Abstract
The BCR-ABL tyrosine kinase inhibitor imatinib is highly effective for chronic myeloid leukemia (CML). However, some patients gradually develop resistance to imatinib, resulting in therapeutic failure. Metabonomic and genomic profiling of patients' responses to drug interventions can provide novel information about the in vivo metabolism of low-molecular-weight compounds and extend our insight into the mechanism of drug resistance. Based on a multi-platform of high-throughput metabonomics, SNP array analysis, karyotype and mutation, the metabolic phenotypes and genomic polymorphisms of CML patients and their diverse responses to imatinib were characterized. The untreated CML patients (UCML) showed different metabolic patterns from those of healthy controls, and the discriminatory metabolites suggested the perturbed metabolism of the urea cycle, tricarboxylic acid cycle, lipid metabolism, and amino acid turnover in UCML. After imatinib treatment, patients sensitive to imatinib (SCML) and patients resistant to imatinib (RCML) had similar metabolic phenotypes to those of healthy controls and UCML, respectively. SCML showed a significant metabolic response to imatinib, with marked restoration of the perturbed metabolism. Most of the metabolites characterizing CML were adjusted to normal levels, including the intermediates of the urea cycle and tricarboxylic acid cycle (TCA). In contrast, neither cytogenetic nor metabonomic analysis indicated any positive response to imatinib in RCML. We report for the first time the associated genetic and metabonomic responses of CML patients to imatinib and show that the perturbed in vivo metabolism of UCML is independent of imatinib treatment in resistant patients. Thus, metabonomics can potentially characterize patients' sensitivity or resistance to drug intervention.
Introduction
In the postgenomic era, the complementary use of high-throughput analytical technologies (such as genomics, proteomics, and metabonomics) in biological systems has revolutionized biological investigations. Genomic variation appears to be an important factor that can enhance or reduce the risk of developing a disease, depending on the specific genetic or epigenetic pathway involved [1], [2]. It has been demonstrated that chronic myeloid leukemia (CML) involves a translocation between chromosomes 9 and 22, which results in the expression of the BCR–ABL fusion protein. The tyrosine kinase activity of oncogenic ABL proteins is known to be essential for its transforming activity [3]. Imatinib mesylate (imatinib) is a small molecular inhibitor of the tyrosine kinase activity of the BCR–ABL fusion protein, and is now a frontline therapy for CML [4]. Despite imatinib's striking efficacy, resistance develops over time in many patients, and is more common in patients with advanced-stage CML [5]. Routine cytogenetic analysis and molecular methodologies can identify resistance or sensitivity to imatinib in CML patients, and are considered the gold standards for evaluating the potential response to imatinib in clinical practice [6], [7]. However, the two methods do not provide further molecular information about the in vivo metabolic perturbation involved, which may clarify the mechanism of resistance, or allow us to metabonomically characterize sensitive CML patients (SCML) and resistant CML patients (RCML). Our understanding of the biological functions involved would benefit greatly from an understanding of the metabolic network, including quantitative measurements of different types of compounds (such as proteins and metabolites) and various biochemical processes (such as gene expression) made in parallel, and preferably combined with other classical phenotypic analyses [8]. Although many researchers have monitored the response to imatinib in CML patients using molecular methodologies and cytogenetic techniques [7], [9], [10], no comprehensive metabonomic investigation has been made of the responses of CML patients to imatinib.
Metabonomics is defined as the quantitative measurement of endogenous low-molecular-weight compounds that reflect the metabolic responses of living systems to diverse stimuli [2], [11], [12]. The metabolic phenotype constitutes the endpoint of various metabolic responses and is influenced by genomic and proteomic factors. It can be used to identify early signals/biomarkers of cellular abnormalities that occur before the appearance of gross phenotypic changes [1]. Metabonomics can be used as a complementary tool, providing information about the metabolic network that cannot be obtained directly from the genotype, gene expression profiles, or even the proteome of an individual [2]. It has been successfully applied to biomedical sciences [2], [11], [13]–[9], and shows promising applications in the exploration of diseases and in the development of personalized drug treatments [2], [18], [19]. Metabonomics can potentially be applied to the discovery of tumor metabolic pathways, the investigation of metabolic responses to treatments [20], and the identification of tumor biomarkers of these responses [11], [12], [15]. In this study, using a metabonomic platform we developed based on gas chromatography/time-of-flight mass spectrometry (GC/TOFMS) and data analysis techniques [21], [22], we integrated metabonomic data with cytogenetic and molecular analyses to profile the metabolic phenotypes of CML patients and differentiate their metabolic responses to imatinib.
Results
CML patients
Fifty-nine patients were enrolled in the study: 53 with chronic-phase (CP) CML and 6 with blast crisis (BC). Two BC patients had a complex karyotype. Of these 59 patients, 26 were untreated (UCML). The other 33 CML patients were treated with imatinib at daily doses of 300–800 mg, and 19 achieved complete cytogenetic remission (0% Philadelphia chromosome; “sensitive CML”, SCML), whereas 14 patients were resistant to imatinib (RCML; Table 1). Seven of the RCML patients displayed primary RCML and seven displayed secondary RCML. Chronic-phase, blast crisis, and primary cytogenetic resistance were determined as described in a recent study [5]. The healthy volunteers included nine men and nine women, with a median age of 35.2 years.
Table 1. Information of the CML patients and the health volunteers.
| Characteristics | HC | UCML | SCML | RCML | |
| Sex | Male | 9 | 17 | 17 | 8 |
| Female | 9 | 9 | 2 | 6 | |
| Age (year) | Mean±SD | 35.2±9.2 | 37.6±13.4 | 38.1±15.7 | 39.2±14.1 |
| range | 18–55 | 18–67 | 23–70 | 18–69 | |
| Recent medicine | No | Hydroxycarbamide | Imatinib | Imatinib | |
| Duration of disease | Mean±SD | 8.3±12.6 | 17.8±24.2 | 48.5±31.6 | |
| (months) | range | 1–33 | 6–84 | 6–168 | |
| Duration of imatinib | Mean±SD | 12.2±23.3 | 25.9±12.7 | ||
| treatment (months) | range | 6–72 | 6–39 | ||
| Daily dose (mg) | Mean±SD | 418±52 | 421±77 | ||
| range | 400–600 | 300–800 | |||
Mutation of the ABL kinase domain in RCML patients
In the 14 resistant patients analyzed, ABL kinase domain mutations were detected in only 1 BC patient, who presented with three types of amino acid changes: L232P, F336L, and C349R.
High-density single nucleotide polymorphism (SNP) array analysis of CML patients
After exclusion of genomic copy number polymorphisms by comparison of the data with recorded copy number polymorphisms in the UCSC Genome Browser (http://genome.ucsc.edu/) databases,a total of 44 deletions, 2 duplication, and 7 regions of loss of heterozygosity (LOH) were identified by SNP array analysis in 9 CML samples, i.e., 1 SCML, 4 primary RCML and 4 secondary RCML (Table 2). In addition to sex chromosome, four of 6 CP RCML patients did not show other abnormal genome. Moreover, Cryptic deletions on chromosome X and Y were found in patients with CP-CML both in SCML and RCML.
Table 2. Lesion detected in CML samples by SNP array analysis.
| Patients ID | Chromosome | Genic status | Starting position | Ending position | Genes identified | Disease status | Response to imatinib |
| R01C02 | 5 | Del | 81629356 | 115815122 | LOC92270 | BC | Resistance |
| R01C02 | 9 | Del | 127196469 | 132665560 | FREQ,CDK9 | BC | Resistance |
| R01C02 | 22 | Del | 22053721 | 22902745 | GSTTP1, LOC388882, IGLL1, C22orf16, MMP11, SLC2A11, MIF, FLJ31568 | BC | Resistance |
| R01C02 | 22 | Dup | 23995985 | 24240879 | LRP5L, ADRBK2 | BC | Resistance |
| R06C01 | 7 | Del | 61862616 | 61921953 | No gene | CP | Resistance |
| R06C01 | 19 | Dup | 32493654 | 33528102 | LOC727780,LOC642290,LOC727771 | CP | Resistance |
| R02C01 | 11 | Del | 50354779 | 51228612 | OR4A5, LOC646813 | CP | Resistance |
| R02C01 | 19 | Del | 20423788 | 20514068 | LOC645490 | CP | Resistance |
| R04C01 | 11 | Del | 46327343 | 47374448 | F2,SPI1,DGKZ,AMBRA1,MADD C11orf49,PACSIN3 | BC | Resistance |
| R04C01 | 17 | Del | 44176361 | 52843822 | >20 genes | BC | Resistance |
| R04C01 | 2 | LOH | 78822450 | 89733531 | >20 genes | BC | Resistance |
| R04C01 | 2 | LOH | 98364441 | 109797238 | >20 genes | BC | Resistance |
| R04C01 | 2 | LOH | 109809150 | 121111631 | >20 genes | BC | Resistance |
| R04C01 | 3 | LOH | 142389922 | 153466638 | >20 genes | BC | Resistance |
| R04C01 | 5 | LOH | 156922335 | 168529461 | >20 genes | BC | Resistance |
| R04C01 | 15 | LOH | 82248349 | 100215359 | >20 genes | BC | Resistance |
| R04C01 | 20 | LOH | 29310063 | 55040991 | >20 genes | BC | Resistance |
| R03C02 | 1 | Del | 240744201 | 241169492 | LOC391183, PLD5 | CP | Sensitivity |
Starting position: start position of genomic lesion; Ending position: end position of genomic lesion;LOH: loss of heterozygosity, Del; deletion, Dup; duplication.BC,blast crisis; CP, chronic-phase.
Acquired GC/TOFMS data and the identification of compounds
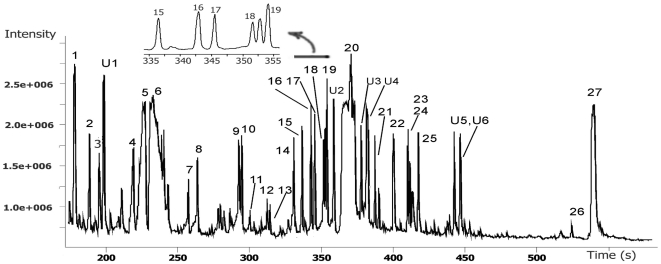
A typical total-ion-current chromatogram showed several hundred peaks in a single analysis (Figure 1). After deconvolution of the chromatograms, much quantitative and qualitative information was obtained. In general, a total of 186 non-targeted peaks/metabolites were detected, and a data matrix of 186×77 (77 observations/sample) was constructed. Altogether, 22 peaks were excluded from further data processing because they were not detected in the normal control and might have been artifacts derived from drug metabolites or other sources. In consequence, 96.97% of the peak areas were validated, with data missing for 382 peak areas (these GC/TOFMS responses are very low) in the whole data matrix of 164×77. By comparison with authentic reference standards or reference compounds in available libraries, 72 compounds were identified, including amino acids, organic acids, amines, saccharides, lipids, fatty acids, etc. (Table S1).
Figure 1. A typical GC/TOFMS chromatogram of blood plasma.
Several hundreds of peaks can be detected at only one analysis and some of the peaks were identified. Part of the chromatogram was zoomed in for clear inspection of the peaks, 15–19, retention time from 335 to 355 s. 1, Lactate; 2, butyamine; 3, 3-hydroxybutyrate; 4, valine; 5, urea; 6, phosphate; 7, serine; 8, threonine; 9, pyroglutamate; 10, hydroxyproline; 11, creatinine; 12, ornithine; 13, phenylalanine; 14, methyl myristate (QC reference standard); 15, glutamine; 16, glycerol-3-phosphate; 17, azelate; 18, citrate; 19, [13C2]-myristic acid (stable isotope internal standard); 20, glucose; 21, palmitic acid; 22, uric acid; 23, linoleic acid; 24, oleic acid; 25, stearic acid; 26, alpha-tocopherol; 27, cholesterol; U1-5, unidentified peaks of retention indices at 1163, 1876, 1986, 2015, 2415, 2448, respectively.
Metabolic patterns of UCML, SCML, and RCML
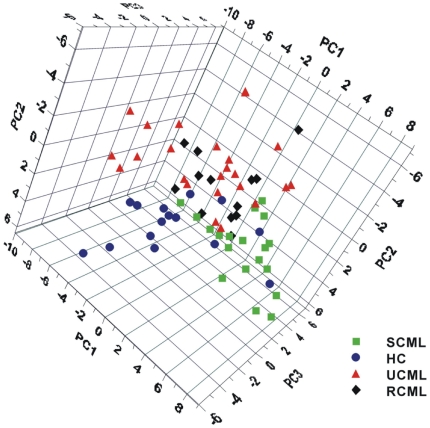
For an overview of the dataset, an unsupervised principal components analysis (PCA) was applied to the data. Three serious outliers were identified, one each from the healthy control group (HC), UCML, and RCML. They were excluded from further data analysis because serious outliers negatively affect and thus bias the model. To calculate a partial least-squares projection to latent structures and discriminant analysis (PLS–DA) model, the remaining samples were classified into four groups (HC, UCML, SCML, and RCML). According to the cross-validation [23], the PLS–DA model of the three principal components explained 30.8% of the variation in X (R2X = 0.308, GC/TOFMS response variables/peaks) and 39.7% of the variation in Y (R2Y = 0.397, sample types), and predicted 23.9% of the sample types. Although the four groups overlapped to some extent on the scores plot (Figure 2), obvious separations were observed for HC, SCML, and UCML. UCML clustered far from SCML and HC, whereas RCML was scattered and primarily overlapped with UCML, suggesting that its metabonomic composition was similar to that of UCML, but different from those of HC and SCML.
Figure 2. The scores plot of PLS-DA model (3 principal components) of the four groups.
UCML, SCML, RCML and the healthy control (HC). This figure shows the difference between UCML and the healthy control and SCML and UCML, respectively. The overlapping of UCML and RCML suggested similar metabolic phenotype and indicated the therapeutic ineffectiveness of imatinib on RCML. The overlapping of SCML and HC suggested similar metabolic phenotype and the therapeutic effectiveness of imatinib on SCML.
A submodel was calculated to further evaluate the metabolic similarities and differences among SCML, HC, and UCML. The two-principal-components PLS–DA model (Figure S1) showed that SCML clustered far from UCML, whereas it overlapped HC, clearly suggesting that the sensitive metabolome was regulated towards the normal profile.
Metabonomic differences between UCML patients and HC
A PLS–DA model between UCML and HC clearly identified the similarities in the two groups and the significant differences between the two groups. To better understand the intrinsic variations, specific compounds were identified. Statistical analysis (one-way analysis of variance, ANOVA) showed higher levels of glutamine, myo-inositol, arabinose, glycine, urea, ornithine, glutamate, pyroglutamate, and an unidentified compound, but lower levels of citrate, high-density lipoprotein (HDL), linoleate, and some unidentified peaks in UCML (P<0.05; Table 3). For the other compounds, such as pipecolate, pseudouridine, urate, α-tocopherol, adipate, total cholesterol, and nonesterified cholesterol (free form), the differences between HC and UCML were obvious but not statistically significant (P>0.05).
Table 3. The relative abundance of the identified compounds/peaks differentiating UCML from HC and the regulation by imatinib.
| Variable Name | Healthy control(HC) | UCML | Fold change | SCML | RCML | |||||||||||||
| Mean | ± | SD | Mean | ± | SD | UCML/HC | Mean | ± | SD | Mean | ± | SD | ||||||
| Pipecolate | 66738 | ± | 27812 | 191019 | ± | 150758 | 2.864 | 113166 | ± | 69717 | 164293 | ± | 105135 | * | ||||
| Glutamine | 397746 | ± | 178241 | 731628 | ± | 477666 | * | 1.839 | 642994 | ± | 256423 | * | 744690 | ± | 508088 | * | ||
| myo-Inositol | 2567687 | ± | 1211442 | 4224531 | ± | 2076166 | * | 1.645 | 2460364 | ± | 851377 | †† | 3201257 | ± | 2305927 | |||
| Arabinose | 160888 | ± | 54011 | 263240 | ± | 134590 | ** | 1.636 | 169357 | ± | 57379 | † | 206307 | ± | 106486 | |||
| Glycine | 4994510 | ± | 1934972 | 7755324 | ± | 3431521 | * | 1.553 | 5697956 | ± | 1783735 | 7589657 | ± | 3502635 | ||||
| Urea§ (mmol/L) | 4.95 | ± | 0.94 | 7.38 | ± | 1.12 | * | 1.491 | 5.10 | ± | 1.21 | 7.49 | ± | 1.41 | * | |||
| Ornithine | 7051554 | ± | 3092520 | 10132171 | ± | 3809942 | * | 1.437 | 9154095 | ± | 4566462 | 10755744 | ± | 5278905 | ||||
| Pseudouridine | 187540 | ± | 93536 | 262470 | ± | 132366 | 1.400 | 123979 | ± | 55572 | † | 230273 | ± | 142358 | ||||
| Glutamate | 486771 | ± | 129371 | 672095 | ± | 230155 | * | 1.381 | 475953 | ± | 94505 | † | 591183 | ± | 252764 | |||
| Pyroglutamate | 12872691 | ± | 3860744 | 17366747 | ± | 3905589 | ** | 1.349 | 14246015 | ± | 3677287 | † | 16553496 | ± | 3610300 | * | ||
| Urate | 8635673 | ± | 5325414 | 9311846 | ± | 5403118 | 1.078 | 11613451 | ± | 3705793 | 9157485 | ± | 3361998 | |||||
| CPU_ DB5_RI1341_P | 66738 | ± | 27812 | 191019 | ± | 99008 | * | 2.862 | 113166 | ± | 69717 | 164293 | ± | 105135 | ||||
| Citrate | 9277095 | ± | 2814476 | 7138455 | ± | 2012240 | * | −1.300 | 8736984 | ± | 1696040 | 7197272 | ± | 2641511 | ||||
| alpha-Tocopherol | 808681 | ± | 165795 | 601033 | ± | 195359 | −1.345 | 683782 | ± | 282667 | 535638 | ± | 240045 | * | ||||
| Linoleate | 8552168 | ± | 3885328 | 5109234 | ± | 2596822 | * | −1.400 | 6439932 | ± | 3154482 | 5584365 | ± | 3110760 | ||||
| Adipate | 202105 | ± | 130022 | 135086 | ± | 105583 | −1.495 | 164819 | ± | 69713 | 168152 | ± | 82656 | |||||
| HDL § (mmol/L) | 1.38 | ± | 0.33 | 0.83 | ± | 0.25 | ** | −1.663 | 1.27 | ± | 0.26 | † | 0.87 | ± | 0.39 | |||
| Total cholesterol§ (mmol/L) | 4.57 | ± | 1.28 | 3.28 | ± | 1.03 | −1.393 | 3.79 | ± | 0.60 | 2.76 | ± | 0.77 | |||||
| Non-esterified cholesterol | 66222510 | ± | 11227958 | 61596173 | ± | 12264833 | −1.075 | 59449993 | ± | 10150252 | 55116351 | ± | 10009437 | |||||
| CPU_ DB5_RI2494_P | 4252017 | ± | 1132727 | 3208128 | ± | 1021383 | * | −1.325 | 4008764 | ± | 1149047 | † | 3019455 | ± | 826266 | ** | ‡ | |
| CPU_ DB5_RI2664_P | 988481 | ± | 549499 | 648226 | ± | 389483 | * | −1.525 | 1080495 | ± | 309271 | † | 728768 | ± | 315890 | ‡ | ||
| CPU_ DB5_RI2124_P | 177008 | ± | 101696 | 105477 | ± | 98053 | −1.678 | 186469 | ± | 97861 | † | 110002 | ± | 57932 | ||||
All peaks areas were normalized by the stable isotope internal standard, [13C2]-myristic acid; CPU_ DB5_RIx_P: Unidentified compounds in plasma samples detected with DB-5 capillary column in GC/TOFMS, China Pharmaceutical university (CPU); RI, retention index; x, retention time index value; P, plasma sample.
,**: statistically significant different from that in HC, p<0.05, 0.01;
, ††: statistically significant different from that in UCML, p<0.05, 0.01;
, ‡‡: statistically significant different from that in SCML, p<0.05, 0.01.
, clinic measurement results.
Metabolic perturbation of CML after imatinib treatment
To facilitate the identification of the compounds that were affected by exposure to imatinib, three separate PLS–DA models were computed between SCML, RCML, and UCML. The first model between SCML and UCML explained 28.3% of the GC/TOFMS response variables/peak areas and 74.2% of the sample types, and also predicted 47.3% of the sample types. The relatively low capacity to explain the GC/TOFMS response variables (28.3%) means that the majority of them were not significantly affected, i.e., not many metabolites were affected in SCML. Another model between SCML and RCML resulted in a clustering similar to that of the previous model, suggesting that metabolic responses to imatinib were evident in these two groups of patients. However, the model between RCML and UCML failed to differentiate the two groups according to cross-validation, which resulted in a negative prediction parameter, Q2Y, of −10.2%. This means that the metabonomic composition of RCML was not significantly different from that of UCML. In summary, in terms of metabonomics, SCML patients differed from UCML, whereas RCML patients were similar to UCML patients.
Compared with UCML patients, significant reductions in myo-inositol, arabinose, pseudouridine, glutamate, and pyroglutamate, and increases in HDL and three unidentified peaks were observed in SCML patients (P<0.05, one-way ANOVA; Table 3). Pipecolate, glutamine, glycine, urea, ornithine, citrate, α-tocopherol, and linoleate levels were also more or less rectified, although not statistically significantly (P>0.05), but most of them were adjusted towards normal levels. Interestingly, none of the metabolites listed in Table 3 were significantly different in RCML and UCML patients. The results both from the general scores plot (Figure 2) and for specific compounds (Table 3) confirmed that RCML was quite similar to UCML.
Metabonomic differences between RCML BC patients and RCML CP patients
RCML BC patients showed a totally different metabonomic phenotype from that of RCML CP patients (Figure S2). It is noteworthy that many compounds, including oleate, palmitate, linoleate, stearate, glycine, pyroglutamate, arabinose, β-d-methylglucopyranoside, and fumarate, showed significant elevation in RCML BC patients compared with those in RCML CP patients (Table S2). Lysine was the only compound occurring at a lower level in RCML BC patients.
Discussion
Although the correlations between the cytogenetic and molecular effects and clinical efficacy have been well established, previous studies have provided little information about the in vivo metabolism of CML patients during imatinib treatment, except in terms of bone and mineral metabolism [24], hyperlipidemia [25], creatine kinase, phosphate, and phosphocholine [26]–[27]. Hundreds of low-molecular-weight endogenous compounds in the plasma were profiled here with a high-throughput GC/TOFMS analysis, and many metabolites that distinguished the different CML treatment groups were identified. Factors such as sex, age, background, disease duration, imatinib treatment duration, and diet can massively affect metabonomics and may muffle the variation in different groups. Despite these confounding factors, our results not only show that UCML differs from HC in terms of metabonomics, but also characterize SCML and RCML, and suggest their similarity to HC and UCML, respectively. This indicates that metabolic profiling can potentially characterize the status of CML in response to imatinib (ineffective or effective).
Metabonomic differences between UCML patients and HC
The significant metabonomic differences between UCML and HC indicate metabolic perturbation in CML patients. Of the metabolites contributing most to the differences between UCML and HC, intermediates of the tricarboxylic acid (TCA) cycle (citrate) and lipid metabolism (cholesterol, HDL, linoleate) were downregulated in UCML, whereas urea cycle metabolites, such as pyrimidines (pseudouridine), amino acids (glycine, ornithine), and purines (urate), were elevated to some extent (Table 3, Figure S3). Consistent with the results for other tumors [16], [28], some amino acids, such as glutamate, ornithine, glycine, and pyroglutamate, were found at higher levels in UCML compared with those in HC, which indicates a cellular requirement for a higher turnover of structural proteins. Purines (urate) were detected at higher levels in UCML, indicative of a higher capacity for DNA synthesis. After the imatinib intervention, we detected a higher level of urate in SCML than in UCML, which is supported by a previous report [27]. The relatively lower level of citrate suggests the downregulation of the TCA cycle and that CML cells require higher-energy metabolism. This is achieved through the upregulation of amino acid transporters, cellular molecule synthesis, and signal transduction by the delivery of carbon backbones. Increased glycolysis has been consistently observed in cancer cells [28], [29], but whether this metabolic shift is a consequence or cause of cancer remains controversial [30].
Metabonomic differences between SCML and RCML
After imatinib treatment, SCML patients underwent an obvious metabolic perturbation, whereas RCML patients were not significantly affected. The relevant changes in regulatory pathways, in response to the drug intervention, can be discerned from the discrete transcriptomic and metabonomic datasets [31]. Based on our metabonomic results, both the mathematical PLS–DA model and the suggested markers offer alternative diagnostic methods to cytogenetics.
To evaluate the genomic effect on metabolic phenotype, SNP array analysis was applied to identify the difference between CP & BC CML, and SCML & RCML. Our data are consistent with a recent SNP array study that submicroscopic alterations were detectable on chromosome 1,9,17 and 22 [32]. Deletions, duplication and LOH on chromosome 17, 9, 22, 5 and 19 were identified in several important chromosomal regions of BC patients. Chromosome 17 was most heavily affected by secondary genomic alterations on development of TKI resistance during disease progression [32], [33]. The deletions and amplifications on chromosomes 9 and 22, which were suggested to be associated with TKI resistance or disease progression [32], were detectable in 1 secondary resistant sample developing imatinib resistance. In contrast to the lesions on chromosome 17, 9 and 22, chromosome 1 may be involved in the initial development of CML rather than associated with TKI resistance [32], just as one of SCML patients showed deletion on chromosome 1 (Table 2).
Of the 14 RCML patients, only one BC patients carried ABL kinase domain mutations, so the different metabolic phenotypes of RCML and SCML are not necessarily dependent on ABL domain mutations. The difference is probably attributable to the activation of other pathways for the survival and proliferation of CML cells [34]. On the other hand, deletion or duplication on chromosome 1, 11 or 22, may lead to abnormal expression of genes which are all involved in cell lipid metabolism, including PLD5, LRP5L and AMBRA1. One of the genes in this deleted region on chromosome 22 was SLC2A11, a class II sugar transport facilitator, which exhibits highest similarity with the fructose transporter GLUT5. Three studies have reported expression signatures of CD34+ cells in relation to CML blast crisis and duration of chronic phase in patients treated with nonimatinib or imatinib therapy, respectively [35], [36], [37]. Comparison with both the Zheng et al.[35] and Yong et al [36] data, McWeeney et al.[37] found five genes (CSTA, RNASE3, PRTN3, PLAUR, and MPO) overlapped between the 3 datasets. Unfortunately, we failed to associate the metabolic data and the published genomic data.
The metabonomic data (Table S2) indicate that BC cells maintain a highly glycolytic status, with increased glucose and urate production, and require more source materials (such as amino acids and lipids) to support cell proliferation. Taken together, it strongly suggested that metabolic variation between BC and CP patients were closely related to genomic alterations.
Conversely, as in other solid cancer cells, the leukemia cells exhibited increased glycolysis, and took advantage of this metabolic pathway for ATP generation as a main source of energy. This phenomenon is known as the “Warburg effect” and is considered one of the most fundamental metabolic changes that occur during malignant transformation [28], [29], [38]. Imatinib reverses the Warburg effect in BCR–ABL-positive cells by switching from glycolysis to mitochondrial glucose metabolism, resulting in a reduction in glucose uptake and a higher energy state [38]. The metabolic phenotype of SCML showed modification of the TCA cycle and lipid metabolism, together with the upregulation of de novo nucleotide triphosphate/deoxynucleotide triphosphate synthesis [38]. In accordance with a previous study, these results suggest that imatinib exerts its effect by arresting DNA replication and the cell-proliferation cycle [11].
In this study, both HDL and total cholesterol decreased in RCML and increased in SCML. Cholesterol is an integral component of all eukaryotic cell membranes and is essential for normal cellular function. It is well known that the major function of HDL is to maintain cholesterol homeostasis in normal cells through the removal of excess cholesterol from intracellular pools. Plasma HDL concentrations reflect the activity of a number of intra- and extracellular metabolic pathways. The plasma levels of this lipoprotein can be determined by the flow of cholesterol through the extracellular pathways [39]. A reduction in HDL correlates with the accumulation of cholesterol esters during cell proliferation [40]. We presume that the low level of HDL observed in our study was associated with increasing BCR–ABL-mediated cell proliferation and a resistance to imatinib in vivo.
Metabonomic differences between SCML and HC
The modification of the metabolome and the genome occurred synchronously during effective treatment with imatinib, followed by a reduction in the burden of CML cells. The metabolic phenotype of SCML was similar to that of HC, but the two groups did not completely overlap, even after years of treatment with imatinib. This result reinforces the observation that SCML patients are in fact genetically different from HC patients. We postulate three reasons why the metabolomes of SCML patients did not completely overlap those of HC. First, although the active CML cells that were sensitive to imatinib shifted towards a normal metabolism, quiescent and residual CML cells may influence the biochemical networks [41]. Second, besides its positive effect on CML, imatinib may affect other metabolic pathways. For example, Rowley proposed that imatinib treatment potentially targets the BCR–ABL fusion gene but may disrupt critical pathways required for normal cell function [42]. Third, many patients who were translocation negative in a standard cytogenetic analysis showed very low amounts of the BCR–ABL fusion on reverse transcriptase–polymerase chain reaction (PCR) analysis [6]. Therefore, we believe that patients with SCML, even those with a normal karyotype, still maintain a diseased status.
In summary, our results not only identify metabonomic differences between UCML and HC, but also demonstrate and correlate the genetic and metabolic responses to the imatinib intervention. Therefore, the metabonomic profiling described here may be useful for monitoring the metabolic changes that occur following imatinib treatment of CML. The analysis of metabolic changes may also be used as an alternative method of characterizing individual responses to imatinib in a population of CML patients, which may ultimately lead to personalized therapeutic strategies. This study opens the way to the identification of gene-to-metabolite networks, and offers a method for the early prediction of responsiveness to imatinib and the personalization of treatment, thus improving clinical and therapeutic decisions.
Materials and Methods
Chemicals
All the authentic reference standards and reagents were of analytical, silylation, or chromatography grades, and were purchased from Sigma-Aldrich (St Louis, MO, USA), Merck (Darmstadt, Germany), Fluka (Buchs, Switzerland), Aldrich (Steinhein, Germany), Serva (Heidelberg, Germany), Sigma-Aldrich (Isotec, USA), and Pierce Chemical Company (USA). Distilled water was produced with a Milli-Q Reagent Water System (Millipore, MA, USA).
Patients and plasma samples
Fifty-nine male and female individuals attending Jiangsu Province Hospital, aged 18 years or older, were eligible for inclusion if they had been diagnosed with CML between January 2007 and September 2008. Of these patients, 26 were untreated or were treated with hydroxycarbamide, and were designated “untreated CML” (UCML), and 33 were treated with 300–800 mg/d imatinib. Eighteen healthy adult volunteers with no genetic, hepatic, renal, or cardiovascular disease were selected by matching them in age and sex to the CML patients. Diet, weight, and levels of physical activity remained unchanged during the course of the imatinib therapy. All the patients and healthy volunteers provided their written informed consent before enrollment in the study. The study was approved by the Institutional Review Board of the Nanjing Medical University.
Cytogenetic analysis
Bone marrow was obtained from the patients. The karyotypes were evaluated with an R-banded analysis of 20 metaphase cells from unstimulated, cultured (24 h) bone-marrow aspirates. The karyotypes were described according to the international system for human cytogenetic nomenclature guidelines [43]. Chronic-phase, blast crisis, and primary cytogenetic resistance were as defined in recent studies [5], [6].
Analysis of mutations in the ABL kinase domain
DNA was extracted from peripheral blood or bone marrow samples with a standard phenol/chloroform extraction method. The primer pairs were designed with primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to amplify the coding sequence of ABL (Table S3). The ABL kinase domain of the BCR–ABL allele was amplified with a PCR–restriction fragment length polymorphism technique, followed by direct sequencing and sequence homology analysis. Sequencing was performed with the BigDye Terminator Cycle Sequencing Ready Reaction Kit (PerkinElmer BioSystems, Foster City, CA, USA) on an ABI Prism 3700 DNA Analyzer (PerkinElmer BioSystems).
SNP array analysis
Genomic DNA isolated from cells was subjected to GeneChip Human mapping microarray (Shanghai SNP-chip, Illumina,Inc, CA, USA). High-quality genomic DNA was processed in accordance with the genomic mapping 250K NspI protocol and hybridized to 250K NspI SNP arrays according to the manufacturer's instructions. Data analysis of deletions, amplifications and loss of heterozygosity (LOH) was carried out using the CNAG (copy number analysis for Affymetrix GeneChips)software with nonmatched references.
Sample preparation and GC/TOFMS analysis
Plasma was isolated and treated as previously reported [22], with a few modifications. Briefly, 100 µL of plasma was thawed, 400 µL of methanol was added to the extract, and the protein was precipitated. [1,2-13C2]-Myristic acid (2 µg) was added as the internal standard. The supernatant (100 µL) was dried, methoximated, trimethylsilylated, and prepared for analysis. To extract more information from the study, triglyceride (TC), urea, total cholesterol (TG), and high- and low-density lipoproteins (HDL and LDL, respectively) were determined with an automatic biochemical analyzer (Olympus AU5400, Japan) at Nanjing Medical University.
To minimize the systematic variation, all plasma samples were randomly analyzed by GC/TOFMS using a previously reported method [22]. To achieve better chromatographic separation, the column temperature was initially maintained at 70°C for 2 min, and then increased at a rate of 35°C/min from 70°C to 305°C, where it was maintained for 2 min. Masses were scanned and the data were acquired from m/z 50–800 at a rate of 30 spectra/s.
Data processing and pattern recognition
Automatic peak detection and mass spectrum deconvolution were performed with the ChromaTOF™ software (version 3.25) and the peak areas were obtained as reported previously [21]. A data matrix of the peak areas was thus constructed, with the observation/samples in the first column and the responses/peaks as variables in the first row. The retention index was calculated by comparing the retention times of the peaks with those of the alkane series C8–C40. The compounds were identified by the comparison of the mass spectra and retention indices of all the detected peaks with authentic reference standards available in the National Institute of Standards and Technology library 2.0 (2005) and an in-house mass spectra library database maintained by Umea Plant Sciences Center, Sweden.
Pattern recognition was performed with the professional data processing software SIMCA-P 11 (Umetrics, Umeå, Sweden). PCA and PLS–DA were used to analyze the data with a standard method [44]. The statistical results of the normalized peak areas were validated with one-way ANOVA.
Supporting Information
The PLS_DA scores plot of SCML, UCML and healthy control (HC). The overlapping of SCML and HC suggested the rectification of endogenous compound in SCML towards normal. While the distinct separation of SCML and UCML suggested metabolic responses to imatinib.
(0.32 MB TIF)
The PLS_DA scores plot of RCML BC (▪, n = 6) and RCML CP (□, n = 8) patients. RCML BC patients showed totally different metabonomic phenotype from that of RCML CP patients.
(0.16 MB TIF)
Metabolic perturbations in UCML and metabolic response to imatinib treatment. The marked metabolites were observed at an abnormal level in UCML, and most of them were regulated towards normal in SCML with imatinib. Red or green blocks represent a statistically significant higher or lower level(p<0.05, one-way ANOVA) in UCML in comparison with the healthy control, respectively. Pink and blue blocks represent a higher or lower (not statistical significance, p>0.05, one-way ANOVA) level in UCML in comparison with healthy control, respectively.
(0.20 MB TIF)
Compounds identified in human blood plasma.
(0.12 MB DOC)
The relative abundance of the identified compounds/peaks differentiating RCML BC from RCML CP patients.
(0.06 MB DOC)
Primer pairs used for PCR.
(0.04 MB DOC)
Acknowledgments
We thank Guangrong Zhu for his assistance in samples collecting and processing.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Natural Science Foundation of the People's Republic of China (No. 81070437,30700461), the National Key New Drug Creation Special Programs (2009ZX09304-001 and 2009ZX09502-004), Jiangsu Province Social Development Foundation (BE2008673), Jiangsu Province's Outstanding Medical Academic Leader Program (LJ200623), and the National Science & Technology Pillar Program of China (Grant No. 2008BAI61B01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fan TW, Higashi RM, Lane AN. Integrating metabolomics and transcriptomics for probing SE anticancer mechanisms. Drug Metab Rev. 2006;38:707–732. doi: 10.1080/03602530600959599. [DOI] [PubMed] [Google Scholar]
- 2.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 3.Kurzrock R, Gutterman J, Talpaz M. The molecular genetics of Philadelphia chromosome-positive leukemias. N Engl J Med. 1988;319:990–998. doi: 10.1056/NEJM198810133191506. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 5.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 6.Kantarjian H, O'Brien S, Cortes J, Shan J, Giles FJ, et al. Complete cytogenetic and molecular responses to interferon-based therapy for chronic myelogenous leukemia are associated with excellent long-term prognosis. Cancer. 2003;97:1033–1041. doi: 10.1002/cncr.11223. [DOI] [PubMed] [Google Scholar]
- 7.Kantarjian H, Schiffer C, Jones D, Cortes J. Monitoring the response and course of chronic myeloid leukemia in the modern era of BCR-ABL tyrosine kinase inhibitors: practical advice on the use and interpretation of monitoring methods. Blood. 2008;111:1774–1780. doi: 10.1182/blood-2007-09-110189. [DOI] [PubMed] [Google Scholar]
- 8.Oksman-Caldentey KM, Saito K. Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr Opin Biotechnol. 2005;16:174–179. doi: 10.1016/j.copbio.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Branford S, Rudzki Z, Harper A, Grigg A, Taylor K, et al. Imatinib produces significantly superior molecular responses compared to interferon plus cytarabine in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Leukemia. 2003;17:2401–2409. doi: 10.1038/sj.leu.2403158. [DOI] [PubMed] [Google Scholar]
- 10.Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morvan D, Demidem A. Metabolomics by proton nuclear magnetic resonance spectroscopy of the response to chloroethylnitrosourea reveals drug efficacy and tumor adaptive metabolic pathways. Cancer Res. 2007;67:2150–2159. doi: 10.1158/0008-5472.CAN-06-2346. [DOI] [PubMed] [Google Scholar]
- 12.Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics. 2007;8:1243–1266. doi: 10.2217/14622416.8.9.1243. [DOI] [PubMed] [Google Scholar]
- 13.Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat. Med. 2002;8:1439–1444. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- 14.Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, et al. Metabonomic Identification of Novel Biomarkers of Myocardial Ischemia. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 15.Xu G, Liebich HM, Lehmann R, Müller-Hagedorn S. Capillary electrophoresis of urinary normal and modified nucleosides of cancer patients. Methods Mol Biol. 2001;162:459–474. doi: 10.1385/1-59259-055-1:459. [DOI] [PubMed] [Google Scholar]
- 16.Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, et al. Metabolite profiling of human colon carcinoma—deregulation of TCA cycle and amino acid turnover. Mol Cancer. 2008;7:72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Wang B, Zhang M, Rantalainen M, Wang S, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 20.Tiziani S, Lodi A, Khanim FL, Viant MR, Bunce CM, et al. Metabolomic Profiling of Drug Responses in Acute Myeloid Leukaemia Cell Lines. Public Library of Sciences (PLoS) 2009;4(1):e4251. doi: 10.1371/journal.pone.0004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson P, Johansson AI, Gullberg J, Trygg J, A J, et al. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem. 2005;77:5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- 22.A J, Trygg J, Gullberg J, Johansson AI, Jonsson P, et al. Extraction and GC/MS Analysis of the human blood plasma metabolome. Anal Chem. 2005;77:8086–8094. doi: 10.1021/ac051211v. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Umetrics AB, Sweden; 2001. Multi- and megavariate data analysis principles and applications. [Google Scholar]
- 24.Berman E, Nicolaides M, Maki RG, Fleisher M, Chanel S, et al. Altered bone and mineral metabolism in patients receiving imatinib mesylate. N Engl J Med. 2006;354:2006–2013. doi: 10.1056/NEJMoa051140. [DOI] [PubMed] [Google Scholar]
- 25.Gottardi M, Manzato E, Gherlinzoni F. Imatinib and hyperlipidemia. N Engl J Med. 2005;353:2722–2723. doi: 10.1056/NEJMc052500. [DOI] [PubMed] [Google Scholar]
- 26.Franceschino A, Tornaghi L, Benemacher V, Assouline S, Gambacorti-Passerini C. Alterations in creatine kinase, phosphate and lipid values in patients with chronic myeloid leukemia during treatment with imatinib. Haematologica. 2008;93:317–318. doi: 10.3324/haematol.11680. [DOI] [PubMed] [Google Scholar]
- 27.Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin Cancer Res. 2004;10:6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 28.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–46. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 29.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Lu W, Garcia-Prieto C, Huang P. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr. 2007;39:267–274. doi: 10.1007/s10863-007-9086-x. [DOI] [PubMed] [Google Scholar]
- 31.Keurentjes JJ, Fu J, de Vos C, Lommen A, Hall RD, et al. The genetics of plant metabolism. Nat Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- 32.Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107:76–94. doi: 10.1159/000046636. [DOI] [PubMed] [Google Scholar]
- 33.Nowak D, Ogawa S, Müschen M, Kato M, Kawamata N, et al. SNP array analysis of tyrosine kinase inhibitor-resistant chronic myeloid leukemia identifies heterogeneous secondary genomic alterations. Blood. 2010;115:1049–1053. doi: 10.1182/blood-2009-03-210377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 35.Zheng C, Li L, Haak M, Brors B, Frank O, et al. Gene expression profiling of CD34+ cells identifies a molecular signature of chronic myeloid leukemia blast crisis. Leukemia. 2006;20:1028–1034. doi: 10.1038/sj.leu.2404227. [DOI] [PubMed] [Google Scholar]
- 36.Yong AS, Szydlo RM, Goldman JM, Apperley JF, Melo JV. Molecular profiling of CD34+ cells identifies low expression of CD7, along with high expression of proteinase 3 or elastase, as predictors of longer survival in patients with CML. Blood. 2006;107:205–212. doi: 10.1182/blood-2005-05-2155. [DOI] [PubMed] [Google Scholar]
- 37.McWeeney SK, Pemberton LC, Loriaux MM, Vartanian K, Willis SG, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010;115:315–325. doi: 10.1182/blood-2009-03-210732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kominsky DJ, Klawitter J, Brown JL, Boros LG, Melo JV, et al. Abnormalities in glucose uptake and metabolism in imatinib-resistant human BCR-ABL-positive cells. Clin Cancer Res. 2009;15:3442–3450. doi: 10.1158/1078-0432.CCR-08-3291. [DOI] [PubMed] [Google Scholar]
- 39.Sviridov D, Nestel P. Dynamics of reverse cholesterol transport: Protection against atherosclerosis. Atherosclerosis. 2002;161:245–254. doi: 10.1016/s0021-9150(01)00677-3. [DOI] [PubMed] [Google Scholar]
- 40.Batetta B, Sanna F. Cholesterol metabolism during cell growth: Which role for the plasma membrane? Eur. J. Lipid Sci. Technol. 2006;108:687–699. [Google Scholar]
- 41.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukemia. Nat Rev Cancer. 2005;5:173–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 42.Rowley JD, Blumenthal T. Medicine. The cart before the horse. Science. 2008;321:1302–1304. doi: 10.1126/science.1163791. [DOI] [PubMed] [Google Scholar]
- 43.Mitelman F. Basel: S. Karger; 1995. ISCN an international system for human cytogenetic nomenclature. [Google Scholar]
- 44.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PLS_DA scores plot of SCML, UCML and healthy control (HC). The overlapping of SCML and HC suggested the rectification of endogenous compound in SCML towards normal. While the distinct separation of SCML and UCML suggested metabolic responses to imatinib.
(0.32 MB TIF)
The PLS_DA scores plot of RCML BC (▪, n = 6) and RCML CP (□, n = 8) patients. RCML BC patients showed totally different metabonomic phenotype from that of RCML CP patients.
(0.16 MB TIF)
Metabolic perturbations in UCML and metabolic response to imatinib treatment. The marked metabolites were observed at an abnormal level in UCML, and most of them were regulated towards normal in SCML with imatinib. Red or green blocks represent a statistically significant higher or lower level(p<0.05, one-way ANOVA) in UCML in comparison with the healthy control, respectively. Pink and blue blocks represent a higher or lower (not statistical significance, p>0.05, one-way ANOVA) level in UCML in comparison with healthy control, respectively.
(0.20 MB TIF)
Compounds identified in human blood plasma.
(0.12 MB DOC)
The relative abundance of the identified compounds/peaks differentiating RCML BC from RCML CP patients.
(0.06 MB DOC)
Primer pairs used for PCR.
(0.04 MB DOC)