induced by nutrient stress, one through relief of repression and the other through trans-activation, remain unknown. However, it is important to note that deletion of the conserved CTAG core nucleotides in the p53-binding element described in this work abrogates SIRT1 promoter activation by nutrient stress (Fig. 3D), suggesting that this element plays a dominant role in nutrient stress-stimulated SIRT1 transcription. It is also important to recognize that prior work examining the effect of p53 on SIRT1 transcription has utilized human and mouse SIRT1 promoter constructs that are at most 200 nucleotides from the transcription start site, or synthetic promoter–reporter constructs (11,15). In contrast, the use of a promoter–reporter construct that encompasses 1266 bp 5′ of the transcription start site is more likely to recapitulate the SIRT1 promoter in the genomic context, reflecting the role of further upstream elements in regulating SIRT1 transcription. Using this promoter construct, we did not observe a significant effect of p53 overexpression or knockdown on SIRT1 promoter activity or expression in the absence of nutrient stress. These findings are not surprising if p53, by virtue of distinct cis regulatory elements in the SIRT1 promoter, functions as both a repressor and an activator of SIRT1 transcription.
The regions of the human and mouse SIRT1 promoters proximal to the transcription start sites are highly conserved. The human and mouse p53 response sequences previously identified as repressor elements are within 200 bp of the transcription start sites and share 95% identity (15). In contrast, there is little homology in the sequences of the mouse and human promoters further distal to the transcription start site. Examination of the distal mouse SIRT1 promoter reveals no putative p53- or HIC1-binding sequences. However, there is a strong conservation of the human and Pan troglodytes SIRT1 promoters, with 100% identity of the human p53-binding element we identified to a sequence in the SIRT1 gene on chromosome 10 of the P.troglodytes genome. This suggests that the precise mechanism by which p53 regulates nutrient stress-induced SIRT1 transcription may have diverged during evolution. In higher mammals, the presence of at least two p53 responsive elements, one functioning as an activator and the other as a repressor, indicates an additional layer of complexity in the regulation of SIRT1 by p53 that may reflect tighter transcriptional control in such highly evolved organisms.
The HIC1 and p53 sites in the human SIRT1 promoter are in proximity to one another (within an 83 bp region in the promoter), but do not overlap. Therefore, it is unlikely that HIC1 and p53 directly compete for occupancy of the SIRT1 promoter. HIC1 forms a co-repressor complex with the adenoviral C-terminal binding protein (CtBP) on the human SIRT1 promoter (16), and CtBP can repress gene transcription in an HDAC-dependent manner (24) by quenching the function of activators that bind to neighboring sites (25). Therefore, HIC1 likely restricts access of p53 to the SIRT1 promoter by a quenching mechanism involving chromatin condensation mediated by CtBP-recruited HDACs. p53-induced antagonism of occupancy of the SIRT1 promoter by HIC1 is also likely to be indirect and may involve competition for protein-modification mechanisms, such as SUMOylation, which occurs on both p53 and HIC1 (26,27), and which are important for the transcriptional activating and repressing functions of p53 and HIC1, respectively (27,28).
Foxo3A binds to p53, and this association is stimulated by nutrient stress (11). Whether this association directly disrupts binding of p53 to DNA and is responsible for nutrient stress-stimulated SIRT1 transcription is not clear. Our finding that active Foxo3A stimulates SIRT1 promoter activity, and this increase is dependent on the p53-binding element (Fig. 3E), indicates that inhibition of p53 binding to this element is unlikely to account for the effect of Foxo3A on SIRT1 transcription. p53 binds to HDAC (29), including SIRT1 itself (30), and deacetylation of p53 by HDAC and SIRT1 negatively impacts on p53-dependent gene activation (29,30). Thus, increasing the pool of acetylated p53 by disrupting p53–HDAC interaction could be an indirect mechanism responsible for Foxo3A-stimulated, p53-mediated, trans-activation of SIRT1.
In addition to AMPKα2 and PGC-1β, we also looked for allele-specific differences in upregulation with CR or CREX of peroxisome proliferator-activated receptor gamma coactivator-1-α (PGC-1α), a well known target of SIRT1 (3). Although skeletal muscle PGC-1α expression increased by 32.8% after 6 months of CR or CREX in subjects with the T allele compared with only a 6.7% increase in those without, statistical significance was not reached (P = 0.17). The lack of statistical difference in the change in expression of PGC-1α (and possibly other SIRT1 target genes) most likely reflects the lack of statistical power offered by the small size of this cohort. In this regard, it is important to recognize that the TT genotype was rare (12.5%) in our cohort, consistent with the T allele being the minor allele in white American and African-American subjects (http://www.hapmap.org/). This low frequency did not allow us to compare subjects homozygous for the C and T alleles in this small sample size, compelling us to group together the CT and TT genotypes. The grouping of genotypes, resulting in allelic dilution, could have masked significant differences, or diminished the magnitude of difference, in expression of SIRT1 and SIRT1 target genes between the C and T alleles.
Variations in the SIRT1 gene are not associated with exceptional human longevity (31). Nevertheless, increasing evidence points toward the association of specific SIRT1 genetic variations with clinical phenotypes. SNPs in SIRT1 have been associated with clinical parameters of human metabolism such as fat mass and visceral obesity (32), insulin sensitivity (33) and energy expenditure (34). Although the rs3758391 SNP is not associated with any particular metabolic profile, there is an association between a haplotype containing this SNP and cardiovascular mortality risk, as well as cognitive function in an aged population (35). In comparison with a reference haplotype with the C allele, a haplotype with the T allele was associated with a trend for lower cardiovascular mortality (P = 0.062). Moreover, heterozygous carriers of the T allele had lower cardiovascular mortality risk (P = 0.018), and homozygosity for the T allele was associated with better cognitive function, with memory preserved best (35). By demonstrating that the C/T variation affects p53 binding to the SIRT1 promoter, and nutrient stress-induced SIRT1 transcription, our observations provide a potential mechanism for the association of this SNP to these phenotypes. This may be particularly relevant when taken in the context of experimental work showing that SIRT1 promotes normal vascular function (36), inhibits cardiac dysfunction (37) and protects against neurodegeneration in models of Alzheimer's disease (38). Furthermore, clinical studies have demonstrated a favorable effect of long-term calorie restriction on cardiovascular parameters (39), as well as improved memory in elderly humans (40). It is important to recognize, however, that in contrast to 6 months of dietary and lifestyle changes, change in skeletal muscle SIRT1 expression after a 3-month period of CR or CREX did not show a significant allele-specific difference, despite a similar increase in SIRT1 expression in the overall group at both 3 and 6 months. Similarly, allele-specific increase in AMPKα2 and PGC-1β did not achieve statistical significance after 3 months of CR or CREX (Supplementary Material, Figs 8 and 9). These results could be explained if one hypothesizes that the C/T SNP has an effect on SIRT1 expression only in the context of robust weight loss, as the weight loss after 3 months of CR or CREX was 8.86 ± 3.79% from baseline, which was appreciably smaller than that achieved after 6 months (11.3 ± 3.3% decline from baseline). This hypothesis may have merit since further analysis of change in SIRT1 expression, when stratified by magnitude of weight loss, showed that the sub-group that achieved a weight loss of >10% from baseline (the majority of which was at 6 months of CR or CREX) showed a significant allele-specific difference in SIRT1 increase, whereas the group that had a <10% weight loss from baseline (the majority of which was at 3 months of CR or CREX) did not (Supplementary Material, Figs 10 and 11). Whatever the reason, the association of this SNP with calorie restriction-stimulated SIRT1 expression has to be interpreted with caution as it may become evident only under conditions of prolonged reduction in calories associated with robust weight loss. Further validation of this association will require studies with larger sample sizes.
We did not observe an allele-specific difference in the magnitude of weight loss with CR or CREX. This indicates that the magnitude of change in SIRT1 with CR or CREX does not confer a metabolic advantage (or disadvantage) with respect to losing weight. This is somewhat surprising given the important role of SIRT1 in regulating metabolic pathways. It is also possible that our study was not sufficiently powered to detect SIRT1-mediated metabolic and physiologic phenotypes. Nevertheless, the association of this SNP with calorie restriction-stimulated SIRT1 expression, combined with the varying prevalence of the C and T alleles among different human populations (http://www.hapmap.org/), suggests that SIRT1-mediated phenotypes in humans, triggered by prolonged reduction in caloric intake, may be more pronounced, or have a wider impact, in certain populations.
MATERIALS AND METHODS
Cell culture
PC12, HEK 293, HepG2 and HeLa cells were purchased from ATCC (Rockville, MD, USA). Cells were maintained in Dulbecco's Modified Essential Media supplemented with 10% fetal calf serum. Nutrient withdrawal consisted of growing cells in DMEM free of serum and glucose for 15 h prior to harvest. PCR-amplified genomic DNA from human cell lines were sequenced with an ABI automated sequencer.
DNA constructs, transfections and reporter assays
A 1403 bp fragment of the human SIRT1 promoter (−1266 to +137 relative to transcription start site) that encompasses the two HIC1-binding elements and the putative p53-binding site was amplified from human genomic DNA, using appropriate primers. The fragment was cloned into the pGL4.1 firefly luciferase reporter vector (Promega). Genotyping of the cloned promoter–reporter revealed the C variation in the putative p53-binding site and was used as a template to introduce the T variation by site-directed mutagenesis using the QuickChange kit (Stratagene). The CTAG deletion was constructed by restriction digestion followed by re-ligation of the promoter–reporter bearing the T variation. All mutations and deletions were verified by sequencing. The SIRT1 promoter–reporter plasmids bearing the T/C variant or the CTAG-deleted mutant were co-transfected with a constitutive renilla reporter plasmid using Lipofectamine2000 (InVitrogen) as per manufacturer's recommendations. Transfections with cDNA constructs were similarly performed using Lipofectamine2000. Firefly and renilla luciferase luminescence were measured 48 h after transfection, with or without nutrient withdrawal, using the Dual Luciferase reporter kit (Promega) as per manufacturer's recommendations. Results presented are from representative experiments performed in triplicate. Firefly luciferase activity was normalized to renilla luciferase control to correct for differences in transfection efficiency.
Electrophoretic mobility shift (gel retardation) assays
Sequences of the double-stranded DNA oligonucleotides were (nucleotides in the putative p53-binding sequence are shaded with the SNP in bold): SIRT1 promoter oligo-C: 5′-GCT CTA GAT CTA CCA CGG GTT ATA TGG G-3′; SIRT1 promoter oligo-T: 5′-GCT CTA GAT CTA CCA TGG GTT ATA TGG G-3′; SIRT1 promoter oligo-CTAG-del: 5′-CAA ACA CTG GCT ATC TAC CAT GGG TTA TAT GGG T-3′; p53 consensus oligo: 5′-GGA CATG CCC C GGG CATG TCC-3′. 32P-labeled oligonucleotides of 106 cpm were incubated with 10 µg of lysate of HEK293 cells expressing p53 or 100 ng of recombinant p53 (Active Motif) for 20 min at room temperature in binding buffer (20 mm Hepes, 25 mm KCl, 10% glycerol, 2 mm MgCl2,10 mm DTT, 0.1 µg dIdC and 0.1 µg BSA). In competition assays, 10-fold excess of unlabeled oligonucleotide was added to the reaction mixture. One microgram of antibody to p53 (p(Ab)421; CalBiochem) was added to the binding mixture for supershift experiments. The mixtures were electrophoresed on a 6% polyacrylamide gel and the gel autoradiographed.
Antibodies and immunoblotting
Immunoblotting of 50 µg of whole-cell lysates was performed using the following antibodies: p53 (1C12; Cell Signaling); HIC1 (D-17; Santa Cruz); SIRT1 (H-300; Santa Cruz); FLAG (F-3165; Sigma); chemiluminescent signal was developed using Super Signal West Femto substrate (Pierce), and blots imaged with a Gel Doc 2000 Chemi Doc system (BioRad).
Quantitative real-time PCR
Total RNA from cells was isolated by the acid guanidinium thiocyanate/phenol/chloroform method. Real-time PCR was performed using the Prism 7000 Sequence Detection System (Applied Biosystems) with the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (InVitrogen). The primer sequences for human SIRT1 were: forward 5′-TCGCAACTATACCCAGAACATAGACA-3′, reverse 5′-CTGTTGCAAAGGAACCATGACA-3′. Human GAPDH was used as an internal control. The primer sequences for human GAPDH were: forward 5′-CCACATCGCTCAGACACCAT-3′, reverse 5′-CCAGGCGCCCAATACG-3′.
ChIP assays
ChIP assays were performed for p53 and HIC1 or species-specific non-immune immunoglobulin (N-IgG) using the appropriate antibodies and a ChIP Assay Kit (Upstate), as per manufacturer's recommendations. p53 was immunoprecipitated by overnight incubation of 1 mg of cross-linked chromatin with 2 mg of p53 antibody (1801X; Santa Cruz). HIC1 was similarly immunoprecipitated using a HIC1-specific antibody (D-17; Santa Cruz). Immunoprecipitations with N-IgG of the same species were performed as controls. A 169 bp fragment of human genomic DNA harboring the p53-binding site was amplified from immunoprecipitates using specific primers (forward 5′-CCC GAG CTC GCA GTA TGG CCA GAA CCC ATA C-3′; reverse 5′-T GAC TCC ATA TCT AAT CTT AAC TAA GAC-3′). In experiments examining HIC1 binding, a 179 bp element that encompasses the two HIC1- and p53-binding sites was PCR-amplified using specific primers (forward 5′-CCC GAG CTC GCA GTA TGG CCA GAA CCC ATA C-3′; reverse 5′-ATT CTG GCA CAC TGT GAC TCC ATA TCT A-3′). A 177 bp region of the human GAPDH gene was PCR-amplified with specific primers (forward 5′-ATG ACA TCA AGA AGG TGG TG-3′; reverse 5′-CAT ACC AGG AAA TGA GCT TG-3′) as an internal non-binding control.
Clinical study
SIRT1, AMPKα2, PGC-1β and PGC-1α mRNA expression in healthy overweight participants in a clinical trial (trial registration: http://www.ClinicalTrials.gov; identifier: NCT00099151) were measured by quantitative real-time PCR in banked skeletal muscle biopsies as described previously (12). Muscle biopsies and amphometric data were obtained at baseline and after 3 and 6 months of dietary and/or lifestyle modifications. The lifestyle and dietary modifications were: calorie restriction (CR; 25% reduction in calories or an 890 kcal/day diet) or CREX (12.5% calorie restriction plus 12.5% increase in energy expenditure by structured exercise). The percent change in mRNA expression and body weights after 3 and 6 months of CR or CREX were calculated. Genotype was determined using whole-blood genomic DNA with an Applied Biosystems automated sequencer.
Statistical methods
Statistical differences between two groups were measured by a t-test. For data that failed a normality test, a Mann–Whitney rank-sum test was used for comparison of two groups.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH grants R01 HL070929, P01 HL065608 and R01 HL094959 and R21 HL098892 to K.I., and U01 AG20478 to E.R.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul M. Hwang for the p53 cDNA, Dominique Leprince for the HIC1 cDNA and Bert Vogelstein for the HCT 116 p53−/− and +/+ cell lines.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. doi:10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boily G., Seifert E.L., Bevilacqua L., He X.H., Sabourin G., Estey C., Moffat C., Crawford S., Saliba S., Jardine K., et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. doi:10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. doi:10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 4.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M.W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. doi:10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerhart-Hines Z., Rodgers J.T., Bare O., Lerin C., Kim S.H., Mostoslavsky R., Alt F.W., Wu Z., Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. doi:10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Zhang S., Blander G., Tse J.G., Krieger M., Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. doi:10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 7.Rodgers J.T., Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl Acad. Sci. USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. doi:10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordone L., Motta M.C., Picard F., Robinson A., Jhala U.S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. doi:10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C., Zhang F., Ge X., Yan T., Chen X., Shi X., Zhai Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell. Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. doi:10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Kessler B., Howitz K.T., Gorospe M., de Cabo R., Sinclair D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. doi:10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto S., Fergusson M.M., Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. doi:10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 12.Civitarese A.E., Carling S., Heilbronn L.K., Hulver M.H., Ukropcova B., Deutsch W.A., Smith S.R., Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. doi:10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara N., Rinaldi B., Corbi G., Conti V., Stiuso P., Boccuti S., Rengo G., Rossi F., Filippelli A. Exercise training promotes SIRT1 activity in aged rats. Rejuvenation Res. 2008;11:139–150. doi: 10.1089/rej.2007.0576. doi:10.1089/rej.2007.0576. [DOI] [PubMed] [Google Scholar]
- 14.Kanfi Y., Peshti V., Gozlan Y.M., Rathaus M., Gil R., Cohen H.Y. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582:2417–2423. doi: 10.1016/j.febslet.2008.06.005. doi:10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Shang L., Zhou H., Xia Y., Wang H., Gao G., Chen B., Liu Q., Shao C., Gong Y. Serum withdrawal up-regulates human SIRT1 gene expression in a p53 dependent manner. J. Cell. Mol. Med. 2009;104:4176–4184. doi: 10.1111/j.1582-4934.2008.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q., Wang S.Y., Fleuriel C., Leprince D., Rocheleau J.V., Piston D.W., Goodman R.H. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc. Natl Acad. Sci. USA. 2007;104:829–833. doi: 10.1073/pnas.0610590104. doi:10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Estep P.W., 3rd, Warner J.B., Bulyk M.L. Short-term calorie restriction in male mice feminizes gene expression and alters key regulators of conserved aging regulatory pathways. PLoS ONE. 2009;4:e5242. doi: 10.1371/journal.pone.0005242. doi:10.1371/journal.pone.0005242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.el-Deiry W.S., Kern S.E., Pietenpol J.A., Kinzler K.W., Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. doi:10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 19.Nagaich A.K., Zhurkin V.B., Sakamoto H., Gorin A.A., Clore G.M., Gronenborn A.M., Appella E., Harrington R.E. Architectural accommodation in the complex of four p53 DNA binding domain peptides with the p21/waf1/cip1 DNA response element. J. Biol. Chem. 1997;272:14830–14841. doi: 10.1074/jbc.272.23.14830. doi:10.1074/jbc.272.23.14830. [DOI] [PubMed] [Google Scholar]
- 20.Chen W.Y., Wang D.H., Yen R.C., Luo J., Gu W., Baylin S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53 dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. doi:10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Heilbronn L.K., de Jonge L., Frisard M.I., DeLany J.P., Larson-Meyer D.E., Rood J., Nguyen T., Martin C.K., Volaufova J., Most M.M., et al. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA. 2006;295:1539–1548. doi: 10.1001/jama.295.13.1539. doi:10.1001/jama.295.13.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canto C., Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. doi:10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feige J.N., Lagouge M., Canto C., Strehle A., Houten S.M., Milne J.C., Lambert P.D., Mataki C., Elliott P.J., Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. doi:10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. doi:10.1016/S1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 25.Nibu Y., Zhang H., Bajor E., Barolo S., Small S., Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. doi:10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahyo T., Nishida T., Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. doi:10.1016/S1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 27.Stankovic-Valentin N., Deltour S., Seeler J., Pinte S., Vergoten G., Guerardel C., Dejean A., Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. doi:10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Megidish T., Xu J.H., Xu C.W. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1) J. Biol. Chem. 2002;277:8255–8259. doi: 10.1074/jbc.C200001200. doi:10.1074/jbc.C200001200. [DOI] [PubMed] [Google Scholar]
- 29.Juan L.J., Shia W.J., Chen M.H., Yang W.M., Seto E., Lin Y.S., Wu C.W. Histone deacetylases specifically down-regulate p53 dependent gene activation. J. Biol. Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. doi:10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 30.Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. doi:10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 31.Flachsbart F., Croucher P.J., Nikolaus S., Hampe J., Cordes C., Schreiber S., Nebel A. Sirtuin 1 (SIRT1) sequence variation is not associated with exceptional human longevity. Exp. Gerontol. 2006;41:98–102. doi: 10.1016/j.exger.2005.09.008. doi:10.1016/j.exger.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Peeters A.V., Beckers S., Verrijken A., Mertens I., Roevens P., Peeters P.J., Van Hul W., Van Gaal L.F. Association of SIRT1 gene variation with visceral obesity. Hum. Genet. 2008;124:431–436. doi: 10.1007/s00439-008-0567-8. doi:10.1007/s00439-008-0567-8. [DOI] [PubMed] [Google Scholar]
- 33.Weyrich P., Machicao F., Reinhardt J., Machann J., Schick F., Tschritter O., Stefan N., Fritsche A., Haring H.U. SIRT1 genetic variants associate with the metabolic response of Caucasians to a controlled lifestyle intervention—the TULIP Study. BMC Med. Genet. 2008;9:100. doi: 10.1186/1471-2350-9-100. doi:10.1186/1471-2350-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. doi:10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Kuningas M., Putters M., Westendorp R.G., Slagboom P.E., van Heemst D. SIRT1 gene, age-related diseases, and mortality: the Leiden 85-plus study. J. Gerontol. A. Biol. Sci. Med. Sci. 2007;62:960–965. doi: 10.1093/gerona/62.9.960. [DOI] [PubMed] [Google Scholar]
- 36.Mattagajasingh I., Kim C.S., Naqvi A., Yamamori T., Hoffman T.A., Jung S.B., DeRicco J., Kasuno K., Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl Acad. Sci. USA. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. doi:10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcendor R.R., Gao S., Zhai P., Zablocki D., Holle E., Yu X., Tian B., Wagner T., Vatner S.F., Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. doi:10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 38.Kim D., Nguyen M.D., Dobbin M.M., Fischer A., Sananbenesi F., Rodgers J.T., Delalle I., Baur J.A., Sui G., Armour S.M., et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. doi:10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fontana L., Meyer T.E., Klein S., Holloszy J.O. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc. Natl Acad. Sci. USA. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. doi:10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witte A.V., Fobker M., Gellner R., Knecht S., Floel A. Caloric restriction improves memory in elderly humans. Proc. Natl Acad. Sci. USA. 2009;106:1255–1260. doi: 10.1073/pnas.0808587106. doi:10.1073/pnas.0808587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.