Abstract
Current treatments have largely failed to slow the rapidly increasing world-wide prevalence of obesity and its co-morbidities. Despite a strong genetic contribution to obesity (40–70%), only a small percentage of heritability is explained with current knowledge of monogenic abnormalities, common sequence variants and conventional modes of inheritance. Epigenetic effects are rarely tested in humans because of difficulties arranging studies that distinguish conventional and transgenerational inheritance while simultaneously controlling environmental factors and learned behaviors. However, growing evidence from model organisms implicates genetic and environmental factors in one generation that affect phenotypes in subsequent generations. In this report, we provide the first evidence for paternal transgenerational genetic effects on body weight and food intake. This test focused on the obesity-resistant 6C2d congenic strain, which carries the Obrq2aA/J allele on an otherwise C57BL/6J background. Various crosses between 6C2d and the control C57BL/6J strain showed that the Obrq2aA/J allele in the paternal or grandpaternal generation was sufficient to inhibit diet-induced obesity and reduce food intake in the normally obesity-susceptible, high food intake C57BL/6J strain. These obesity-resistant and reduced food intake phenotypes were transmitted through the paternal lineage but not the maternal lineage with equal strength for at least two generations. Eliminating social interaction between the father and both his offspring and the pregnant dam did not significantly affect food intake levels, demonstrating that the phenotype is transmitted through the male germline rather than through social interactions. Persistence of these phenotypes across multiple generations raises the possibility that transgenerational genetic effects contribute to current metabolic conditions.
INTRODUCTION
The prevalence of obesity is increasing worldwide, causing an increase in the incidence of co-morbidities that include type 2 diabetes, cardiovascular disease and cancer (1). Therapeutic interventions involving life style, medication and surgery have largely failed to reduce the prevalence of obesity. With epidemiological evidence suggesting that genetic factors control 40–70% of inter-individual variation in body weight, uncovering the genetic basis of obesity may lead to improved treatment strategies based on predictive risk markers and targeted therapeutic designs (2). To this end, rare cases of extreme obesity have been shown to result from mutations in genes such as leptin, leptin receptor and melanocortin 4 receptor (MC4R) as well as multi-gene copy number variants (3–6). In addition, genome-wide association studies (GWASs) identified common variants that influence body weight in fat mass and obesity-associated (FTO) and MC4R, among others (7,8). However, the genetic variants identified to date collectively account for only a small portion of the overall heritability of body weight (2).
Missing heritability can result from several factors (9). First, heritability may be over-estimated (10). Secondly, rare variants, closely linked genes, small effect sizes and non-additive effects are usually difficult to detect because of limited statistical power (11). Thirdly, both untested classes of genetic variants and unexplored regions of the genome could contribute to missing heritability (12,13). Finally, parental and transgenerational effects resulting from epigenetic mechanisms have also been largely untested (14).
Current genetic approaches usually focus on associations between genotypes and phenotypes within individuals in the study population (15). However, without specific study designs and appropriate tests, transgenerational effects can easily elude discovery. In these cases, genetic or environmental factors acting in previous generations transmit their effects epigenetically without inheriting the original causative genetic variants or recurrent exposure to the environmental factor (16). These modes of inheritance can be distinguished by referring to them, respectively, as transgenerational genetic effects or transgenerational environmental effects. Despite the difficulty in testing for these classes of transgenerational effects, increasing evidence from humans and model organisms suggests that they contribute to disease risk (17). For example, male or female rats exposed to dexamethasone have F1 and F2 male offspring with reduced birth weight and a metabolic syndrome phenotype (18). In addition, metastable alleles such as Agoutiviable yellow (Avy) and Axin1fused have expression levels that are inherited transgenerationally with effects on coat color and adiposity or tail kinking, respectively (19–21). Other examples of transgenerational genetic effects include paramutation (22–25) and interacting genes in different generations (26–28). Although considerably less information is available for humans owing to experimental difficulties, two recent GWASs found regions of the genome that control susceptibility to cancer and type 1 and 2 diabetes in a parent-of-origin-dependent manner (29,30). Additional studies identified multiple chromosomal locations with parent-of-origin effects on obesity probably due to the imprinted genes (31–33). Indeed, imprinting is a prototypic example of epigenetic inheritance, where inherited allelic expression differences depend on the parent of origin. However, other possible mechanisms underlying epigenetic inheritance include DNA methylation, histone modification or inheritance of molecules such as RNAs (14,16).
Our laboratory and others have used mouse chromosome substitution strains (CSSs) and congenic strains derived from them to identify quantitative trait loci (QTLs) that regulate complex traits (34–38). The CSS paradigm differs from segregating crosses and heterogeneous stocks in ways that enable detection of both conventional and transgenerational genetic effects (37). We sought to take advantage of the CSS paradigm and test for transgenerational genetic effects in a mouse model of diet-induced obesity and related metabolic conditions. In particular, we focused on the Obrq2a QTL in an established model of diet-induced obesity and metabolic disease (38,39–41). Obrq2a is a body weight- and insulin-resistant QTL that was previously mapped to a 3.2 Mb interval between single nucleotide polymorphism (SNP) markers rs29927775 and rs30221945 using the CSS mapping strategy (D. Buchner, personal communication). 6C2d is the obesity-resistant, insulin-sensitive congenic strain that defines Obrq2a. In this report, we show that strain 6C2d has both reduced body weight and reduced food intake relative to the obese control strain C57BL/6J (B6) and that this phenotypic effect is inherited for multiple generations through the paternal lineage, independent of inheriting the causative Obrq2aA/J genetic variant.
RESULTS
Parental effect of Obrq2a on body weight
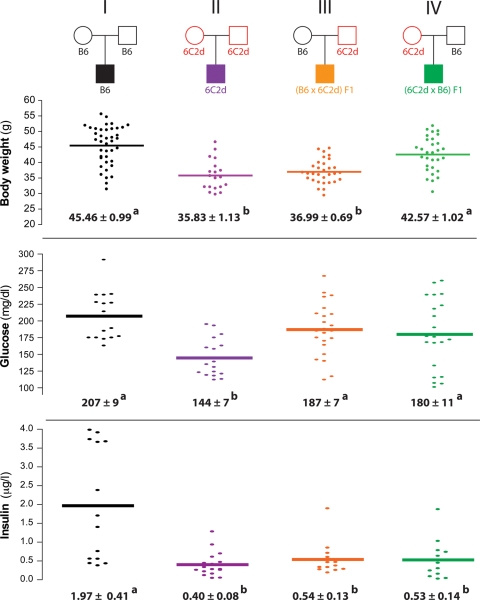
To test whether the Obrq2a QTL affects body weight in a parent-of-origin manner, F1 offspring of reciprocal crosses between the B6 obesity-sensitive strain and the 6C2d obesity-resistant congenic strain were tested for response to a high-fat, simple carbohydrate (HFSC) diet. For this test, B6 females were crossed to 6C2d males to obtain (B6 × 6C2d)F1 hybrids (Fig. 1, Cross III) and 6C2d females were crossed to B6 males to obtain the reciprocal (6C2d × B6)F1 hybrids (Fig. 1, Cross IV). Five-week-old (B6 × 6C2d)F1 and (6C2d × B6)F1 males were then fed the HFSC diet for 100 days. At the end of the study, (B6 × 6C2d)F1 males weighed significantly less than both the B6 males and the reciprocal (6C2d × B6)F1 hybrid males (P < 0.001; Fig. 1). These results suggest that inheritance through the paternal but not the maternal lineage accounted for most of the diet-induced effect of Obrq2a on body weight.
Figure 1.
A parental effect accounts for the body weight regulation by Obrq2a. Body weight, fasting glucose levels and fasting insulin levels are shown for control B6, control 6C2d and reciprocal F1 male mice following 100 days on the HFSC diet. Circles represent females, squares represent males. Mean values ± SEM are shown. Mean values with different superscript letters correspond to groups that are significantly different (P < 0.05 following Tukey's multiple testing correction), whereas mean values with identical superscript letters correspond to groups that are not significantly different (P > 0.05 following Tukey's multiple testing correction). Red indicates homozygosity for the Obrq2aA/J allele.
In contrast, no differences were detected in levels of fasting insulin and fasting glucose in the reciprocal F1 hybrids. Levels of insulin were similar to those of the 6C2d parental strain, suggesting that the Obrq2aA/J allele has dominant effects on insulin homeostasis, whereas glucose levels were similar to those of the B6 parental strain, suggesting that the Obrq2aA/J allele has recessive effects on glucose homeostasis (Fig. 1). Thus, these three metabolically related traits are inherited in distinct manners, perhaps because of the action of multiple genes at the Obrq2a locus or because of physiological interactions between genetic variants at Obrq2a and the B6 host background.
Care was taken in husbandry to separate genetic and diet effects. In all cases, breeder mice were raised on the conventional diet (5010) and were not exposed to the HFSC diet. Only test mice were maintained on the HFSC diet, and these mice were never used for breeding.
Transgenerational inheritance of the Obrq2a parental effect
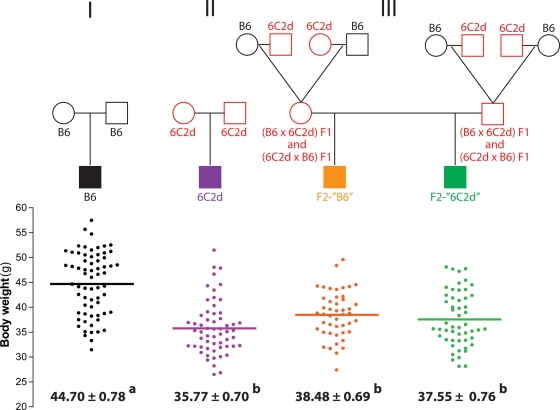
Differing body weights in reciprocal (B6 × 6C2d)F1 and (6C2d × B6)F1 males may result from a parent-of-origin effect such as imprinting, or from a heritable epigenetic effect that is independent of inheriting the Obrq2a allele. To test these possibilities, (B6 × 6C2d)F1 mice were intercrossed, as were (6C2d × B6)F1 mice, to generate offspring with the following Obrq2a genotypes in a Mendelian 1 : 2 : 1 ratio: Obrq2aB6/Obrq2aB6, Obrq2aB6/Obrq2aA/J and Obrq2aA/J/Obrq2aA/J (Fig. 2). We note that Obrq2aB6/Obrq2aB6 mice are genetically identical to B6 mice, and Obrq2aA/J/Obrq2aA/J mice are genetically identical to 6C2d mice, despite differences in parental genotype. These reconstituted Obrq2aA/J/Obrq2aA/J males are hereafter referred to as F2-‘6C2d’ and the reconstituted Obrq2aB6/Obrq2aB6 males as F2-‘B6’ in contrast to the 6C2d and B6 control strains that are bred in a conventional manner.
Figure 2.
Obesity resistance conferred by Obrq2aA/J does not depend on inheritance of the Obrq2aA/J allele. Body weight is shown for control B6, control 6C2d, F2-‘B6’ and F2-‘6C2d’ male mice following 100 days on the HFSC diet. Circles represent females, squares represent males. Mean values ± SEM are shown. Mean values with different superscript letters correspond to groups that are significantly different (P < 0.05 following Tukey's multiple testing correction), whereas mean values with identical superscript letters correspond to groups that are not significantly different (P > 0.05 following Tukey's multiple testing correction). Red indicates heterozygosity or homozygosity for the Obrq2aA/J allele.
To test diet response, F2-‘6C2d’, F2-‘B6’ and control males were fed the HFSC diet for 100 days and the resulting body weights were examined. Interestingly, although phenotypic differences were found between the reciprocal F1 hybrids (see above, and Fig. 1), the response of the F2-‘B6’ and F2-‘6C2d’ males to the high-fat diet did not differ between the (B6 × 6C2d)F1 or (6C2d × B6)F1 intercrosses, or between F2-‘B6’ males housed either in the same cage as F2-‘6C2d’ males or without F2-‘6C2d’ males (data not shown). We therefore pooled data from each of these experiments. After 100 days on the HFSC diet, F2-‘B6’ mice showed a 14% (6.26 g) reduction in body weight relative to B6 (Fig. 2, Crosses I versus III). Intriguingly, body weight did not differ between F2-‘B6’ and either control 6C2d or F2-‘6C2d’ mice (Fig. 2, Crosses II versus III). These results demonstrate that the parental effect on body weight did not depend on inheriting the Obrq2aA/J variant and that parental heterozygosity for the Obrq2aA/J allele protected F2-‘B6’ males from diet-induced obesity.
Epigenetic inheritance of the Obrq2a effect across multiple generations
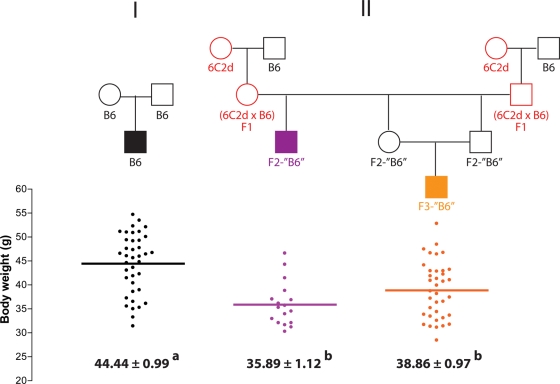
Having generated F2-‘B6’ males that are resistant to diet-induced obesity despite being isogenic with obesity-susceptible B6 males that are offspring of conventional brother–sister matings, we sought to test whether this epigenetic phenomenon was transmissible to the ‘F3’ generation. For this test, F2-‘B6’ mice were intercrossed and the body weight of the resulting F3-‘B6’ offspring was analyzed after 100 days on the HFSC diet. Body weight of the F3-‘B6’ males did not differ from that of F2-‘B6’ males, but was significantly less than that of control B6 mice (P < 0.001) (Fig. 3). Therefore, the Obrq2aA/J allele demonstrates a transgenerational effect by reducing body weight in the ‘F3’ generation even though neither F2 nor F3 generations inherited the Obrq2aA/J allele.
Figure 3.
Multi-generational inheritance of obesity resistance. Body weight is shown for control B6, F2-‘B6’ and F3-‘B6’ male mice following 100 days on the HFSC diet. Circles represent females, squares represent males. Mean values ± SEM are shown. Mean values with different superscript letters correspond to groups that are significantly different (P < 0.05 following Tukey's multiple testing correction), whereas mean values with identical superscript letters correspond to groups that are not significantly different (P > 0.05 following Tukey's multiple testing correction). Red indicates heterozygosity or homozygosity for the Obrq2aA/J allele.
Male but not female germline transmits epigenetic protection from diet-induced obesity
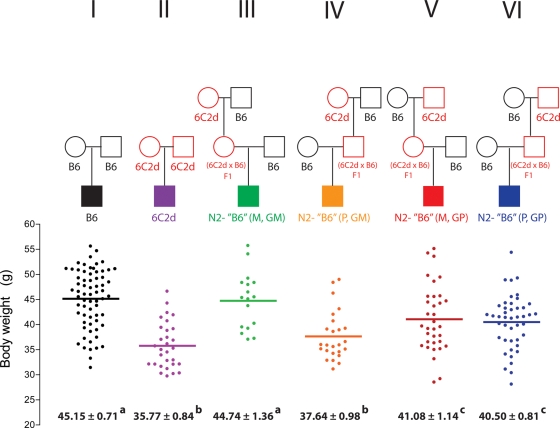
To test whether the transgenerational effect of Obrq2aA/J was inherited through the maternal or paternal lineage, four reciprocal backcrosses were made that involved all four parental and grandparental combinations of Obrq2aA/J heterozygosity, namely P,GM, P,GP, M,GP and M,GM, where M and P denote maternal or paternal heterozygosity and GM and GP denote grandmaternal and grandpaternal heterozygosity, respectively (Fig. 4, Crosses III–VI). For each cross, the standard protocol was used to test susceptibility to diet-induced obesity among genetically identical ‘B6’ segregants. Interestingly, the only cross where ‘B6’ segregants were susceptible to diet-induced obesity involved both maternal and grandmaternal heterozygosity for the Obrq2aA/J QTL. In contrast, ‘B6’ male segregants were resistant to diet-induced obesity for the three other crosses. In each of these three crosses, at least one instance of paternal or grandpaternal heterozygosity for the Obrq2aA/J QTL was involved. Thus, transmission of the epigenetic effect of Obrq2aA/J through either the paternal or grandpaternal lineage confers resistance to diet-induced obesity to ‘B6’ males, despite their genetic identity with control B6 males. Importantly, reversal of the transgenerational effect occurred only after transmission through the maternal germline for two consecutive generations.
Figure 4.
Obesity resistance is inherited through the paternal, but not maternal lineage. Body weight is shown for control B6, control 6C2d and N2-‘B6’ male mice following 100 days on the HFSC diet. N2-‘B6’ mice of all four parental and grandparental combinations of Obrq2aA/J heterozygosity are shown, namely P,GM, P,GP, M,GP and M,GM, where M and P denote maternal or paternal heterozygosity, and GM and GP denote grandmaternal or grandpaternal heterozygosity. Circles represent females, squares represent males. Mean values ± SEM are shown. Mean values with different superscript letters correspond to groups that are significantly different (P < 0.05 following Tukey's multiple testing correction), whereas mean values with identical superscript letters correspond to groups that are not significantly different (P > 0.05 following Tukey's multiple testing correction). Red indicates heterozygosity or homozygosity for the Obrq2aA/J allele.
Paternal transgenerational effects on body weight are due to polymorphisms within the Obrq2a interval
To confirm that the paternal transgenerational effects of Obrq2a were due to the A/J-derived sequence within the 6C2d interval, two 6C2d mice from different parents were genotyped by microarray hybridization for 556 698 SNPs, of which 113 184 were polymorphic between B6 and A/J. These genotypes were compared with those of B6, A/J and 72 other inbred strains that were previously genotyped (42). No evidence was found for a genetic contribution in strain 6C2d from strains other than A/J. Among the 113 184 polymorphic SNPs between B6 and A/J, strain 6C2d carried the B6-derived allele for 113 104 of the SNPs and the A/J-derived allele for 80 of the SNPs. Seventy-nine of the A/J-derived SNPs were located within the Obrq2a interval on chromosome 6 and collectively represent every SNP within that interval that is polymorphic between B6 and A/J. The remaining SNP (JAX00572617/rs31078310) was on chromosome 5 and therefore potentially represented the A/J-derived sequence in strain 6C2d located outside of the Obrq2a interval. This SNP was therefore amplified by PCR, re-sequenced and found not to differ between B6 and 6C2d. Thus, the microarray-generated genotype for that SNP probably represented a genotyping error. Therefore, the genome of strain 6C2d is entirely derived from strain B6 except for the A/J-derived sequence within the Obrq2a interval. In addition, two N2-‘B6’ (P,GP) mice were also genotyped using the same microarray platform, and no evidence was found for any genetic differences between the ‘B6’ mice and the control B6 mice, which is consistent with an epigenetic mode of inheritance for the obesity resistance in ‘B6’ mice.
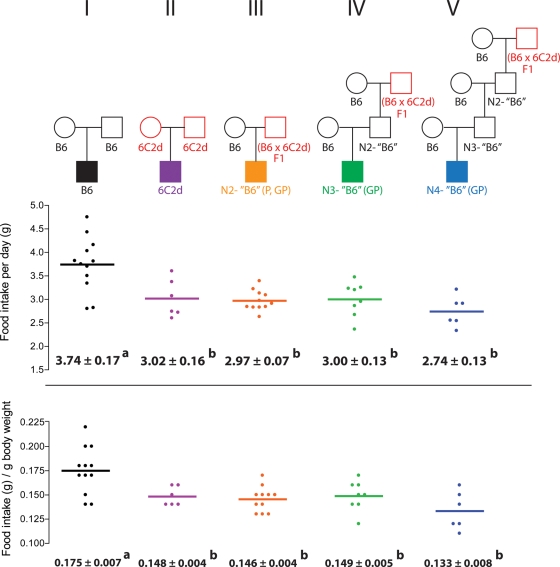
Transgenerational effect on food intake
To address mechanisms that underlie differences in response to the HFSC diet, we measured 24 h food intake for control B6 (Fig. 5, Cross I) and isogenic N2-‘B6’, N3-‘B6’ and N4-‘B6’ derived from backcrosses that involve either paternal, grandpaternal or great-grandpaternal heterozygosity for Obrq2aA/J (Fig. 5, Crosses III, IV and V, respectively). Regardless of whether daily food intake was calculated as per gram of body weight or as grams per day, ‘B6’ males from Crosses III, IV and V showed significantly reduced intake relative to control B6 males (Cross I) and were indistinguishable from the level in 6C2d males (Cross II). Thus, daily food consumption was significantly reduced in ‘B6’ males through transgenerational transmission of Obrq2aA/J effects through either the paternal, grandpaternal or great-grandpaternal males compared with isogenic control B6 from conventional crosses.
Figure 5.
A transgenerational genetic effect on food intake. Food intake per day and food intake per gram of body weight is shown for control B6, control 6C2d, N2-‘B6’, N3-‘B6’ and N4-‘B6’ male mice. Circles represent females, squares represent males. Mean values ± SEM are shown. Mean values with different superscript letters correspond to groups that are significantly different (P < 0.05 following Tukey's multiple testing correction), whereas mean values with identical superscript letters correspond to groups that are not significantly different (P > 0.05 following Tukey's multiple testing correction). Red indicates heterozygosity or homozygosity for the Obrq2aA/J allele.
Effect of Obrq2a on food intake is epigenetically transmitted through the germline
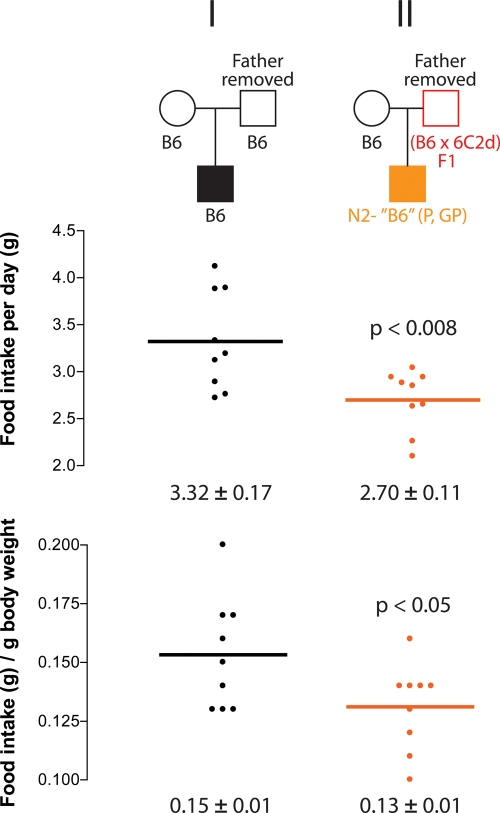
The transgenerational paternal effect on food intake may be transmitted through an epigenetic (germline) mechanism or through a behavioral mechanism whereby the male parent reduces the body weight and food intake of his offspring through social interactions (43). Results from Cross IV (Fig. 4) support an epigenetic mechanism, where obesity resistance persists in the N2-‘B6’ offspring, despite being raised in a shared environment with an obesity-susceptible B6 father. Nonetheless, we sought to formally test this hypothesis by crossing either a control B6 male or a (B6 × 6C2d)F1 male with a B6 female; however, the male mouse was removed from the cage after a single night. This design eliminated social interactions between the father and his offspring as well as with the pregnant dam beyond a single night. Only genetically B6 offspring from both crosses were tested so that all mice were genetically identical. Food intake was decreased in the offspring of (B6 × 6C2d)F1 males relative to offspring of B6 control males, demonstrating that the epigenetic effect of Obrq2a on food intake is inherited through the male germline and not the result of social interactions (Fig. 6).
Figure 6.
The paternal influence of Obrq2a on food intake does not require social interactions. Food intake per day and food intake per gram of body weight are shown for B6 and ‘B6’ offspring from control B6 males crossed to B6 females and (B6 × 6C2d)F1 males crossed to B6 females, respectively. Fathers were removed from the cage following a single night. Circles represent females, squares represent males. Mean values ± SEM are shown. P-values shown are relative to the control B6 cross. Red indicates heterozygosity for the Obrq2aA/J allele.
Transgenerational paternal effect is not due to differences in litter size
Litter sizes have been shown to affect obesity susceptibility in adult mice, with smaller litters associated with more obese offspring and larger litters associated with leaner offspring (44). However, litter sizes did not differ between any of the strains analyzed in any of the crosses undertaken (Supplementary Material, Figs S1–S6), demonstrating that the effects of Obrq2a were not due to differences in litter size.
Statistical analysis to account for parental mode of inheritance confirms epigenetic transgenerational regulation of body weight and food intake
The data analysis conducted above was performed in a similar manner to conventional genetic tests by analyzing the phenotypes of the offspring. These results collectively suggest that an epigenetic modification inherited through the paternal germline regulates body weight and food intake. Compared with models of conventional genetic inheritance, the epigenetic model differs in that offspring inherit an epigenetic mark rather than a genetic variant. Therefore, given the nature of the proposed paternal inheritance, it remains possible that offspring from an individual father may inherit the same epigenetic modifications and not represent independent replicates. Therefore, the data were re-analyzed using the parents rather than the offspring as the unit of analysis. For this analysis, all data from the offspring of individual parents were averaged to yield a single value. The averages for each parental unit were then tested in the same manner that the individual offspring were analyzed.
The parental analysis and the offspring analysis demonstrated similar trends for all crosses (Supplementary Material, Figs S1–S6). The results of the F2-‘B6’ cross (Fig. 2) remained statistically significant (Supplementary Material, Fig. S2), confirming a parental effect on body weight for Obrq2a that was independent of inheriting the A/J-derived sequence. In addition, the results of the F3-‘B6’ cross (Fig. 3) also remained statistically significant (Supplementary Material, Fig. S3), confirming the multi-generational inheritance of the Obrq2a obesity-resistant phenotype. Because of the multi-generational inheritance in this cross, these data were re-analyzed using the grandparent as the unit of analysis, and similar statistically significant results were obtained (Supplementary Material, Fig. S7). Furthermore, the results of the food intake analysis in both the ‘B6’ mice (generations N2–N4) (Supplementary Material, Fig. S5) and ‘father removed from the cage’ study (Supplementary Material, Fig. S6) remained statistically significant, demonstrating that the Obrq2a phenotype depended on the paternal genotype and was transmitted through the germline rather than behaviorally. The N4-‘B6’ parents were each derived from separate grandparents, and therefore the analysis of food intake remains statistically significant using the grandparent as the unit of analysis (Supplementary Material, Fig. S5). Although the reduction in power associated with using the parent as the unit of analysis prevented the results of the reciprocal F1 (Supplementary Material, Fig. S1) and ‘N2’ (Supplementary Material, Fig. S4) crosses from reaching statistical significance, the trends were consistent with those previously observed (Figs 1 and 4). Thus, the re-analysis of these data using the parent and, when appropriate, the grandparent as the unit of analysis collectively supports the conclusion, based on using the offspring as the unit of analysis, that the obesity resistance of Obrq2a is epigenetically inherited through the paternal lineage over multiple generations.
DISCUSSION
The discovery that resistance to diet-induced obesity and reduced food intake can be epigenetically inherited addresses two important questions in studies of transgenerational genetics (14,45), namely do these effects persist for multiple generations after exposure to the original genetic variant, and what is the relative magnitude of transgenerational and conventional genetic effects? We showed that these phenotypes were transmitted epigenetically through the paternal germline for at least three generations (Figs 3 and 5), and that the obesity phenotype reverted to a conventional mode of inheritance after transmission through the maternal germline for at least two consecutive generations (Fig. 4). Remarkably, the magnitude of these phenotypic effects was comparable for epigenetic and conventional inheritance (Figs 2 and 5). Other studies show that these transgenerational genetic effects are, in general, as common and as strong as conventional effects across a variety of traits (46). Together, these results suggest that transgenerational genetic effects might rival conventional effects in their impact on phenotypic variation.
The presence of the Obrq2aA/J allele in the paternal or grandpaternal generation can be considered as an initial genetic insult, as this is the only variable in the designed experiments. Therefore, the effects on body weight and food intake are attributable to DNA sequence variation at this locus.
These sequence differences initiate multi-generation epigenetic transmission of resistance to diet-induced obesity and reduced food intake. The use of 6C2d, a CSS-6-derived congenic strain, narrows the causative allele to at least one of the 19 candidate genes in the variable interval and excludes direct influence of host strain background. Using CSS-2 from the same B6- and A/J-derived CSS panel (38), Lesscher et al. (47) also found evidence of grandparental genotype influencing ethanol preference. The difficulty in attaining multi-generation pedigrees and controlling for environmental exposure in humans results in circumstantial evidence for parental and transgenerational effects. In contrast, CSSs and congenic strains derived from them provided definitive evidence implicating a limited number of genes and provided a model for focused mechanistic studies.
Transgenerational genetic effects are probably not unique to the Obrq2a locus. This QTL was selected for study for two simple reasons: its phenotypic effects were consistently strong and its small size (3 Mb) meant recombination was unlikely to disrupt its integrity during these multi-generational studies. Importantly, genetic content was not a consideration and indeed none of the genes at Obrq2a is known to be directly involved in DNA methylation, histone modification, RNA biology or translation control, the usual explanations for transgenerational effects (14). Preliminary results suggest that the Obrq1 QTL also shows evidence for transgenerational effects, whereas parental effects that contribute to Obrq3 phenotypes have not been detected (40, data not shown). Thus several but not all segments of mouse chromosome 6 show transgenerational genetic effects. An important task is now to map the locations of these segments and to characterize their epigenetic features based on the use of the CSSs and derived congenic strains.
Epigenetic inheritance through the paternal germline strongly suggests that the molecular basis for transgenerational genetic effects is present in sperm. During spermatogenesis, the paternal genome undergoes dramatic and crucial epigenetic changes, where nucleosomal histones are replaced with protamines that are essential for tight packaging of DNA in sperm (48). However, ∼4% of the haploid genome in sperm retains histones, with modifications such as H3K4me3, especially at genes involved in embryo development, imprinted gene clusters and miRNA loci (49). Interestingly, of the 12 human genes within the syntenic region corresponding to the Obrq2a locus, eight escape the histone–protamine–histone transitions (49). These genes are aldo keto reductase 1 (AKR1B1), plexin 4 (PLXNA4), solute receptor 35b4 (SLC35B4), exocyst complex protein 4 (EXOC4), coiled-coil-helix–coiled-coil-helix domain containing 3 (CHCHD3), 2,3-bisphosphoglycerate mutase (BPGM), leucine-rich repeats and guanylate kinase domain (LRGUK) and caldesmon 1 (CALD1). LRGUK carried the H3K27me3 mark that was frequently associated with genes that were repressed early in development. The remaining seven genes carried the H3K4me3 mark associated with developmental genes, non-coding RNAs and paternally expressed imprinted genes. PLXNA4 was unique in that it maintained multiple epigenetic marks (H3K27me3, H3K4me3, H3K4me2, methylated promoter), suggesting that its expression pattern is tightly regulated at the epigenetic level. Two sub-QTLs within the Obrq2a locus (Obrq2a5 and Obrq2a6) affect body weight, but not insulin resistance, similar to the paternal effect of Obrq2a (D. Buchner, personal communication). Testing the congenic strains that define these QTLs for transgenerational paternal effects should further define which of these candidate genes underlie the transgenerational effects.
Although circumstantial evidence implicates several mechanisms, the molecular basis for epigenetic inheritance remains to be discovered. Perhaps the most provocative evidence involves small RNAs and mRNAs. These RNAs have been detected in both eggs and sperm, where they have been shown to induce epigenetically heritable phenotypes through both the maternal and paternal germlines (14). For example, exposure of fertilized mouse eggs to miRNAs that target Kit mRNA induces heritable changes in tail color as well as decreased levels of Kit mRNA and non-polyadenylated Kit transcripts (24). Similar phenomena have also been reported for miRNA effects on embryogenesis, growth and cardiac development (22,23). In addition, partial deficiency for the Kit ligand as well as genes involved in RNA editing and miRNA access to their target mRNAs lead to transgenerational effects on testicular cancer risk (26,50). The presence of both miRNAs and small non-coding RNAs in human and mouse sperm has been reported (51–54). However, none of the genes at the Obrq2a locus is involved in DNA methylation, histone modifications and RNA biology. As a result, genetic variants at Obrq2a must lead directly to epigenetic changes in the germline, or indirectly to the germline via the soma.
Studies of complex traits typically focus on associations between genotypes and phenotypes within each individual in the study population. Many important discoveries have been made based on the functions of genes that are discovered with this paradigm (15). However, most of the genetic variation has to date eluded discovery, a phenomenon known as ‘missing heritability’ (9). Transgenerational genetic effects could explain some of the missing heritability because the genetic basis occurs in previous generations and is not necessarily present in the study population. In particular, genetically identical B6 males show distinct phenotypes depending on the genetic constitution of individuals in preceding generations. Moreover, increasing evidence shows that these transgenerational genetic effects are common, strong and long-lasting (46). Determining the molecular mechanism, whether similar effects act in humans and whether they contribute to ‘missing heritability’ remain important challenges.
MATERIALS AND METHODS
Husbandry
Mice were housed in ventilated racks at 21°C in a 12 h light–dark cycle. The mice were weaned at 3 weeks of age and fed Labdiet 5010 chow (PMI Nutrition International, Richmond, OH, USA) unless otherwise noted. For the high-fat diet experiments, mice were fed an HFSC diet (D12331; Research Diets, Inc, New Brunswick, NJ, USA) for 100 days, beginning at 5 weeks of age. The HFSC diet derives 58% of its kilocalories from saturated fat (soybean and coconut oil), 25.5% from simple carbohydrate (sucrose and maltodextrin) and 16.4% from protein (casein). Mice had access to food and water ad libitum. All animal protocols were approved by the Institutional Animal Care and Use Committee of Case Western Reserve University.
Mice
B6 and 6C2d congenic mice were obtained from our breeding colonies at Case Western Reserve University. Strain 6C2d carries the A/J-derived allele at Obrq2a on an otherwise B6 background, including the X and Y chromosomes as well as the mitochondria. All test and control mice used for each experiment were born within 3 months of each other.
Genotyping
Mice were genotyped for the B6- and A/J-derived alleles of Obrq2a using polymorphic SNP markers at the proximal (rs30468068) and distal (rs30320124) ends of the QTL interval. PCR primers that were used to amplify the rs30468068 SNP were: 5′-GAAGG GACCT TCTGA GCAAA TA-3′ and 5′-GTGTG GACAT GTATG TCTGT GC-3′. PCR primers for the rs30320124 SNP were: 5′-TGGGG TGATT TTTGT TGTTG-3′ and 5′-CCAGG GGACA TTTTC TGTTG-3′. Both PCR products were digested with the restriction enzyme RsaI (New England Biolabs, MA, USA) and analyzed with agarose gel electrophoresis. In addition, two 6C2D mice and two ‘B6’ mice (mice that are genetically identical to B6 but have genetically different parents) were genotyped using the Mouse Diversity Genotyping Array (Affymetrix, Santa Clara, CA, USA) at the Dana-Farber Cancer Institute Microarray Core (Dr Edward Fox, Director). These data were compared with genotyping data from additional inbred strains (42) (http://cgd.jax.org/tools/diversityarray.shtml). To exclude genotyping errors in the genotype analysis, SNPs for which there was no genotype determined, that were not called as homozygotes or for which the two B6 control samples differed from each other were excluded from analysis. An additional SNP, JAX00572617 (also referred to as rs31078310), was amplified by PCR using the following primers: 5′-CCAGC CTGAC AGGAT TTAAT G-3′ and 5′-CCTGC GACGT TCTAG AGTTT G-3′. The resulting PCR product was sequenced at the CWRU DNA sequencing core (Dr Mark Adams, Director).
Plasma glucose and insulin measurements
Mice were anesthetized with isoflurane, and blood was collected from the retro-orbital sinus using heparin-coated capillary tubes into EDTA microtainers. Whole-blood glucose was measured using a hand-held glucometer (OneTouch Ultra, Life Scan Inc., Milpitas, CA, USA). Plasma was then centrifuged and stored at −80°C. Insulin levels were determined using an ultrasensitive mouse ELISA kit (Mercodia, Uppsala, Sweden).
Food intake
Thirty-five day old mice were acclimated to the HFSC diet for 2 days. On the third day, mice were placed in a clean cage and the amount of food was weighed. After 24 h, food was again weighed to determine daily food intake and the mice were weighed to calculate food intake per gram of body weight. Mice were housed two to three per cage to avoid the stress of individual housing. Therefore, each data point represents an average value per cage.
Statistics
Data were analyzed using a one-way ANOVA followed by Tukey's multiple comparison test to correct for multiple observations or by an unpaired t-test when only two groups were compared.
SUPPLEMENTARY MATERIAL
FUNDING
This research was supported by the NIH NCRR grant RR12305, the NIH NCI Transdisciplinary Research on Energetics and Cancer (TREC) grant U54 CA116867 and the NIH NHLBI grant F32-HL-82213.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Morgan and Jonathan Phan for genotyping assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kelly T., Yang W., Chen C.S., Reynolds K., He J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 2008;32:1431–1437. doi: 10.1038/ijo.2008.102. doi:10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- 2.Walley A.J., Asher J.E., Froguel P. The genetic contribution to non-syndromic human obesity. Nat. Rev. Genet. 2009;10:431–442. doi: 10.1038/nrg2594. doi:10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 3.Ranadive S.A., Vaisse C. Lessons from extreme human obesity: monogenic disorders. Endocrinol. Metab. Clin. North Am. 2008;37:733–751. doi: 10.1016/j.ecl.2008.07.003. doi:10.1016/j.ecl.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S., et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. doi:10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K., Saeed S., Hamilton-Shield J., Clayton-Smith J., O'Rahilly S., et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 463:666–670. doi: 10.1038/nature08689. doi:10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sha B.Y., Yang T.L., Zhao L.J., Chen X.D., Guo Y., Chen Y., Pan F., Zhang Z.X., Dong S.S., Xu X.H., et al. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J. Hum. Genet. 2009;54:199–202. doi: 10.1038/jhg.2009.10. doi:10.1038/jhg.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R.B., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. doi:10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loos R.J.F., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. doi:10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. doi:10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visscher P.M., Hill W.G., Wray N.R. Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet. 2008;9:255–266. doi: 10.1038/nrg2322. doi:10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 11.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. doi:10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 12.Kruglyak L. The road to genome-wide association studies. Nat. Rev. Genet. 2008;9:314–318. doi: 10.1038/nrg2316. doi:10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. doi:10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadeau J.H. Transgenerational genetic effects on phenotypic variation and disease risk. Hum. Mol. Genet. 2009;18:R202–R210. doi: 10.1093/hmg/ddp366. doi:10.1093/hmg/ddp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. doi:10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youngson N.A., Whitelaw E. Transgenerational epigenetic effects. Annu. Rev. Genomics Hum. Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. doi:10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 17.Gluckman P.D., Hanson M.A., Beedle A.S. Non-genomic transgenerational inheritance of disease risk. BioEssays. 2007;29:145–154. doi: 10.1002/bies.20522. doi:10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- 18.Drake A.J., Walker B.R., Seckl J.R. Intergenerational consequences of fetal programming by in utero exposure to glucocorticoids in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R34–R38. doi: 10.1152/ajpregu.00106.2004. [DOI] [PubMed] [Google Scholar]
- 19.Morgan H.D., Sutherland H.G., Martin D.I., Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 20.Rakyan V.K., Chong S., Champ M.E., Cuthbert P.C., Morgan H.D., Luu K.V., Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl Acad. Sci. USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. doi:10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterland R.A., Travisano M., Tahiliani K.G., Rached M.T., Mirza S. Methyl donor supplementation prevents transgenerational amplification of obesity. Int. J. Obes. 2008;32:1373–1379. doi: 10.1038/ijo.2008.100. doi:10.1038/ijo.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner K.D., Wagner N., Ghanbarian H., Grandjean V., Gounon P., Cuzin F., Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev. Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. doi:10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Grandjean V., Gounon P., Wagner N., Martin L., Wagner K.D., Bernex F., Cuzin F., Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. doi:10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 24.Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. doi:10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 25.Suter C.M., Martin D.I.K. Paramutation: the tip of an epigenetic iceberg? Trends Genet. 2010;26:9–14. doi: 10.1016/j.tig.2009.11.003. doi:10.1016/j.tig.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam M.-Y.J., Heaney J.D., Youngren K.K., Kawasoe J.H., Nadeau J.H. Trans-generational epistasis between Dnd1Ter and other modifier genes controls susceptibility to testicular germ cell tumors. Hum. Mol. Genet. 2007;16:2233–2240. doi: 10.1093/hmg/ddm175. doi:10.1093/hmg/ddm175. [DOI] [PubMed] [Google Scholar]
- 27.Parkhurst S.M., Ish-Horowicz D. wimp, a dominant maternal-effect mutation, reduces transcription of a specific subset of segmentation genes in Drosophila. Genes Dev. 1991;5:341–357. doi: 10.1101/gad.5.3.341. doi:10.1101/gad.5.3.341. [DOI] [PubMed] [Google Scholar]
- 28.Xing Y., Shi S., Le L., Lee C.A., Silver-Morse L., Li W.X. Evidence for transgenerational transmission of epigenetic tumor susceptibility in Drosophila. PLoS Genet. 2007;3:1598–1606. doi: 10.1371/journal.pgen.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., Besenbacher S., Jonasdottir A., Sigurdsson A., Kristinsson K.T., Jonasdottir A., et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. doi:10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace C., Smyth D.J., Maisuria-Armer M., Walker N.M., Todd J.A., Clayton D.G. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 2010;42:68–71. doi: 10.1038/ng.493. doi:10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong C., Li W.D., Geller F., Lei L., Li D., Gorlova O.Y., Hebebrand J., Amos C.I., Nicholls R.D., Price R.A. Possible genomic imprinting of three human obesity-related genetic loci. Am. J. Hum. Genet. 2005;76:427–437. doi: 10.1086/428438. doi:10.1086/428438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorlova O.Y., Amos C.I., Wang N.W., Shete S., Turner S.T., Boerwinkle E. Genetic linkage and imprinting effects on body mass index in children and young adults. Eur. J. Hum. Genet. 2003;11:425–432. doi: 10.1038/sj.ejhg.5200979. doi:10.1038/sj.ejhg.5200979. [DOI] [PubMed] [Google Scholar]
- 33.Lindsay R.S., Kobes S., Knowler W.C., Hanson R.L. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of birth weight. Hum. Genet. 2002;110:503–509. doi: 10.1007/s00439-002-0718-2. doi:10.1007/s00439-002-0718-2. [DOI] [PubMed] [Google Scholar]
- 34.Gelegen C., Collier D.A., Campbell I.C., Oppelaar H., van den Heuvel J., Adan R.A., Kas M.J. Difference in susceptibility to activity-based anorexia in two inbred strains of mice. Eur. Neuropsychopharmacol. 2007;17:199–205. doi: 10.1016/j.euroneuro.2006.04.007. doi:10.1016/j.euroneuro.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Hill A.E., Lander E.S., Nadeau J.H. Chromosome substitution strains: a new way to study genetically complex traits. Methods Mol. Med. 2006;128:153–172. doi: 10.1385/1-59745-159-2:153. [DOI] [PubMed] [Google Scholar]
- 36.Leussis M.P., Frayne M.L., Saito M., Berry E.M., Aldinger K.A., Rockwell G.N., Hammer R.P., Jr, Baskin-Hill A.E., Singer J.B., Nadeau J.H., et al. Genomic survey of prepulse inhibition in mouse chromosome substitution strains. Genes Brain Behav. 2009;8:806–816. doi: 10.1111/j.1601-183X.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadeau J.H., Singer J.B., Matin A., Lander E.S. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 2000;24:221–225. doi: 10.1038/73427. doi:10.1038/73427. [DOI] [PubMed] [Google Scholar]
- 38.Singer J.B., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O'Brien W.E., Conti D.V., Witte J.S., Lander E.S., et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. doi:10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 39.Buchner D.A., Burrage L.C., Hill A.E., Yazbek S.N., O'Brien W.E., Croniger C.M., Nadeau J.H. Resistance to diet-induced obesity in mice with a single substituted chromosome. Physiol. Genomics. 2008;35:116–122. doi: 10.1152/physiolgenomics.00033.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao H., Burrage L.C., Sinasac D.S., Hill A.E., Ernest S.R., O'Brien W., Courtland H.W., Jepsen K.J., Kirby A., Kulbokas E.J., et al. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl Acad. Sci. USA. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. doi:10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surwit R.S., Feinglos M.N., Rodin J., Sutherland A., Petro A.E., Opara E.C., Kuhn C.M., Rebuffe-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44:645–651. doi: 10.1016/0026-0495(95)90123-x. doi:10.1016/0026-0495(95)90123-X. [DOI] [PubMed] [Google Scholar]
- 42.Yang H., Ding Y., Hutchins L.N., Szatkiewicz J., Bell T.A., Paigen B.J., Graber J.H., de Villena F.P.-M., Churchill G.A. A customized and versatile high-density genotyping array for the mouse. Nat. Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. doi:10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peter D.G., Mark A.H., Alan S.B. Non-genomic transgenerational inheritance of disease risk. BioEssays. 2007;29:145–154. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt I., Schoelch C., Ziska T., Schneider D., Simon E., Plagemann A. Interaction of genetic and environmental programming of the leptin system and of obesity disposition. Physiol. Genomics. 2000;3:113–120. doi: 10.1152/physiolgenomics.2000.3.2.113. [DOI] [PubMed] [Google Scholar]
- 45.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. doi:10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson V., Nadeau J. Transgenerational genetic effects of the paternal Y chromosome on daughter's phenotypes. Epigenomics. 2010;2:513–521. doi: 10.2217/epi.10.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesscher H.M., Kas M.J., van der Elst S., van Lith H.A., Vanderschuren L.J. A grandparent-influenced locus for alcohol preference on mouse chromosome 2. Pharmacogenet. Genomics. 2009;19:719–729. doi: 10.1097/FPC.0b013e3283311320. doi:10.1097/FPC.0b013e3283311320. [DOI] [PubMed] [Google Scholar]
- 48.Wykes S.M., Krawetz S.A. The structural organization of sperm chromatin. J. Biol. Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. doi:10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 49.Hammoud S.S., Nix D.A., Zhang H., Purwar J., Carrell D.T., Cairns B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A., et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. doi:10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 51.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 52.Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. doi:10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krawetz S.A. Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 2005;6:633–642. doi: 10.1038/nrg1654. doi:10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 54.Yan W., Morozumi K., Zhang J., Ro S., Park C., Yanagimachi R. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and microRNAs in the sperm nuclei. Biol. Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. doi:10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.