Abstract
Reduced expression of the survival motor neuron (SMN) gene causes the childhood motor neuron disease spinal muscular atrophy (SMA). Low levels of ubiquitously expressed SMN protein result in the degeneration of lower motor neurons, but it remains unclear whether other regions of the nervous system are also affected. Here we show that reduced levels of SMN lead to impaired perinatal brain development in a mouse model of severe SMA. Regionally selective changes in brain morphology were apparent in areas normally associated with higher SMN levels in the healthy postnatal brain, including the hippocampus, and were associated with decreased cell density, reduced cell proliferation and impaired hippocampal neurogenesis. A comparative proteomics analysis of the hippocampus from SMA and wild-type littermate mice revealed widespread modifications in expression levels of proteins regulating cellular proliferation, migration and development when SMN levels were reduced. This study reveals novel roles for SMN protein in brain development and maintenance and provides the first insights into cellular and molecular pathways disrupted in the brain in a severe form of SMA.
INTRODUCTION
Proximal spinal muscular atrophy (SMA), a leading genetic cause of infant mortality, is an autosomal-recessive neuromuscular disorder with a carrier frequency of 1:50 and an annual incidence of around 1:10 000 live births (1). SMA is classically characterized by degeneration and loss of large alpha motor neurons in the ventral horn of the spinal cord and paralysis of proximal muscles in the limbs and trunk, resulting from denervation at the neuromuscular junction (2–5). The most severe and common form of SMA, accounting for ∼50% of SMA diagnoses (SMA type I; Werdnig–Hoffmann syndrome), leads to disease onset in the first 6 months of life, no achievement of sitting without support and death within the first 2 years of life (5,6).
The vast majority of SMA cases are caused by homozygous deletion of the survival motor neuron 1 (SMN1) gene, leading to reduced expression levels of full-length SMN protein (7). SMN protein is known to play a critical role in several core cellular pathways, including snRNP biogenesis and pre-mRNA splicing (reviewed in 8,9). Previous studies have suggested that motor neuron-specific changes in splicing pathways may be responsible for tissue-specific targeting in SMA (10), although recent evidence has shown that these changes are likely to occur as a consequence, rather than a cause, of the disease (11).
Although SMN protein is ubiquitously expressed in tissues and organs across a range of different species, including rats, monkeys and humans (12), the extent to which SMN deficiency leads specifically to targeted pathological changes in the neuromuscular system remains unclear. For example, the fact that significant levels of SMN protein have previously been identified in the brain (12,13) raises the possibility that reduced levels of expression may have effects on non-motor regions of the nervous system. Here, we show that reduced levels of SMN protein result in regionally selective, impaired brain development in a mouse model of severe SMA [Smn−/−;SMN2 mice (14)]. In highly affected regions, such as the hippocampus, SMN deficiency resulted in lower cell density, decreased cell proliferation and decreased postnatal neurogenesis. Proteomic analysis of the hippocampus from SMA mice revealed changes in molecular pathways involved in cellular proliferation, migration and development.
RESULTS
Regionally specific modifications in brain morphology reveal abnormal postnatal brain development in a mouse model of severe SMA
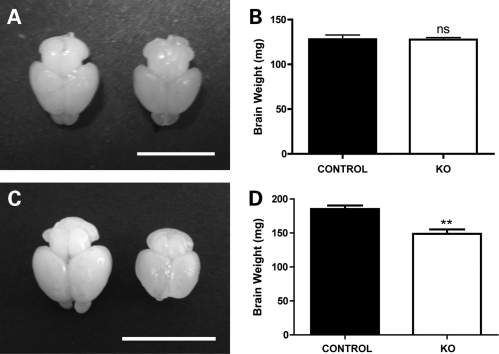
To establish whether reduced levels of SMN protein influence brain morphology, we examined postnatal brain development in an established mouse model of severe SMA [Smn−/−;SMN2 mice (14)]. A comparison of whole, freshly dissected brains from Smn−/−;SMN2 mice with brains from non-affected, wild-type littermates (Smn+/+;SMN2 mice) at a late-symptomatic disease time point (postnatal Day 5; P5) revealed a significant reduction in brain size and weight (Fig. 1C and D). Equivalent analysis at a pre-symptomatic time point (P1) revealed that brains from Smn−/−;SMN2 mice were indistinguishable from those of control littermates and were of an almost identical weight (Fig. 1A and B). Thus, gross morphological changes were apparent in the brains of mice with a severe form of SMA at late-symptomatic ages.
Figure 1.
Modified brain size and weight in mice with severe SMA. (A) Photograph showing two representative brains, one from an Smn−/−;SMN2 mouse (right) and the other from a wild-type littermate mouse (left) at a pre-symptomatic time point (postnatal Day 1; P1). There were no observable differences in the gross appearance or size of the brain between genotypes at this age. (B) Bar chart (mean ± SEM) showing mean brain weights from Smn−/−;SMN2 (−/−) and wild-type littermate (+/+) mice at P1 (+/+ n = 4 mice; −/− n = 3). There was no significant difference between the two genotypes (P > 0.05, unpaired t-test). (C) Photograph showing two representative brains, one from an Smn−/−;SMN2 mouse (right) and the other from a wild-type littermate mouse (left) at a late-symptomatic time point (P5). At this age, brains from Smn−/−;SMN2 mice were noticeably smaller than those from wild-type mice. (D) Bar chart showing mean brain weights from Smn−/−;SMN2 and wild-type littermate mice at P5 (n = 6 per genotype). There was a significant reduction in brain weight in the Smn−/−;SMN2 mice (**P < 0.01; unpaired t-test). Scale bar = 0.75 cm (A), 1 cm (C).
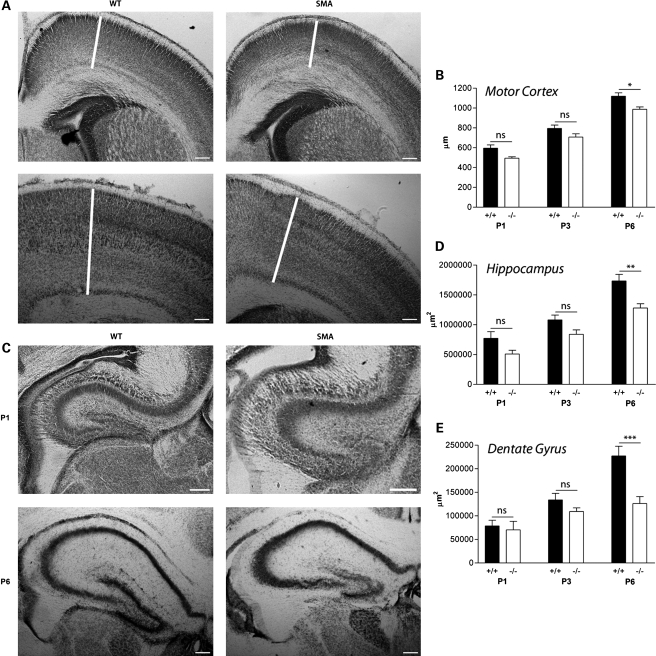
Higher-resolution morphological assessment of Nissl-stained coronal brain sections from Smn−/−;SMN2 mice at pre- and late-symptomatic time points revealed region-specific effects of reduced SMN levels. Qualitatively, the most striking morphological changes were observed in the hippocampus and particularly the hippocampal dentate gyrus. This region was noticeably smaller in Smn−/−;SMN2 mice at late-symptomatic time points (Fig. 2). Quantitative assessment confirmed our qualitative observations, as by P6 the area of the hippocampal dentate gyrus was ∼50% smaller in Smn−/−;SMN2 mice compared with wild-type littermates (Fig. 2E). Statistical comparisons of hippocampal dentate gyrus area in Smn−/−;SMN2 and littermate control mice confirmed a significant effect of the reduction in SMN levels [two-way ANOVA; P < 0.001; F(1,35) = 20.45]. Interestingly, previous reports have cited the hippocampus as a brain region likely to be particularly susceptible to SMA pathology in human patients with severe forms of SMA (15).
Figure 2.
Modified morphological development of the hippocampus and primary motor cortex in severe SMA mice. (A) Primary motor cortex (M1; white line) in Nissl-stained brain sections from wild-type (WT) and Smn−/−;SMN2 (SMA) mice at pre-symptomatic (P1) and late-symptomatic (P6) ages. Modest reductions were apparent in M1 thickness of Smn−/−;SMN2 mice at both pre- and late-symptomatic ages. (B) Bar chart (mean ± SEM) showing M1 thickness in wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P1, P3 and P6. There were modest reductions in M1 thickness at all ages, becoming statistically significant at P6 (*P < 0.05; two-way ANOVA with Bonferroni post hoc test). (C) Hippocampi from Nissl-stained brains at P1 and P6. (D) Bar chart showing total hippocampal area in wild-type and Smn−/−;SMN2 mice at P1, P3 and P6. The hippocampal area in Smn−/−;SMN2 mice was smaller than in wild-type littermates at all ages examined, but only became statistically significant at P6 (**P < 0.01). (E) Bar chart showing the hippocampal dentate gyrus area in wild-type and Smn−/−;SMN2 mice at P1, P3 and P6. The area of the dentate gyrus in Smn−/−;SMN2 mice was no different from that of wild-type littermates at P1 (ns P > 0.05). However, by P6, the area of the dentate gyrus was ∼50% smaller in Smn−/−;SMN2 mice compared with wild-type littermates (***P < 0.001). P1, n = 3 mice per genotype; P3, n = 4; P6, n = 3/4. Scale bar = 160 µm.
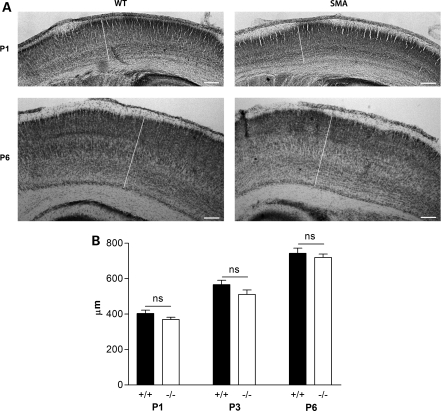
More modest morphological reductions were observed in the primary motor cortex (M1) of Smn−/−;SMN2 mice. These subtle morphological differences were present at the time of birth (pre-symptomatically) and persisted through late-symptomatic stages where they became statistically significant (Fig. 2A and B). Despite this decrease in motor cortex thickness, the primary somatosensory cortex (S1BF) remained unaffected in Smn−/−;SMN2 mice at all stages of disease progression [Fig. 3; two-way ANOVA; P > 0.05; F(1,36) = 3.93], suggesting that distinct cortical areas were differentially affected. Importantly, the finding that not all brain regions were equally affected in Smn−/−;SMN2 mice confirmed that the developmental changes identified were likely to represent specific vulnerability of regions such as the hippocampus, rather than simply occurring as a result of generalized shrinkage caused by the reduced body size of late-symptomatic Smn−/−;SMN2 mice (14).
Figure 3.
Morphological development occurs normally in the primary somatosensory cortex in severe SMA mice. (A) Photomicrographs showing representative primary somatosensory cortices (S1BF; indicated by the white line in each micrograph) from Nissl-stained sections of brains from wild-type (WT; left panels) and Smn−/−;SMN2 mice (right panels) at pre-symptomatic (P1) and late-symptomatic (P6) ages. No qualitative differences were observed in S1BF thickness between genotypes at either age. (B) Bar chart (mean ± SEM) showing average S1BF thickness in wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P1, P3 and P6. There was no significant difference observed between genotypes at any age examined (P > 0.05 for all comparisons; two-way ANOVA with Bonferroni post hoc test. P1, n = 3 mice; P3, n = 4; P6, n = 4/6). Scale bar = 160 µm.
These data suggest that perinatal growth and development of a subset of brain regions are disrupted when SMN levels are reduced to those known to cause a severe form of SMA in mice, with disruption of the hippocampal formation being particularly apparent.
Reduced density and proliferation of cells in the hippocampi of mice with severe SMA
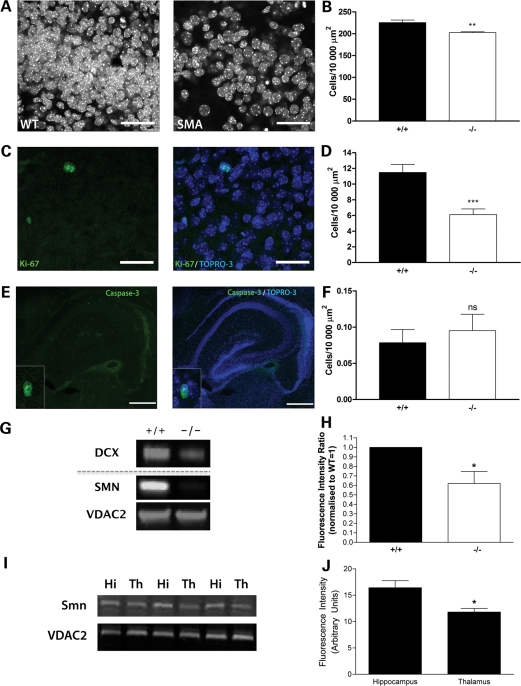
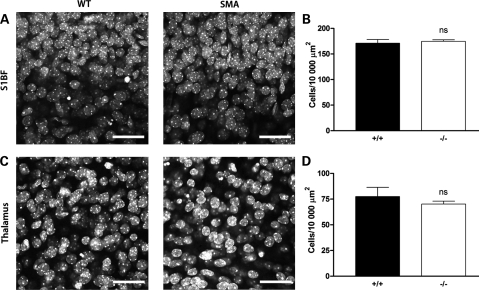
Next, we asked whether changes in cell density and/or proliferation could account for the morphological changes observed in the brains of Smn−/−;SMN2 mice. We decided to focus on the hippocampus for these experiments, as this region showed the most dramatic morphological changes (see above). Smn−/−;SMN2 mice had significantly fewer cell nuclei in the hippocampal dentate gyrus at late-symptomatic time points compared with wild-type littermate mice (Fig. 4A and B; P < 0.01, unpaired t-test). This reduced cell density correlated with a significant reduction in the numbers of proliferating cells in the hippocampus in Smn−/−;SMN2 mice, identified using Ki67 as an immunocytochemical marker of proliferation (Fig. 4C and D; P < 0.001, unpaired t-test). These experiments suggested that attenuated cell proliferation was leading to reduced numbers of cells in the hippocampus. A comparison of cell densities in two unaffected brain regions—the primary somatosensory cortex and ventral thalamic nuclei (VPM/VPL nuclei)—showed no difference in Smn−/−;SMN2 mice (Fig. 5). Similarly, cell proliferation markers were not altered in unaffected brain regions (Fig. 6).
Figure 4.
Modified brain development in the hippocampus is associated with reduced cell density and cell proliferation, but not an increase in apoptosis. (A) Confocal micrographs showing TOPRO3-labelled nuclei in the hippocampal dentate gyrus from wild-type (WT) and Smn−/−;SMN2 (SMA) mice at late-symptomatic (P5) ages. (B) Bar chart (mean ± SEM) showing cell density in the hippocampal dentate gyrus (granular layer) in wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P5. There was a significant reduction in cell density in Smn−/−;SMN2 mice (**P < 0.01; unpaired t-test). (C) Ki-67 immuno-labelling (green; marking proliferating cells) and TOPRO3-labelled nuclei (blue) in the hippocampus from a wild-type mouse. (D) Bar chart showing Ki-67 cell proliferation in the hippocampal dentate gyrus (granular layer) of wild-type and Smn−/−;SMN2 mice at P5. There was a significant reduction in the number of Ki-67-positive cells in Smn−/−;SMN2 mice (***P < 0.001; unpaired t-test). (E) Caspase-3 staining (green) and TOPRO3-labelled nuclei (blue) in the hippocampus from a P5 Smn−/−;SMN2 mouse. The high-power insets show examples of a cell positive for caspase-3 staining. (F) Bar chart showing no significant difference in the numbers of caspase-3 positive cells between Smn−/−;SMN2 and wild-type littermate mice at P5 (ns P > 0.05; unpaired t-test). (G) Representative fluorescent western blots showing reduced expression of the neurogenesis marker doublecortin (DCX) in Smn−/−;SMN2 mice (−/−). SMN levels are also shown as are those of VDAC2 as a loading control. (H) Bar chart showing quantitative analysis of fluorescent western blots revealing significantly reduced levels of DCX in the hippocampus from Smn−/−;SMN2 mice (−/−) compared with wild-type littermates (+/+) (*P < 0.05, Mann–Whitney test). (I) Triplicate fluorescent western blots showing expression levels of SMN and the VDAC2 loading control in the hippocampus (Hi) and thalamus (Th) of three wild-type mice. (J) Bar chart showing quantitative analysis of fluorescent western blots revealing significantly higher levels of SMN in the hippocampus than in the thalamus (*P < 0.05, Mann–Whitney test). n = 4 mice per genotype. Scale bar = 35 µm (A and C) and 930 µm (E).
Figure 5.
No change in cell density or cell proliferation markers in the primary somatosensory cortex or unaffected thalamic nuclei in severe SMA mice. (A) Representative confocal micrographs showing TOPRO3-labelled nuclei of all cells in the primary somatosensory cortex (S1BF) from wild-type (WT; left panel) and Smn−/−;SMN2 (right panel) mice at late-symptomatic (P5) ages. No qualitative differences in cell density were observed in Smn−/−;SMN2 mice. (B) Bar chart (mean ± SEM) showing cell density measured in the S1BF region (layers IV and V) of the brain in wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P5. There was no significant difference between the genotypes (ns P > 0.05; unpaired t-test. n = 4 mice per genotype). (C) Representative confocal micrographs showing TOPRO3-labelled nuclei of all cells in non-affected VPM/VPL thalamic nuclei from wild-type (left panel) and Smn−/−;SMN2 (right) mice at late-symptomatic (P5) ages. No qualitative differences in cell density were observed in Smn−/−;SMN2 mice. (D) Bar chart showing cell density measured in the thalamus in wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P5. There was no significant difference between the genotypes (ns P > 0.05; unpaired t-test; n = 4 mice per genotype). Scale bar = 35 µm.
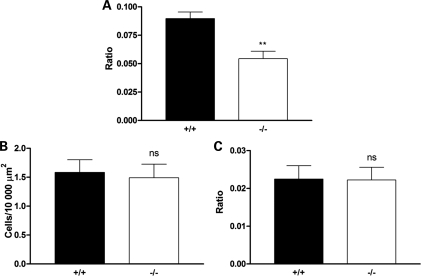
Figure 6.
Reduced cell proliferation in the hippocampus of severe SMA mice was confirmed using a ratio analysis but no change in cell proliferation markers was observed in unaffected thalamic nuclei. (A) Bar chart (mean ± SEM) showing cell proliferation identified by Ki-67 staining in the hippocampus of wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P5, analysed using a ratio analysis (ratio of Ki-67-positive cells to the total number of cells identified with TOPRO3 labelling; cf. with density analysis shown previously). There was a significant reduction in the number of Ki-67-positive cells analysed using both approaches in the Smn−/−;SMN2 mice (**P < 0.01; unpaired t-test. +/+ n = 3 mice; −/− n = 4 mice). (B and C) Bar charts showing cell proliferation identified by Ki-67 staining in the thalamus (VPM and VPL nuclei) of wild-type (+/+) and Smn−/−;SMN2 (−/−) mice at P5, analysed using both a density analysis (number of cells per 10 000 µm2; B) and a ratio analysis (ratio of Ki-67-positive cells to the total number of cells identified with TOPRO3 labelling; C). In contrast to the hippocampus, there was no reduction in the number of Ki-67-positive cells in the thalamus in Smn−/−;SMN2 mice (ns P > 0.05. +/+ n = 3 mice; −/− n = 4 mice). Scale bar = 35 µm.
To test whether the reduced cell density in the hippocampus was occurring as a result of increased postnatal apoptotic cell death, we compared caspase 3 expression in the hippocampus of late-symptomatic Smn−/−;SMN2 and wild-type littermate mice. Cells labelling positive for caspase 3 were found in very low numbers in the hippocampus of both SMN−/−;SMN2 and wild-type littermate mice (Fig. 4E), and no significant difference was observed between the genotypes (Fig. 4F; P > 0.05, unpaired t-test). This suggested that reduced cell densities were the result of abnormal proliferation rather than a widespread loss of cells through activation of apoptotic cell death pathways. Experiments comparing levels of cell proliferation markers and apoptotic markers in SMN−/−;SMN2 and wild-type littermate mice were also analysed as the ratio of the total number of cells present, with the same results obtained as for those analysed using cells per unit area (Fig. 6).
As the hippocampus is one of the few regions where neurogenesis can occur in the postnatal mammalian brain (16,17), we next examined whether changes in expression levels of established neurogenic markers were modified in the hippocampus of SMN−/−;SMN2 mice. Expression levels of the neurogenesis marker doublecortin were quantified using fluorescent western blotting and revealed an ∼40% decrease in SMN−/−;SMN2 mice compared with wild-type littermate mice at P5 (Fig. 4G and H). We also observed an ∼25% reduction in the expression levels of the neurogenesis marker TUC-4 in the hippocampus of SMN−/−;SMN2 mice, although variability in the expression levels between individual mice of both genotypes [consistent with previous reports (18)] meant that this did not reach statistical significance (data not shown). This finding suggests that postnatal hippocampal neurogenesis was significantly impaired in SMN−/−;SMN2 mice and was likely to contribute to the reductions in cell density and proliferation previously observed.
Modifications in morphology, cell density and proliferation were most pronounced in brain regions with higher normal expression levels of SMN protein
Quantitative analysis of basal SMN protein levels in brains from wild-type mice confirmed higher expression levels in affected regions such as the hippocampus compared with non-affected regions such as thalamic VPM/VPL nuclei (Fig. 4I and J). These data are supported by previous reports of SMN transcript levels in the mammalian brain, where particularly high expression was noted in the hippocampus (12). Taken together, this suggests that regions found to be most affected in the brains of SMN−/−;SMN2 mice (e.g. the hippocampus) were those normally associated with higher basal expression of SMN in the healthy postnatal brain.
Proteomic analysis of the hippocampus in mice with severe SMA reveals widespread modifications in expression levels of proteins required for cellular proliferation, migration and development
In an attempt to identify molecular correlates of the morphological and cellular changes observed in the brains of mice with severe SMA, a quantitative proteomics analysis was undertaken on isolated hippocampal tissues from SMN−/−;SMN2 and wild-type littermate mice at a late-symptomatic age (P5). Hippocampi from six Smn−/−;SMN2 and six wild-type littermate mice were harvested for the analysis. Using a quantitative proteomics approach (iTRAQ; see Materials and Methods for details), 219 347 individual peptide sequences were submitted to MASCOT for identification. As an indication of identification certainty, the false discovery rate for peptide matches above the identity threshold was 2.06%. In order for inclusion and further analysis, a protein had to be identified by at least two peptides. In order to increase stringency in reporting, a minimum cut-off of 20% change against wild-type littermates was used.
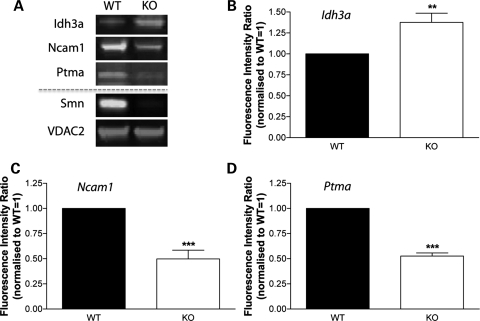
A direct comparison of the hippocampal proteome from Smn−/−;SMN2 and wild-type littermate mice revealed that 39 out of the 144 (27%) individual proteins identifiable across all samples had modified expression levels >20% in the hippocampus from Smn−/−;SMN2 mice (Table 1). Thirteen proteins had increased expression levels in Smn−/−;SMN2 mice compared with wild-type littermates and 26 proteins had reduced expression levels (Table 1). Quantitative fluorescent western blotting was undertaken on freshly prepared hippocampal tissues to validate the proteomics data (Fig. 7A). Three individual proteins were selected for validation because they showed a relatively large magnitude of change and because they were identified by low numbers of unique peptides: isocitrate dehydrogenase (NAD) subunit alpha (Idh3a) as a protein with increased expression in the hippocampus of Smn−/−;SMN2 mice, and prothymosin alpha (Ptma) and neural cell adhesion molecule 1 (Ncam1) as proteins with decreased expression. All three proteins showed similar, significant changes in expression in the hippocampus from Smn−/−;SMN2 mice when assessed using western blotting, confirming the validity and reliability of the proteomics data (Fig. 7B–D).
Table 1.
Proteins with a change in expression levels >20% in the hippocampus of Smn−/−;SMN2 mice (KO) compared with littermate controls (WT)
| Gene name | Protein name | Accession number | Mol weight (Da) | Average ratio (KO/WT) ± SD | Peptides used/unique | Ion score (Mascot) | P-value |
|---|---|---|---|---|---|---|---|
| Proteins upregulated in SMA mice | |||||||
| Idh3a | Isoform 2 of isocitrate dehydrogenase subunit α mitochondrial | IPI00608078.1 | 34 924 | 2.486 ± 0.067 | 3/1 | 44 | <0.0001 |
| Idh3a | Isoform 1 of isocitrate dehydrogenase subunit α mitochondrial | IPI00459725.2 | 43 816 | 2.486 ± 0.067 | 3/1 | 44 | <0.0001 |
| Atp5b | Synthase subunit beta, mitochondrial ATP | IPI00468481.2 | 59 724 | 1.475 ± 0.374 | 21/4 | 277 | <0.0001 |
| Try4 | Trypsin 4 | IPI00128108.1 | 28 382 | 1.437 ± 0.080 | 8/1 | 114 | <0.0001 |
| Pgam1 | Phosphoglycerate mutase 1 | IPI00457898.3 | 31 666 | 1.396 ± 0.161 | 3/2 | 60 | 0.0065 |
| Gm5803 | Predicted gene 5803 | IPI00947561.1 | 37 371 | 1.341 ± nn | 4/1 | 53 | nn |
| Tpm3 | Isoform 2 of tropomyosin alpha-3 chain | IPI00230044.5 | 32 689 | 1.335 ± 0.185 | 3/1 | 51 | 0.0175 |
| Mapt | Microtubule-associated protein | IPI00648564.1 | 47 178 | 1.258 ± nn | 22/5 | 285 | nn |
| Dbi | Diazepam binding inhibitor isoform 1 | IPI00222430.5 | 17 726 | 1.254 ± 0.127 | 10/2 | 182 | <0.0001 |
| Sod1 | Superoxide dismutase | IPI00458744.1 | 13 979 | 1.251 ± 0.100 | 3/1 | 55 | 0.0061 |
| Mapt | Isoform Tau-C of microtubule-associated protein tau | IPI00230408.2 | 40 793 | 1.247 ± nn | 25/6 | 326 | nn |
| Ywhah | 14-3-3 protein eta | IPI00227392.5 | 31 391 | 1.238 ± 0.085 | 74/4 | 763 | <0.0001 |
| Mtap2 | Microtubule-associated protein 2 | IPI00118075.1 | 221 307 | 1.237 ± nn | 12/4 | 175 | nn |
| Proteins downregulated in SMA mice | |||||||
| Ptma | Prothymosin alpha | IPI00224784.4 | 13 544 | 0.444 ± 1.123 | 3/1 | 66 | 0.2197 |
| Gm9034 | Similar to glyceraldehyde-3-phosphate dehydrogenase | IPI00753102.2 | 40 633 | 0.541 ± 0.060 | 5/1 | 36 | <0.0001 |
| Gm10290 | Similar to glyceraldehyde-3-phosphate dehydrogenase isoform 1 | IPI00752289.2 | 40 199 | 0.617 ± 0.099 | 20/2 | 236 | <0.0001 |
| Cct2 | T-complex protein 1 subunit beta | IPI00320217.9 | 63 259 | 0.644 ± 0.304 | 4/1 | 113 | 0.0288 |
| Gm12141 | similar to heat shock protein 1 | IPI00749932.1 | 65 138 | 0.657 ± nn | 4/1 | 131 | nn |
| Rplp1 | 60S acidic ribosomal protein P1 | IPI00113377.1 | 12 735 | 0.662 ± 0.063 | 4/1 | 73 | <0.0001 |
| Hspd1 | Isoform 1 of 60 kDa heat shock protein, mitochondrial | IPI00308885.6 | 69 014 | 0.667 ± 0.161 | 15/2 | 454 | <0.0001 |
| Aldoa | Fructose-bisphosphate aldolase | IPI00856379.1 | 49 583 | 0.674 ± nn | 7/2 | 135 | nn |
| Gm7293 | Similar to glyceraldehyde-3-phosphate dehydrogenase | IPI00751677.3 | 40 369 | 0.677 ± nn | 7/3 | 50 | nn |
| Gm14072 | Hypothetical protein isoform 3 | IPI00125787.4 | 18 543 | 0.692 ± 0.282 | 2/1 | 42 | n/a |
| Eif5a | Eukaryotic translation initiation factor 5A-1 | IPI00108125.4 | 19 067 | 0.693 ± 0.231 | 11/2 | 128 | <0.0001 |
| Gm3222 | Similar to glyceraldehyde-3-phosphate dehydrogenase | IPI00849879.1 | 44 463 | 0.694 ± nn | 24/5 | 269 | nn |
| Rps19 | 15 kDa protein | IPI00857038.1 | 17 428 | 0.704 ± 0.173 | 3/2 | 46 | 0.0207 |
| Gm8683 | Hypothetical protein | IPI00875929.1 | 18 520 | 0.704 ± 0.173 | 3/2 | 46 | 0.0207 |
| Sncb | Beta-synuclein | IPI00131614.1 | 15 916 | 0.728 ± 0.133 | 4/2 | 38 | 0.0032 |
| H2afv | 14 kDa protein | IPI00555055.3 | 15 761 | 0.771 ± 0.122 | 3/1 | 41 | 0.0156 |
| Marcksl1 | MARCKS-related protein | IPI00281011.7 | 22 948 | 0.773 ± 0.127 | 14/2 | 181 | <0.0001 |
| Calm1 | Calmodulin | IPI00761696.2 | 18 124 | 0.774 ± 0.143 | 20/4 | 167 | <0.0001 |
| Ncam1 | Isoform 1 of neural cell adhesion molecule 1 | IPI00122971.2 | 130 527 | 0.776 ± 0.113 | 4/2 | 38 | 0.0037 |
| Marcks | Myristoylated alanine-rich C-kinase substrate | IPI00229534.5 | 33 159 | 0.778 ± 0.097 | 8/1 | 121 | <0.0001 |
| Prdx2 | 16 kDa protein | IPI00831115.3 | 17 619 | 0.78 ± 0.105 | 22/4 | 177 | <0.0001 |
| Arf4 | ADP-ribosylation factor 4 | IPI00331663.17 | 22 227 | 0.782 ± 0.051 | 2/2 | 51 | n/a |
| Gap43 | Neuromodulin | IPI00128973.1 | 28 200 | 0.789 ± 0.157 | 28/3 | 361 | <0.0001 |
| Nrgn | Neurogranin | IPI00380227.1 | 8728 | 0.795 ± 0.123 | 8/1 | 126 | 0.0002 |
| Rbm31y | MCG53108 | IPI00136573.1 | 66 759 | 0.796 ± 0.068 | 14/1 | 128 | <0.0001 |
| Tmsb4x | Isoform short of thymosin beta-4 | IPI00228757.1 | 6491 | 0.796 ± 0.123 | 18/3 | 104 | <0.0001 |
P-values were calculated using an unpaired two-tailed t-test. nn, data not normally distributed so SD and P-value not available. n/a, P-value not calculated due to n < 3 peptides.
Figure 7.
Protein expression changes identified in the proteomics screen of SMA mouse hippocampus were validated using quantitative fluorescent western blotting. (A) Representative fluorescent western blots showing expression levels of Idh3a, Ncam1 and Ptma in the hippocampus of Smn−/−;SMN2 (KO) mice and wild-type littermates (WT) at P5. Smn protein levels are shown to confirm the genetic status of the Smn−/−;SMN2 mice and VDAC2 protein is shown as a loading control. Note the increased level of expression of Idh3a in the Smn−/−;SMN2 mice, but decreased levels of both Ncam1 and Ptma, in agreement with data from our proteomics experiments (Table 1). (B–D) Bar charts (mean ± SEM) showing relative expression levels of Idh3a (B), Ncam1 (C) and Ptma (D) in wild-type (black bars) and Smn−/−;SMN2 (white bars) mice at P5. Note the significant increase in expression of Idh3a (**P < 0.01; Mann–Whitney test), and significant decrease in expression of both Ncam1 (***P < 0.001) and Ptma (***P < 0.001), in Smn−/−;SMN2 mice. n = minimum of three mice per genotype.
In order to confirm that the protein changes identified in the hippocampus of Smn−/−;SMN2 mice were specific responses to reduced SMN levels in affected tissues, we also quantified expression levels of Ptma, Idh3a and Ncam1 in brain tissues containing less affected regions as well as liver and skin (Supplementary Material, Fig. S1). None of the proteins examined showed changes in expression levels of a similar magnitude or direction to those observed in the hippocampus.
Finally, to identify specific molecular pathways and networks disrupted in the hippocampus of Smn−/−;SMN2 mice, we undertook an in silico pathway analysis of our proteomics data set using Ingenuity Pathway Analysis software. This software incorporates information extracted by experts from the full text of the scientific literature, allowing analysis of candidate proteins with respect to their known roles in biological functions. This analysis revealed that many of the proteins found to be modified in the hippocampus from Smn−/−;SMN2 mice are involved in pathways related to known genetic disorders including neurological, skeletal and muscular disorders (Table 2). For example, 13 of our candidate proteins (representing 43.3% of our total candidate list) have previously been shown to have a role in neurological disorders. Importantly, many of the proteins modified in the hippocampus in response to low SMN levels are known to be the key regulators of cell proliferation, growth, migration and morphological development (Table 2).
Table 2.
In silico functional pathways analysis of proteomics data shown in Table 1
| Functional annotation | Number of proteins | % of candidate list | Proteins |
|---|---|---|---|
| Genetic disorder | 17 | 56.7 | ALDOA, DBI, GAP43, HSPD1, MAPT, MARCKS, MARCKSL1, NCAM1, NRGN, PGAM1, PRDX1, PRDX2, PTMA, RPS19, SNCB, SOD1, TMSB4X, TPM3 |
| Neurological disorder | 13 | 43.3 | GAP43, HSPD1, MAPT, MARCKS, MARCKSL1, NCAM1, NRGN, PGAM1, PRDX1, PRDX2, SNCB, SOD1, TMSB4X, TPM3 |
| Cell death | 13 | 43.3 | ALDOA, CCT2, EIF5A, HSPD1, MAPT, NCAM1, NRGN, PRDX2, PTMA, SNCB, SOD1, TMSB4X, TPM3 |
| Skeletal and muscular disorder | 10 | 33.3 | ALDOA, GAP43, MAPT, NRGN, PRDX2, PTMA, SNCB, SOD1, TMSB4X, TPM3 |
| Proliferation of cells | 9 | 30.0 | EIF5A, HSPD1, MAPT, MARCKSL1, NCAM1, PRDX2, PTMA, RPS19, TPM3 |
| Growth of cells | 9 | 30.0 | ATP5B, CALR, CCT2, EIF5A, GAP43, HNRNPD, HSPD1, MAPT, PTMA, SOD1, TMSB4X |
| Migration of cells | 8 | 26.7 | HSPD1, MAPT, MARCKS, NCAM1, PPIB, RPS19, TMSB4X, TPM3 |
| Morphology of cells | 7 | 23.3 | GAP43, MAPT, MARCKS, MARCKSL1, PRDX2, TMSB4X, TPM3 |
| Quantity of calcium | 6 | 20.0 | MARCKSL1, NCAM1, NRGN, PPIB, PRDX2, SOD1 |
| Developmental disorder | 6 | 20.0 | GAP43, MAPT, MARCKS, MARCKSL1, SOD1, TPM3 |
| Development of cells | 6 | 20.0 | GAP43, HSPD1, MARCKS, NCAM1, PRDX2, SOD1 |
| Differentiation of cells | 6 | 20.0 | GAP43, HSPD1, MAPT, NCAM1, PRDX2, RPS19 |
| Cell movement | 6 | 20.0 | HSPD1, MARCKS, RPS19, SOD1, TMSB4X, TPM3 |
| Adhesion of cells | 5 | 16.7 | MARCKS, NCAM1, PPIB, PRDX2, TMSB4X |
| Outgrowth of neurites | 5 | 16.7 | GAP43, MAPT, MARCKS, NCAM1, TPM3 |
| Transport of protein | 4 | 13.3 | ARF4, CALR, EIF5A, MAPT |
| Formation of filaments | 4 | 13.3 | GAP43, MAP1A, MAPT, TPM3 |
DISCUSSION
Data presented in the current study show for the first time that high levels of SMN protein are required for normal brain development in vivo. As a result, reduced expression of SMN protein—down to levels sufficient to cause a severe form of SMA in mice—caused abnormal brain development, particularly affecting regions such as the hippocampus. We have determined that reduced levels of cellular proliferation and postnatal hippocampal neurogenesis, as well as modified expression levels of proteins regulating proliferation, migration and developmental processes, underlie these abnormalities and are likely to contribute to disease pathogenesis in severe forms of SMA. The identification of downstream molecular consequences of reduced SMN expression in the brain also provides a significant opportunity to gain new insights into the cell biology of the SMN protein and its roles during development of the nervous system. Taken together, these findings suggest that attempts to develop novel therapeutic approaches for SMA will need to take into account changes occurring in numerous regions of the nervous system, including the brain, when considering treatment targets and delivery options.
Our finding that low SMN levels disrupt cell proliferation and neurogenesis in vivo provides the first direct evidence of modified developmental processes in the brain during severe forms of SMA. The observation that the hippocampus was particularly severely affected is in keeping with previous reports showing high levels of SMN expression in hippocampal neurons (12,19). It would now be of interest to examine cell-type-specific responses in the hippocampus given published data showing differential expression of SMN (19). Our novel findings also provide indirect support to preliminary reports suggesting that some lower motor neurons may be vulnerable in SMA due to aberrant developmental processes. For example, microarray analysis of the spinal cord from mice with a severe form of SMA has suggested that normal proliferative pathways may be affected (11). Subsequent preliminary morphological investigations of proliferating cells in the ventral horn of the spinal cord suggested that numbers may be reduced in SMA mice (11). In addition, Simic and colleagues (20,21) have proposed a model whereby SMA pathogenesis in lower motor neurons may occur via a combination of impaired axonogenesis, disrupted synapse formation and abnormal motor neuron differentiation and migration. Although recent studies have demonstrated that impaired axonogenesis and disrupted synapse formation of lower motor neuron processes are unlikely to be a major factor in SMA pathogenesis in vivo (22), the finding that populations of heterotopic lower motor neurons, which are migratory and displaced, are dramatically increased in SMA spinal cord (20,21), alongside data from the current study, suggest that pathways regulating cellular proliferation and development may be influencing pathogenesis in both motor and non-motor regions of the nervous system. What remains unclear is whether non-neuronal cells are affected alongside neuronal cells in affected brain regions as well as in the spinal cord. It will now be important to establish whether specific populations of cells (e.g. neurons, oligodendrocytes and/or microglia) are particularly susceptible to the effects of low SMN levels in the affected brain regions identified in this study.
The current study also adds a significant new dimension to the hypothesis that low levels of SMN protein can have effects on cells and tissues beyond the neuromuscular system. It has previously been shown that SMA patients have an increased incidence of congenital bone fractures, and mice with a severe form of SMA are known to have decreased levels of osteoblast differentiation markers (23). Similarly, the SMN protein might be required for normal cardiogenesis, as approximately three out of four patients with a single copy of the SMN2 gene were shown to have haemodynamically relevant atrial or ventricular septal defects (24), and SMN levels have been reported to influence myoblast proliferation (25). From a neurobiological perspective, previous reports have demonstrated that populations of neurons outside the motor system, notably sensory neurons in the periphery, are affected when SMN levels are severely depleted in both humans and mice (26,27). Taken together, these findings led to the generation of a ‘threshold’ hypothesis concerning the vulnerability of different tissues and cells to pathological changes in response to low SMN levels, where lower motor neurons are the most sensitive cell type, but other cells and tissues sit along a sliding scale of vulnerability. As the mouse model employed in the current study recapitulates the physical and genetic characteristics of a very severe form of SMA (equivalent to type 0 or type I SMA in human patients), it is possible that the pathological changes observed during brain development have been revealed because the affected regions are towards the vulnerable end of a resistance–vulnerability spectrum. As a result, it will now be of interest to establish whether similar pathological changes can be observed, probably occurring at lower levels (as patients with less severe forms of SMA show no evidence of brain dysfunction or cognitive impairment), in mouse models with a less severe form of the disease [e.g. Smn+/− mice (28)].
Finally, data presented here support sporadic reports in the literature detailing low-level cytopathological changes in the brain of human SMA patients (15,29–36), suggesting that pathological changes in the nervous system of human patients with the severest forms of SMA may extend beyond the neuromuscular system. Taken together, these findings suggest that detailed neuropathological and/or neuroimaging studies on human patients with the severest forms of SMA are now required to accurately determine the long-term effects of low SMN levels on human brain development. The likelihood of undertaking such experiments on human post-mortem material alone remains low because of a lack of available tissues resulting from genetic diagnosis rather than a diagnosis at autopsy. Moreover, many of the currently available brain samples of the SMA patient do not have the required associated level of genetic information needed to accurately correlate pathological changes with SMN expression levels (e.g. SMN1 mutation analysis and SMN2 copy number). Detailed studies of human SMA patients utilizing modern neuroimaging techniques are therefore likely to be required.
MATERIALS AND METHODS
Mice
Breeding pairs of Smn+/−;SMN2 mice (14) were originally obtained from Jackson Laboratories. Breeding was carried out under licence from the UK Home Office. Mice were bred to produce litters that contained animals which were wild-type (Smn+/+;SMN2), heterozygous for the SMN mutation (Smn+/−;SMN2) and homozygous for the mutation (Smn−/−;SMN2). Data were obtained from tissues harvested from mice between postnatal Day 1 (P1; pre-symptomatic) and P5/6 (late-symptomatic). A minimum of three mice per genotype were used in each experiment. Non-affected wild-type littermate mice (Smn+/+;SMN2) were used as controls throughout the study. Neonatal mice were sacrificed by ice-bath chilling and decapitation, in accordance with the guidance and rules of the UK Home Office. All mice were genotyped using standard PCR methods (14,37).
Tissue sections
Whole brains were removed from the exposed cranial cavity, immediately collected into 4% paraformaldehyde solution and fixed overnight before being transferred into 0.1 m PBS and stored at 4°C. Prior to histological sectioning, brains were cryoprotected overnight in distilled water containing 30% sucrose at 4°C. Each brain was weighed and photographed before being mounted on a freezing microtome in OCT Embedding Matrix and cut coronally at a thickness of 48 µm for Nissl staining and 24 µm for immunohistochemical staining. Nissl stain was realized using a standard protocol and mounted in distyrene/plasticizer/xylene (DPX). For cell proliferation analysis, coronal sections were immunohistochemically labelled with primary antibody (anti-Ki-67 polyclonal antibody, 1:200, Abcam Cat No. 15580), secondary antibody (FITC-conjugated Ig, 1:50, DAKO Cat No. F0205) and co-stained with fluorescent nuclear dye (TOPRO-3, 1:100, Invitrogen Molecular Probes Cat No. T3605). Only Ki-67-positive cells directly corresponding to TOPRO-labelled cell nuclei were included in cell proliferation counts. For cell degeneration analysis, coronal sections were immunohistochemically labelled with primary antibody (anti-active Caspase-3 monoclonal antibody, 1:100 dilution, BD Biosciences Cat No. 559565), secondary antibody (FITC-conjugated Ig, 1:50) and co-stained with fluorescent nuclear dye (TOPRO-3, 1:100).
Microscopy
Nissl-stained sections were imaged on an inverted bright-field microscope (Olympus IX71S1F microscope) with a chilled CCD camera (Hamamatsu C4742-95). Calibrated photomicrographs (×4 magnification, ×1 binning) were collected using Improvision Openlab software and imported into Image J for morphological analysis. Immunostained sections were mounted in Mowoil® (Calbiochem) and imaged on a laser scanning confocal microscope (Zeiss LSM 510) using a ×63 magnification, oil-immersion lens (Plan-Apochromat 63×, NA 1.4).
The precise depth of coronal brain sections selected for morphological analyses was standardized using robust, easily identifiable neuroanatomical landmarks illustrated in the Paxinos Mouse Brain Atlas. All depth measurements are described relative to Bregma. The decussation of the corpus callosum (CC) at Bregma 1.10 mm and the obliquely oriented fasciola corneum (FC) at Bregma −1.70 mm were employed as neuroanatomical landmarks to identify sections containing the primary motor cortex, hippocampal formation and primary somatosensory cortex. The thickness of the primary motor cortex was measured on three consecutive sections from each side of the brain. In addition to the section in which the CC first decussates, the sections immediately before and after were also selected for measurement, approximating to Bregma 1.15, 1.10 and 1.05 mm in the Paxinos Mouse Brain Atlas. On each section, measurements were taken at three equidistant points 250, 500 and 750 µm lateral to the angle of the CC. The average of nine thickness measurements (three measurements from three sections) taken on each side of the brain was used to calculate the average thickness. A similar approach was taken to measure the thickness of primary somatosensory cortex, using the section in which the FC was first observed in an oblique orientation, as well as sections immediately before and after, approximating to Bregma −1.65, −1.70 and −1.75 mm. On the same sections, the complete hippocampal area (excluding the fimbria) and the complete dentate gyrus area were measured on each side of the brain. All measurements were performed on randomized photomicrographs using Image J software by a single researcher blinded to specimen genotype.
Protein extraction and quantitative western blot analysis
Brains were rapidly removed and briefly chilled in ice-cold ACSF (125 mm NaCl, 26 mm NaHCO3, 25 mm glucose, 2.5 mm KCl, 1.25 mm NaH2PO4, 1 mm CaCl2, 4 mm MgCl2) before regions such as the VPM/VPL thalamic nuclei and hippocampus were microdissected out. Quantitative fluorescent western blotting was performed as previously described (38–40). Membranes were incubated with primary antibodies according to the manufacturer's instructions [N-CAM1, Sigma; SMN, doublecortin and PTα (N-18), Santa Cruz; IDH3A, Lifespan Biosciences; VDAC2, β-actin and βIII-tubulin, Abcam; TUC-4, Chemicon]. Odyssey secondary antibodies were added according to the manufacturer's instructions (goat anti-rabbit IRDye 680 and goat anti-mouse IRDye 800). Blots were imaged using an Odyssey Infrared Imaging System (Li-COR Biosciences) at 169 µm resolution.
iTRAQ proteomics analysis
Protein was extracted in an MEBC buffer (50 mm Tris, 100 mm NaCl, 5 mm NaEDTA, 5 mm NaEGTA, 40 mm β-glycerophosphate, 100 mm sodium fluoride (NaF), 100 mm sodium orthovanadate, 0.25% NP-40, 1 Roche ‘complete’ protease inhibitor tablet, pH 7.4) before acetone precipitation and labelling for iTRAQ analysis.
Hippocampal samples (n = 6 mice per genotype) were precipitated with −20°C chilled acetone (1:4, v/v) and stored at −20°C overnight. The precipitates were spun at 4°C for 10 min then washed with an acetone:water mixture (4:1,v/v) twice prior to air-drying. The pellets were then resuspended in an iTRAQ sample buffer [25 µl of 500 mm TEAB, 1 µl of denaturant (2% SDS) and 2 µl of reducing agent (TCEP)]. The samples were allowed to incubate for 1 h at 60°C prior to protein estimation in triplicate (3 × 1 µl) by microBCA assay (Pierce).
Aliquots of each sample equivalent to 100 µg were made up to 28 µl using the iTRAQ sample buffer minus denaturant. To each sample, 1 µl of 84 mm indole-3-acetic acid (IAA) was added, and the samples were mixed and spun prior to incubation at room temperature in the dark for 30 min. To each sample, 10 µl of a 1 µg/µl solution of trypsin (sequencing grade, Roche) in water was added and the samples incubated overnight on a shaking platform at 30°C. To each vial of iTRAQ reagent, 70 µl of ethanol was added, mixed and spun prior to transfer to each sample vial (WT-116, SMA-114). The pH value was checked for each sample to ensure pH was >8.0 prior to incubation for 1 h at room temperature. One hundred microlitres of water were added to each sample to quench the reaction prior to pooling of the two iTRAQ-labelled samples and subsequent drying by vacuum centrifugation.
The pooled iTRAQ sample was re-suspended in 25% acetonitrile/0.1% formic acid (50 ml) prior to loading through a home-made SCX ziptip using a 10% slurry of Poros HS (10 ml) in a methanol:water mixture (50:50). The SCX ziptip was equilibrated with 25% acetonitrile/0.1% formic acid (3 × 25 ml) prior to loading of the pooled iTRAQ sample. After sample loading, the SCX ziptip was washed with 25% acetonitrile in 0.1% formic acid (3 × 25 ml). The bound iTRAQ peptides were eluted from the SCX ziptip using increasing millimolar concentrations of sodium chloride (5, 10, 20, 50, 100,150, 200, 150, 300, 400, 800) in 25% acetonitrile/0.1% formic acid (2 × 25 ml). A final elution of the SCX ziptip with 200 mm NH4OH in 50% propanol was used to remove most of the hydrophobic peptides. Each fraction from the SCX ziptip was then dried by vacuum centrifugation and stored until analysed by nano liquid chromatography-mass spectrometry/mass spectrometry (nLC-MS/MS). Prior to the analysis, each dried SCX fraction was re-suspended in 35 ml of 1% formic acid and 10 ml aliquots were injected onto an Agilent 6520 Q-TOF using an Agilent 1200 series nanoLC system with microfluidic interface.
Raw data files were converted to mascot generic file (mgf) by MassHunter workstation software prior to merging of the files with Mascot Daemon and subsequent database (IPI Mouse) searching with the Mascot search engine (Matrix Science, Version 2.2). In keeping with recent recommendations for publication of proteomics data (41), full details of iTRAQ methodology can be found in Supplementary Methods.
Statistical analysis
All statistical analyses were completed using GraphPad Prism software. P-values of <0.05 were considered to be statistically significant for all analyses (*P < 0.05; **P < 0.01; ***P < 0.001).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from BDF Newlife (to T.H.G., K.T.), the SMA Trust (to T.H.G.), the Wellcome Trust (to T.H.G., T.M.W.) and the Anatomical Society of Great Britain & Ireland (to L.M.M., T.H.G.). J.P.-W.H. was supported by the Advanced Medical Sciences program from the University of Melbourne.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to members of the Gillingwater and Parson Laboratories for advice and assistance with this study, and Mr Kenneth Beattie and Dr Wenzhang Chen for assistance with proteomics experiments.
Conflict of Interest statement. The authors have no potential conflicts of interest to declare.
REFERENCES
- 1.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. doi:10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 2.Ferri A., Melki J., Kato A.C. Progressive and selective degeneration of motoneurons in a mouse model of SMA. Neuroreport. 2004;15:275–280. doi: 10.1097/00001756-200402090-00013. doi:10.1097/00001756-200402090-00013. [DOI] [PubMed] [Google Scholar]
- 3.Swoboda K.J., Prior T.W., Scott C.B., McNaught T.P., Wride M.C., Reyna S.P., Bromberg M.B. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann. Neurol. 2005;57:704–712. doi: 10.1002/ana.20473. doi:10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray L.M., Talbot K., Gillingwater T.H. Neuromuscular synaptic vulnerability in motor neurone disease: amyotrophic lateral sclerosis and spinal muscular atrophy. Neuropathol. Appl. Neurobiol. 2010;36:133–156. doi: 10.1111/j.1365-2990.2010.01061.x. [DOI] [PubMed] [Google Scholar]
- 5.Lunn M.R., Wang C.H. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. doi:10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 6.Han J.J., McDonald C.M. Diagnosis and clinical management of spinal muscular atrophy. Phys. Med. Rehabil. Clin. N. Am. 2008;19:661–680. doi: 10.1016/j.pmr.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. doi:10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 8.Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. doi:10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Burghes A.H., Beattie C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. doi:10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. doi:10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bäumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. doi:10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battaglia G., Princivalle A., Forti F., Lizier C., Zeviani M. Expression of the SMN gene, the spinal muscular atrophy determining gene, in the mammalian central nervous system. Hum. Mol. Genet. 1997;6:1961–1971. doi: 10.1093/hmg/6.11.1961. doi:10.1093/hmg/6.11.1961. [DOI] [PubMed] [Google Scholar]
- 13.Briese M., Richter D.U., Sattelle D.B., Ulfig N. SMN, the product of the spinal muscular atrophy-determining gene, is expressed widely but selectively in the developing human forebrain. J. Comp. Neurol. 2006;497:808–816. doi: 10.1002/cne.21010. doi:10.1002/cne.21010. [DOI] [PubMed] [Google Scholar]
- 14.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. doi:10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 15.Higashi K., Nakagawa M., Higuchi I., Saito K., Osame M. Genetically confirmed spinal muscular atrophy type III with epilepsy, cerebral hypoperfusion, and parahippocampal gyrus atrophy. Rinsho Shinkeigaku. 2000;40:334–338. [PubMed] [Google Scholar]
- 16.Eriksson P.S., Perfilieva E., Björk-Eriksson T., Alborn A.M., Nordborg C., Peterson D.A., Gage F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998;4:1313–1317. doi: 10.1038/3305. doi:10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 17.Cameron H.A., McKay R.D. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J. Comp. Neurol. 2001;435:406–417. doi: 10.1002/cne.1040. doi:10.1002/cne.1040. [DOI] [PubMed] [Google Scholar]
- 18.Jin K., Peel A.L., Mao X.O., Xie L., Cottrell B.A., Henshall D.C., Greenberg D.A. Increased hippocampal neurogenesis in Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2004;101:343–347. doi: 10.1073/pnas.2634794100. doi:10.1073/pnas.2634794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. doi:10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 20.Simic G., Mladinov M., Seso Simic D., Jovanov Milosevic N., Islam A., Pajtak A., Barisic N., Sertic J., Lucassen P.J., Hof P.R., et al. Abnormal motoneuron migration, differentiation, and axon outgrowth in spinal muscular atrophy. Acta Neuropathol. 2008;115:313–326. doi: 10.1007/s00401-007-0327-1. doi:10.1007/s00401-007-0327-1. [DOI] [PubMed] [Google Scholar]
- 21.Simic G. Pathogenesis of proximal autosomal recessive spinal muscular atrophy. Acta Neuropathol. 2008;116:223–234. doi: 10.1007/s00401-008-0411-1. doi:10.1007/s00401-008-0411-1. [DOI] [PubMed] [Google Scholar]
- 22.Murray L.M., Lee S., Bäumer D., Parson S.H., Talbot K., Gillingwater T.H. Pre-symptomatic development of lower motor neuron connectivity in a mouse model of severe spinal muscular atrophy. Hum. Mol. Genet. 2010;19:420–433. doi: 10.1093/hmg/ddp506. doi:10.1093/hmg/ddp506. [DOI] [PubMed] [Google Scholar]
- 23.Shanmugarajan S., Swoboda K.J., Iannaccone S.T., Ries W.L., Maria B.L., Reddy S.V. Congenital bone fractures in spinal muscular atrophy: functional role for SMN protein in bone remodeling. J. Child Neurol. 2007;22:967–973. doi: 10.1177/0883073807305664. doi:10.1177/0883073807305664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudnik-Schöneborn S., Heller R., Berg C., Betzler C., Grimm T., Eggermann T., Eggermann K., Wirth R., Wirth B., Zerres K. Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J. Med. Genet. 2008;45:635–638. doi: 10.1136/jmg.2008.057950. doi:10.1136/jmg.2008.057950. [DOI] [PubMed] [Google Scholar]
- 25.Shafey D., Côté P.D., Kothary R. Hypomorphic Smn knockdown C2C12 myoblasts reveal intrinsic defects in myoblast fusion and myotube morphology. Exp. Cell Res. 2005;311:49–61. doi: 10.1016/j.yexcr.2005.08.019. doi:10.1016/j.yexcr.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Rudnik-Schöneborn S., Goebel H.H., Schlote W., Molaian S., Omran H., Ketelsen U., Korinthenberg R., Wenzel D., Lauffer H., Kreiss-Nachtsheim M., et al. Classical infantile spinal muscular atrophy with SMN deficiency causes sensory neuronopathy. Neurology. 2003;60:983–987. doi: 10.1212/01.wnl.0000052788.39340.45. [DOI] [PubMed] [Google Scholar]
- 27.Jablonka S., Karle K., Sandner B., Andreassi C., von Au K., Sendtner M. Distinct and overlapping alterations in motor and sensory neurons in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2006;15:511–518. doi: 10.1093/hmg/ddi467. doi:10.1093/hmg/ddi467. [DOI] [PubMed] [Google Scholar]
- 28.Jablonka S., Schrank B., Kralewski M., Rossoll W., Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: an animal model for spinal muscular atrophy type III. Hum. Mol. Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. doi:10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 29.Araki S., Hayashi M., Tamagawa K., Saito M., Kato S., Komori T., Sakakihara Y., Mizutani T., Oda M. Neuropathological analysis in spinal muscular atrophy type II. Acta Neuropathol. 2003;106:441–448. doi: 10.1007/s00401-003-0743-9. doi:10.1007/s00401-003-0743-9. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi M., Arai N., Murakami T., Yoshio M., Oda M., Matsuyama H. A study of cell death in Werdnig Hoffmann disease brain. Neurosci. Lett. 1998;243:117–120. doi: 10.1016/s0304-3940(98)00105-0. doi:10.1016/S0304-3940(98)00105-0. [DOI] [PubMed] [Google Scholar]
- 31.Hsu C.F., Chen C.Y., Yuh Y.S., Chen Y.H., Hsu Y.T., Zimmerman R.A. MR findings of Werdnig–Hoffmann disease in two infants. AJNR Am. J. Neuroradiol. 1998;19:550–552. [PMC free article] [PubMed] [Google Scholar]
- 32.Ito Y., Kumada S., Uchiyama A., Saito K., Osawa M., Yagishita A., Kurata K., Hayashi M. Thalamic lesions in a long-surviving child with spinal muscular atrophy type I: MRI and EEG findings. Brain Dev. 2004;26:53–56. doi: 10.1016/s0387-7604(03)00075-5. doi:10.1016/S0387-7604(03)00075-5. [DOI] [PubMed] [Google Scholar]
- 33.Oka A., Matsushita Y., Sakakihara Y., Momose T., Yanaginasawa M. Spinal muscular atrophy with oculomotor palsy, epilepsy, and cerebellar hypoperfusion. Pediatr. Neurol. 1995;12:365–369. doi: 10.1016/0887-8994(95)00058-n. doi:10.1016/0887-8994(95)00058-N. [DOI] [PubMed] [Google Scholar]
- 34.Shishikura K., Hara M., Sasaki Y., Misugi K. A neuropathologic study of Werdnig–Hoffmann disease with special reference to the thalamus and posterior roots. Acta Neuropathol. 1983;60:99–106. doi: 10.1007/BF00685353. doi:10.1007/BF00685353. [DOI] [PubMed] [Google Scholar]
- 35.Towfighi J., Young R.S., Ward R.M. Is Werdnig–Hoffmann disease a pure lower motor neuron disorder? Acta Neuropathol. 1985;65:270–280. doi: 10.1007/BF00687008. doi:10.1007/BF00687008. [DOI] [PubMed] [Google Scholar]
- 36.Yohannan M., Patel P., Kolawole T., Malabarey T., Mahdi A. Brain atrophy in Werdnig–Hoffmann disease. Acta Neurol. Scand. 1991;84:426–428. doi: 10.1111/j.1600-0404.1991.tb04982.x. doi:10.1111/j.1600-0404.1991.tb04982.x. [DOI] [PubMed] [Google Scholar]
- 37.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. doi:10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 38.Kielar C., Wishart T.M., Palmer A., Dihanich S., Wong A.M., Macauley S.L., Chan C.H., Sands M.S., Pearce D.A., Cooper J.D., et al. Molecular correlates of axonal and synaptic pathology in mouse models of Batten disease. Hum. Mol. Genet. 2009;18:4066–4680. doi: 10.1093/hmg/ddp355. doi:10.1093/hmg/ddp355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray L.M., Thomson D., Conklin A., Wishart T.M., Gillingwater T.H. Loss of translation elongation factor (eEF1A2) expression in vivo differentiates between Wallerian degeneration and dying-back neuronal pathology. J. Anat. 2008;213:633–645. doi: 10.1111/j.1469-7580.2008.01007.x. doi:10.1111/j.1469-7580.2008.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wishart T.M., Paterson J.M., Short D.M., Meredith S., Robertson K.A., Sutherland C., Cousin M.A., Dutia M.B., Gillingwater T.H. Differential proteomics analysis of synaptic proteins identifies potential cellular targets and protein mediators of synaptic neuroprotection conferred by the slow Wallerian degeneration (Wlds) gene. Mol. Cell. Proteomics. 2007;6:1318–1330. doi: 10.1074/mcp.M600457-MCP200. doi:10.1074/mcp.M600457-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor C.F., Paton N.W., Lilley K.S., Binz P.A., Julian R.K., Jr, Jones A.R., Zhu W., Apweiler R., Aebersold R., Deutsch E.W., et al. The minimum information about a proteomics experiment (MIAPE) Nat. Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. doi:10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.