Abstract
Fluorescence molecular tomography (FMT) systems coupled to conventional imaging modalities such as magnetic resonance imaging (MRI) and computed tomography provide unique opportunities to combine data sets and improve image quality and content. Yet, the ideal approach to combine these complementary data is still not obvious. This preclinical study compares several methods for incorporating MRI spatial prior information into FMT imaging algorithms in the context of in vivo tissue diagnosis. Populations of mice inoculated with brain tumors that expressed either high or low levels of epidermal growth factor receptor (EGFR) were imaged using an EGF-bound near-infrared dye and a spectrometer-based MRI-FMT scanner. All data were spectrally unmixed to extract the dye fluorescence from the tissue autofluorescence. Methods to combine the two data sets were compared using student’s t-tests and receiver operating characteristic analysis. Bulk fluorescence measurements that made up the optical imaging data set were also considered in the comparison. While most techniques were able to distinguish EGFR(+) tumors from EGFR(-) tumors and control animals, with area-under-the-curve values=1, only a handful were able to distinguish EGFR(-) tumors from controls. Bulk fluorescence spectroscopy techniques performed as well as most imaging techniques, suggesting that complex imaging algorithms may be unnecessary to diagnose EGFR status in these tissue volumes.
Keywords: biomedical optics, image reconstruction, magnetic resonance imaging, fluorescence spectroscopy, tomography
Introduction
The identifiable biochemical changes associated with cancer pathologies have made imaging cancer a major focus of efforts to realize the potential of fluorescence molecular tomography (FMT). These efforts have included imaging elevated enzymatic activity using activatable molecular probes,1, 2, 3 enhanced permeability and retention (EPR) effects using unspecific fluorophores,4 deoxyglucose uptake,5 and cellular protein status using antibody, ligand, or other protein-bound fluorophores.6, 7 Imaging strategies that target a biological process, such as receptor status, may help identify an effective therapy and subsequently monitor the biological changes induced by therapy. These strategies may be employed in preclinical research with the goal of better understanding molecular signaling or toward clinical translation for diagnostic imaging.
Since scattering dominates near-infrared (NIR) photon propagation in tissue, quantifying emitted signals through more than a few millimeters of tissue is a challenging problem. However, the availability and sensitivity of optical probes used in biological research and the spectral separation capabilities unique to luminescent compounds has sustained efforts to address the complex imaging problem. To account for tissue scattering, most FMT systems operate by algorithmically fitting the measured data to the diffusion model. Efforts to improve the imaging performance of FMT have included refining imaging geometries,8 exploiting time-dependent measurements,9, 10, 11, 12, 13, 14, 15, 16 extracting highly resolved spectral data,17, 18 developing more accurate photon propagation models,19 and incorporating information from supplemental measurements into the imaging paradigm.3, 12, 17, 20, 21 The latter approach is particularly attractive, and experiments with new hybrid FMT instruments that operate concurrently or in sequence with computed tomography (CT) or magnetic resonance imaging (MRI) have been reported, along with associated imaging algorithms for combining these bimodal data sets. These prior-information-type algorithms were originally introduced for absorption and scattering tomography and showed promise for improved accuracy with the additional information.22, 23, 24, 25, 26 The most common methods to combine data sets involves segmenting the CT or MRI images into general regions based on tissue type and then using the identified regions to guide the recovery of optical images. Segmented information may be incorporated differently depending on the accuracy of the segmentation and how closely the fluorescence activity corresponds to the segmented tissue types. In some applications, it may be appropriate to assume that fluorescence activity is homogeneous in each segmented region and force this constraint in the imaging algorithm. While this approach makes no assumptions about the actual values in the segmented regions, it does limit the ability to recover more subtle changes throughout the imaging volume. An alternative approach introduces the segmented information as a filter matrix in the image reconstruction process and thus is less rigid in its application of spatial priors. While improvements in imaging performance have been implied and anecdotally reported using these techniques, a full systematic examination comparing data analysis strategies from a diagnostic perspective has not been reported and so is examined here.
In a previous report,7 we demonstrated that preclinical MRI-FMT could be used to diagnose gliomas in vivo based on epidermal growth factor receptor (EGFR) activity. This transmembrane protein is overexpressed in many cancers, and its activation by the epidermal growth factor (EGF) ligand is associated with increased cell proliferation and reduced apoptosis and has thus been the focus of substantial cancer research. In our previously reported study, a fluorophore bound to EGF was used to distinguish between tumors with high and low EGFR status. The analysis was completed using one embodiment of an MR-guided FMT algorithm—namely, the soft-priors approach, which introduces the internal tissue structures by implementing a spatially dependent regularization parameter in the image reconstruction algorithm. In the study reported herein, we reexamine these data to assess diagnostic performance of the optical data using a variety of imaging and bulk spectroscopy approaches and attempt to quantify improvements in diagnostic capacity provided by spatially guided FMT.
Methods
Cell Lines
The two cell lines investigated in this study were the rat 9L gliosarcoma cell line transfected with green fluorescent protein (GFP) and the human glioma cell line U251. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Mediatech, Inc., Manassas, Virginia, Cat. # 10-013-CV) supplemented with penicillin (100 units∕ml)-streptomycin, 100 μg∕ml (HyClone, Logan, Utah, Cat. # SV30010). In vitro studies of the tumor cells have shown that U251 cells have a 20-fold higher expression of EGFR than 9L cells.27 In this study, U251 tumors that overexpress EGFR are referred to as EGFR(+) tumors, while the negative control 9L line tumors are termed EGFR(-) tumors. Cells were grown to 80% confluency in culture, trypsinized, and brought into solution in phosphate buffered saline (PBS) at 5×107 cells∕ml in preparation for injection.
Animal Preparation
All procedures using animals were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Dartmouth. Animal subject preparations for the nude mice used in this study have been described previously7 and are summarized here. Mice were 6 to 7 week-old male athymic NCr-nu∕nu nude mice (strain 01B74) purchased through the NIH Animal Procurement Program. All mice underwent intracranial surgery prior to imaging. During the surgical procedure, the skin on the skull was prepared using betadine. Five μl of the cell suspension were injected slowly via a 1-mm burr hole in the skull with a 25-ga needle at 2 mm anterior and 3 mm to the left of the bregma. Control mice underwent sham-surgery and were injected with PBS only. Tumors grew for 14 to 23 days, and each tumor-bearing animal was imaged with gadolinium-enhanced MRI (Gd-MRI) at least two days prior to the MRI-FMT scans. Only animals with visible Gd-enhanced features in the brain were included in the study. The final distribution of animals used in the study was six mice with U251 tumors, five with 9L tumors, and four in the control group. Forty-eight hours prior to MRI-FMT scanning, each animal was administered 1 nmole of Licor IRDye 800CW EGF (Licor Biosciences, Lincoln, Nebraska) reconstituted per manufacturer’s directions into the tail vein. This imaging agent consists of an NIR fluorescence dye conjugated to the EGF ligand, which has high affinity for the associated EGFR.
MRI-FMT Imaging System
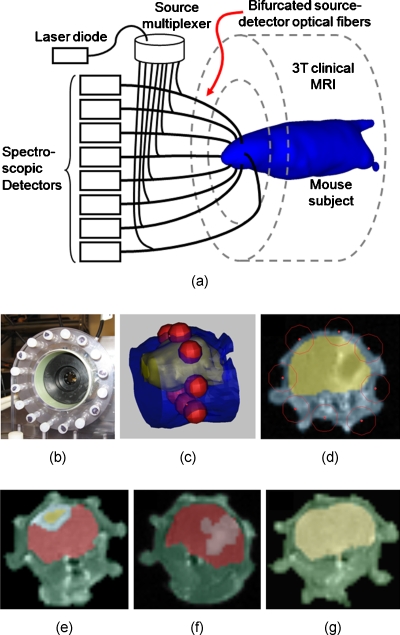
The hybrid MRI-optical system has been detailed in previous publications,7, 17, 18 and a brief overview is included here. A diagram of the system is provided in Fig. 1a, which illustrates the spectroscopic FMT system integrated into a Philips 3 T clinical MRI. This integrated platform allows simultaneous optical and MR scanning that facilitates straightforward integration of data between the modalities. The optical detection component consists of eight spectrometers, each with cooled imaging CCD sensors, which are coupled to the tissue surface through long optical fiber bundles. A specialized rodent MRI coil, shown in Fig. 1b, positions the optical fibers in a circular ring around the animal’s head. The bifurcated branch of the fibers was used to illuminate each channel sequentially with a 690-nm laser diode, while the remaining seven channels act as light pickups to the spectrometers. Both fluorescence emission and excitation spectra were measured for each source–detector pair. In this configuration, a total of 56 emission and 56 excitation measurements (eight sources by seven detectors) were acquired for each animal.
Figure 1.
Diagram of the MRI-FMT scanner illustrates the contact-mode configuration of eight optical fibers around the head (a). The fiber array installed in the RF coil is shown in (b). A close-up 3-D rendering of the head (blue) and brain (yellow) of one of the animals is shown in (c). The red spheres mark the location of the eight spectroscopy fibers encircling the head in a single plane. A T1-weighted coronal MR image corresponding to this fiber plane is shown in (d), overlain with blue and yellow regions to emphasize the location of the brain. Once this plane is selected for a given animal, the image is segmented into tissue regions broadly defined as brain, abnormal structures in the brain, and the region outside the brain. Examples of segmented regions for a mouse with a U251 tumor, a 9L tumor, and no tumor are shown in (e), (f), and (g), respectively. (Color online only.)
During MRI-FMT scanning, an anesthetized (1.5% isoflurane, 1 L∕min oxygen) animal was positioned in the specialized rodent coil and the fiber terminals moved inward to contact the tissue. Scout MRI scans were used to locate the plane of the fibers with respect to the brain and∕or tumor (if visible pre-Gd contrast). This procedure was repeated until the animal alignment was satisfactory. Once the animal was positioned, both optical and MRI scans were initiated. The MRI acquisition protocol included a T1-weighted turbo-spin-echo (TSE) pre-Gd contrast followed by T2-weighted TSE, intraperitoneal gadolinium-DTPA (Magnevist, Bayer Healthcare Pharmaceuticals, Wayne, New Jersey) administration (0.03 ml), and finally another T1-weighted TSE post-Gd contrast. Besides scout scans, all MRI image slices were in the coronal direction. The optical measurements were completed within the 23 min required for the MRI sequences, and the entire procedure including anesthetization could be completed in about 30 min.
Figure 1c shows a portion of a mouse head rendered from an MR image stack, with the brain shown as a rendered surface in yellow. The red spheres surrounding the head lie in a coronal plane and mark the location of the optical fibers. The 2-D image slice corresponding to this plane, shown in Fig. 1d, represents the MRI-FMT image plane. With the exception of control mice, only animals with Gd contrast-enhancement in this plane were included in the analysis. For MRI-FMT imaging, the image was segmented into regions of broadly defined tissue types using MIMICS image processing software (Materialise Medical Software, Leuven, Belgium). Tissues were delineated as brain, the area outside the brain, any gadolinium-enhanced features in the brain, and any other abnormal-looking features not associated with Gd-enhancement. Since no abnormal features were evident in the brains of control animals, only the brain and area surrounding the brain were included in the segmentation. Examples of segmented MR images from each group (U251, 9L, and control) are shown in Figs. 1e, 1f, 1g, respectively, where the color overlays delineate different regions. Similar segmented masks were produced for each animal and used to generate finite element method (FEM) meshes compatible with the imaging algorithms.
Optical Image Reconstruction
FMT image recovery involves fitting calculated data from a light propagation model to the measured data. The diffusion approximation for photon propagation in tissue is the most commonly used model for FMT imaging and was used in this study. These algorithms recover images of fluorescence yield, which is the product of the fluorophore quantum yield, η, and the absorption coefficient of the fluorescence compound at the excitation wavelength, μaf. While early FMT systems were designed as stand-alone imaging systems, the latest generation of research FMT instruments are hybrid systems integrated with established imaging modalities. Paralleling the advances in hardware design, several methods to synthesize the dual data sets have been explored. Two of these methods are considered herein.
In this study, four diffusion-based recovery techniques were investigated, each representing a different degree to which the segmented MRI masks guide the imaging algorithm. The general formulation used to match the measured and calculated fluence rates, and , respectively, and thus recover fluorescence yield is
| (1) |
where J is the Jacobian matrix with dimensions of number of nodes in the FEM mesh (NN) by number of measurements, λ is the regularization parameter, and I is the identity matrix.17 The regularization parameter, λ, is a fixed value multiplied to the maximum of the diagonal of JTJ (Ref. 17) and was determined empirically with a small set of animal images. Once determined, the same initial value for λ was used for all animals in the study for consistency. The classical image reconstruction technique assumes that only the shape of the tissue surface is known. A variety of methods can be used to render the surface features that do not require medical imaging devices,5, 28, 29 but in this study, the surfaces were extracted directly from the MR images. For the purposes of this report, we have termed this classical FMT approach that makes no use of internal tissue information the “unguided” reconstruction technique.
Internal tissue structure was incorporated into the algorithm in two ways. The hard-priors technique assumes that all segmented regions have homogeneous values of fluorescence yield and thus puts complete trust in the quality of the segmented MR images. The formulation is straightforward and here is accomplished by reducing the Jacobian matrix based on tissue regions;
| (2) |
where K is a matrix with dimensions NN×NR, NN is the total number of nodes in the mesh, and NR is the number of tissue regions in the segmented mask. For regions 1 to N, K is written:
| (3) |
where
| (4) |
and Rn represents region n. The dimension of the reduced Jacobian matrix, , is number of measurements by number of regions. The update equation for fluorescence yield is then modified to the following:
| (5) |
Another spatially guided approach introduces the segmented information through a spatially varying regularization parameter and is termed the “soft-priors” or “L-matrix” approach.17, 30 The updated formulation is written as
| (6) |
where the elements in matrix L associated with nodes in a given tissue region are assigned the same value. The L-matrix represents an NN×NN Laplacian-type structure, the diagonal of which is Li,i=1, where i and j are nodal indices. If matrix elements are associated with nodes in the same tissue region (i.e., nodes i and j are in the same region) containing n nodes, Li,j=−1∕n. Elements associated with nodes that are not in the same tissue region are assigned Li,j=0.
The fourth diffusion model–based technique investigated in this study is formulated on the assumption that the fluorescence yield value in the entire head is homogeneous. Like the unguided technique, the geometry of the imaging domain is assumed known, and internal tissue information is not included in the algorithmic reconstructions; however, all nodal values of fluorescence yield are locked to the same value. This “homogeneous fitting” technique is analogous to a single-region hard-priors reconstruction.
All four diffusion-based reconstruction techniques used in the study are summarized in the first four rows of Table 1. A critical consideration when implementing these techniques is the estimation of the optical properties (absorption and reduced scattering coefficients, μa and ) that form the foundation of the light propagation model. Typically, these values are not recovered explicitly and thus must be estimated. For the hard- and soft-priors techniques, the segmented MR images were used to assign heterogeneously distributed optical properties based on two broad tissue types, the brain and all tissue outside the brain. The optical properties in the brain region were estimated from values for rat brain published in the literature31: μax=0.03 mm−1, at the excitation wavelength and μam=0.03 mm−1, at the emission wavelength. Suspected tumor regions in the brain were assigned these values as well. The tissue surrounding the brain is a mixture of bone, muscle, adipose tissue, and skin, all of which were lumped into a single homogeneous region with estimates of μax,m=0.01 mm−1 and at the exciting and emitting wavelengths. Since the internal distribution of tissue types in the unguided and homogeneous fitting algorithms were assumed unknown, the optical properties for these techniques were assumed to be homogeneous throughout the head (μax=0.02 mm−1, , μam=0.02 mm−1, and ). These values are the averages of the properties in the two tissue regions used in the hard- and soft-priors implementations.
Table 1.
Descriptions of the eight data processing techniques considered in this study.
| Analysis technique | Diffusion model? | Description |
|---|---|---|
| Hard-priors reconstruction | Yes | Segmented tissue regions from the MR image are assumed homogeneous. |
| Soft-priors reconstruction | Yes | Segmented tissue regions from the MR image are introduced as a discontinuous regularization matrix in the imaging algorithm. |
| Unguided reconstruction | Yes | No internal structure from the segmented MR image is introduced in the imaging algorithm. The outer boundary of the tissue is known. |
| Homogeneous fitting | Yes | The fluorescence distribution is assumed to be homogeneous in the entire domain, and the outer boundary of the tissue is known. This is essentially a single-region hard-prior reconstruction. |
| Fluorescence-to- excitation ratio (mean) | No | Mean of the fluorescence-to-excitation measurement ratios for all source-detector positions. |
| Fluorescence-to- excitation ratio (max) | No | Maximum value of all fluorescence- to-excitation ratios for a given animal. |
| Fluorescence-to- autofluorescence ratio (sum) | No | Sum of the fluorescence-to- autofluorescence measurement ratios for all source-detector positions. |
| Fluorescence-to- autofluorescence ratio (single measurement) | No | A single fluorescence-to- autofluorescence measurement ratio. The ratio for the source-detector measurement from the top of the head to the bottom of the head was chosen (dorsal to ventral source-detector pair). |
Spectra measured for each source–detector combination were calibrated and then preprocessed with a simple spectral unmixing algorithm described in previous publications.17, 18 This procedure fits premeasured fluorescence spectra of the imaging agent and tissue autofluorescence to the measured data and thus decouples these signals from one another. Once decoupled, the unmixed dye fluorescence spectra were integrated and input into the imaging algorithms.
Bulk Fluorescence Spectroscopy
To compare the diagnostic capacity of MRI-FMT to simpler optical spectroscopy methods, the same optical data used in the image recovery algorithms were also analyzed independent of the imaging problem. These spectroscopy techniques consider the measured spectra alone and therefore do not require geometry-specific FEM modeling or diffusion approximation computations. The four different approaches considered in this study involved calculating the ratios of the unmixed dye fluorescence intensity with either the excitation intensity or the unmixed autofluorescence signal. Descriptions of each method are included in the last four rows of Table 1. The diagnostic performance of the four diffusion model–based techniques and the four bulk spectroscopy techniques were compared using one-tailed student’s t-tests and receiver operating characteristic (ROC) curves.
Results
MRI-FMT Imaging
Examples of MRI-FMT images for three mice are shown in Fig. 2. The first column shows images of a mouse with a U251 tumor, the second column shows a mouse with a 9L tumor, and the third column shows images from a control mouse. Each row corresponds to images recovered using a different reconstruction technique. Images in row I were recovered using hard-priors reconstructions; row II, soft-priors reconstructions; and row III, unguided reconstructions. For illustrative purposes, the color scales are consistent for each example animal (columns) but vary between subjects.
Figure 2.
Representative images of fluorescence yield overlying the corresponding MR images. Each column provides results for a single animal from one of the three mouse populations (U251 tumors, 9L tumors, and controls, respectively). The rows show images recovered using the hard-prior technique (I), the soft-prior technique (II), and an unguided reconstruction technique that operates under the assumption that only the outer boundary was known (III). Given the wide range of recovered values between subjects, the color scales were kept consistent only within the image set for each animal. Transparency levels were varied between images to ensure that the MR images are easily discerned through the optical image overlay. (Color online only.)
Examining the images collectively reveals dramatic difference between the U251 animal and the other two animals for all three imaging techniques. Considering the images in row I, the maximum fluorescence yield value in the U251 animal is approximately tenfold higher than in either the 9L or control animals. The elevated fluorescence activity was well localized in the Gd-enhanced regions of the brain with the U251 tumor, indicating that the fluorescence activity is primarily in the tumor region. This is consistent for the hard- and soft-priors reconstructions for this mouse, which produced similar qualitative results. The unguided reconstruction image for the U251 animal shown in row III produced elevated levels of fluorescence in the same general region, although the spatial resolution and contrasts were substantially lower than produced by the hard- and soft-priors algorithms. Levels of fluorescence yield in the 9L tumor were lower than in the head, indicating poor tumor-to-normal tissue contrast, even when MRI data was used in the imaging algorithms. The unguided solution indicated no tendency toward elevated fluorescence in the Gd-enhanced region, unlike the results in the U251 animal. Images of the control animal also show relatively low overall levels of fluorescence activity and low contrasts throughout the entire head.
The examples in Fig. 2 are representative of the entire image set, demonstrated by Fig. 3, which presents images of fluorescence yield recovered using the soft-priors reconstruction for all animals included in the study. The images are plotted on a consistent color scale, which reveals the large differences between the animal groups. According to the images, U251 tumors produce much higher levels of fluorescence activity than found in 9L or control animals, while there is no noticeable difference between the 9L and control animals themselves. Similar image sets were generated for the hard-priors and unguided reconstruction techniques but are not included here for the sake of brevity.
Figure 3.
Images of fluorescence yield for all animals in the study recovered using the soft-priors image reconstruction technique. The columns correspond to the U251, 9L, or control mouse populations. All images are plotted on the same color scale. (Color online only.)
The fluorescence activity was highest in the Gd-enhanced region of the head in all but one of the animals in the U251 group. In this animal (the last image in the U251 column of Fig. 3), the elevated region of fluorescence activity corresponded to a dark, abnormal feature in the brain that was segmented during the MR image preprocessing. The Gd-enhanced region, circled in blue, produced lower fluorescence activity than the maximum value, but higher activity than the brain background. (Color online only.) This value was also higher than all recovered values in the 9L and control animals.
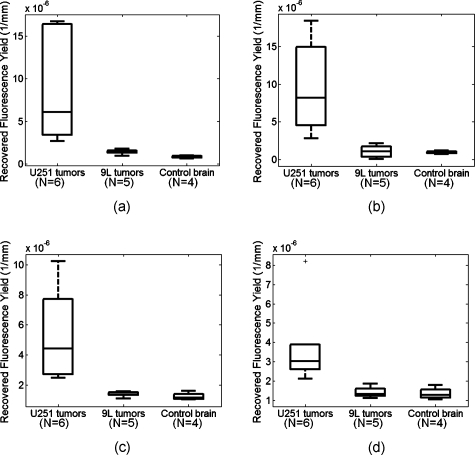
To quantify the imaging results, the mean fluorescence yield value in the region defined by Gd-enhancement was calculated for all tumor-bearing animals. This corresponds to the light blue region in the example shown in Fig. 1e and the light pink region in the example in Fig. 1f. (Color online only.) Since no Gd-enhancement was observed in the brains of control mice, the mean value in the entire brain region was used. These values are compiled in the box and whisker plots presented in Fig. 4 for the hard-priors, soft-priors, and unguided reconstructions [Figs. 4a, 4b, 5c, respectively]. The values plotted in Fig. 4d were recovered using the homogeneous fitting approach, which essentially averages the fluorescence activity over the entire domain. These plots show reasonable separation between U251 and the other two groups, although the difference between 9L tumors and control animals is less obvious. P-values between the animal groups calculated using a one-tailed student’s t-test are tabulated in the first four rows of Table 2. The differences between U251 tumors and the other two groups were statistically significant for all four diffusion-based recovery algorithms. However, only the hard-priors imaging technique revealed a statistically significant difference between the 9L tumor-bearing and control groups.
Figure 4.
Box and whisker plots of fluorescence yield calculated using diffusion-based reconstruction techniques. The values in (a) through (c) were determined by calculating the mean value of fluorescence yield in the region corresponding to gadolinium enhancement for animals with tumors and the mean value in the entire brain region for control animals that showed no gadolinium enhancement. The panels show the results for hard-prior reconstructions (a), soft-prior reconstructions (b), and the unguided reconstruction (c). Values in (d) were recovered using homogeneous fitting, which assumes that the fluorescence activity distribution is homogeneous through the entire head of a mouse subject. Note that the y-axis values are plotted on a different scale for each panel.
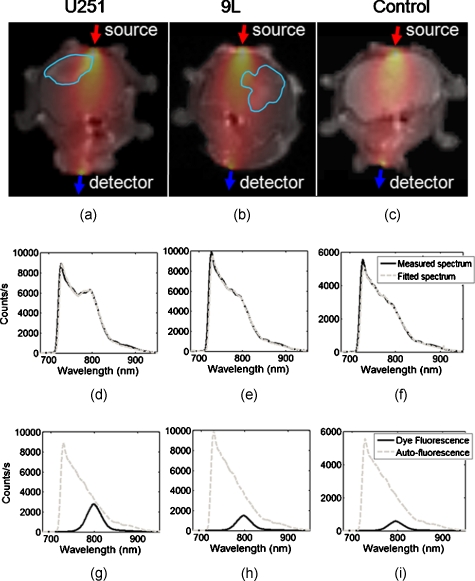
Figure 5.
Raw optical spectra measured with the MRI-FMT are preprocessed using a simple spectral fitting algorithm. Examples of measured fluorescence spectra are shown in (d) through (f) (solid line) for U251, 9L, and control mice, respectively. These spectra correspond to a source-detector pair that transmits light from the top of the head to the bottom, as shown in (a) through (c). For illustrative purposes, the sensitivity values between the source and detector are plotted on the corresponding MR images, and an outline of the tumor region as defined by the gadolinium contrast enhancement is included to demonstrate the extent to which the tumor lies in the sensitivity field. Prerecorded spectra of the fluorescent probe and tissue autofluorescence from mice are used to decouple the signals originating from tissue autofluorescence and the optical probe itself. The results of this process are shown in (g) through (i), and the fitted spectra are compared to the measured spectra in (d) through (f).
Table 2.
p-values from one-tailed student’s t-test.
| Data processing technique | U251 (6) versus 9L (5) | U251 (6) versus controls (4) | 9L (5) versus controls (4) |
|---|---|---|---|
| Hard-priors reconstruction | 0.019* | 0.024* | 0.0053* |
| Soft-priors reconstruction | 0.0067* | 0.012* | 0.40 |
| Unguided reconstruction | 0.010* | 0.016* | 0.12 |
| Homogeneous fitting | 0.021* | 0.032* | 0.35 |
| Fluorescence-to- excitation ratio (mean) | 0.023* | 0.022* | 0.043* |
| Fluorescence-to- excitation ratio (max) | 0.017* | 0.017* | 0.04* |
| Fluorescence-to- autofluorescence ratio (sum) | 0.020* | 0.0084* | 0.052 |
| Fluorescence-to- autofluorescence ratio (single measurement) | 0.012* | 0.0067* | 0.090 |
Asterisks indicate statistical significance (p-value below 0.05).
Bulk Fluorescence Spectroscopy
Figure 5 shows the results of the spectral unmixing algorithm in three example animals, each from a different group. The source–detector measurement pair used in these examples was between the top of the head and the opposing detector below the jaw (dorsal to ventral source–detector pair), illustrated in panels (a) through (c) for the three mice. For illustrative purposes, the sensitivity fields for the source–detector pair overlie the MR images. Also included are outlines of the tumor region as defined by Gd-contrast. The solid lines in the graphs in panels (d) through (f) show the measured spectrum for each animal. The sharp jump in intensity at 720 nm corresponds to the cut-on wavelength of the long-pass filter positioned at the entrance of the spectrometers. The results of the spectral unmixing for each animal are plotted in (g) through (i) and illustrate the relative contributions of the imaging agent fluorescence and the tissue autofluorescence. In all cases, the tissue autofluorescence is the dominant signal, revealing the importance of the spectral unmixing process. The sums of the unmixed basis spectra are plotted as fitted spectra in (d) through (f) (dashed lines), demonstrating an excellent match between measured and fitted data.
Visual inspection of the results in Fig. 5g, 5h, 5i suggests that the spectra alone contain diagnostic information, without the need for light modeling and image reconstruction. While the absolute intensities of the measured spectra are not correlated with the presence of tumors in these examples, the relative intensity of the dye fluorescence to the autofluorescence, or perhaps the excitation intensity (not shown), may well correlate with the presence of a particular tumor type.
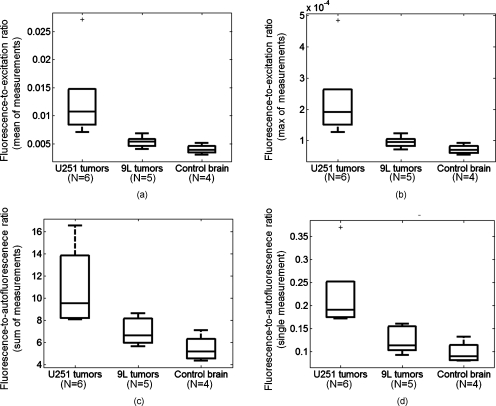
The box and whisker plots and corresponding p-value calculations in Fig. 6 and Table 2, respectively, confirm these observations. In all cases, significantly higher ratio values were calculated for the U251 group relative to the 9L and control groups. Also, two ratio-based techniques produced statistically significant differences between 9L and control animals.
Figure 6.
Box and whisker plots of different measurement ratios. The plots in the top panels were determined by calculating either the sum (a) or average (b) of all fluorescence-to-excitation ratios for a given animal. The plot in (c) contains data determined by calculating the sum of the fluorescence-to-autofluorescence ratios for each animal. Using this ratio eliminates the need to measure the excitation intensity separately and ensures that the numerator and quotient are perfectly calibrated to one another. Last, the fluorescence-to-autofluorescence ratio of single measurement point (shown in Fig. 2) for each animal was used to produce the results plotted in (d). Note that the y-axis values are plotted on a different scale for each panel.
ROC Analysis
The results compiled in Tables 2, 3 indicate that nearly all methods identify the presence of U251 tumors versus 9L tumors or controls. Area-under-the-curve (AUC) values determined from ROC analysis for U251 versus control animals show perfect diagnostic performance for all imaging methods and bulk spectroscopy methods. Similar performance was observed between U251 and 9L tumors, with one exception. The AUC determined from the sum of the fluorescence-to-autofluorescence ratios slipped to 0.93, suggesting that this simple data processing method is a less powerful diagnostic technique.
Table 3.
Area-under-curve values from ROC analysis.
| Data processing technique | U251 (6) versus 9L (5) | U251 (6) versus controls (4) | 9L (5) versus controls (4) |
|---|---|---|---|
| Hard-priors reconstruction | 1 | 1 | 0.95 |
| Soft-priors reconstruction | 1 | 1 | 0.55 |
| Unguided reconstruction | 1 | 1 | 0.65 |
| Homogeneous fitting | 1 | 1 | 0.65 |
| Fluorescence-to- excitation ratio (mean) | 1 | 1 | 0.85 |
| Fluorescence-to- excitation ratio (max) | 1 | 1 | 0.9 |
| Fluorescence-to- autofluorescence ratio (sum) | 0.93 | 1 | 0.85 |
| Fluorescence-to- autofluorescence ratio (single measurement) | 1 | 1 | 0.8 |
Differentiating subjects with 9L tumors from healthy subjects proved to be more difficult for all methods. Three techniques produced statistically significant differences between these groups; the hard-priors imaging technique and the maximum and mean of the fluorescence-to-excitation spectroscopy ratios. These statistical differences translated into reasonably reliable diagnostic performance with AUC values of 0.95, 0.9, and 0.85 for the imaging technique, and maximum and mean fluorescence-to-excitation ratio spectroscopy methods, respectively. The soft-priors imaging technique reported the poorest diagnostic performance between these groups, as the AUC value was just over 0.55, indicating that the groups were nearly indistinguishable. The unguided and homogeneous fitting reconstructions also demonstrated poor diagnostic characteristics with an AUC=0.65 for both methods. All spectroscopic methods produced AUC values over 0.8 for the 9L versus control tests.
Discussion
All techniques considered in this study provided excellent discrimination between animals with U251 tumors and controls, and between animals with the two different tumor lines. These results are consistent with in vitro studies demonstrating significant differences in EGFR expression between U251 and 9L tumor lines.27 Diagnosing 9L tumors from controls was more difficult for most methods, and only three of the eight techniques produced statistically significant differences between these groups. While EGFR expression in 9L tumor cells has been shown in vitro to be relatively low compared to the U251 line, 9L tumors removed from animals after fluorescence-tagged EGF administration do show fluorescence contrast compared to normal brain tissue. Thus, differences between these groups, however modest, are expected. Identifying these differences was highly dependent on the data processing technique used, suggesting that some techniques provide more sensitive and specific characterization. Specifically, the hard-priors imaging technique provided the best discrimination between these animals. Also effective were two bulk spectroscopy methods that used the ratio of the fluorescence emission to the excitation intensity.
The power of the bulk spectroscopy techniques is illustrated in the examples shown in Fig. 5. While the absolute intensity of the measured spectra varies widely between the animals, the absolute and relative intensities of the unmixed dye fluorescence from visual inspection alone seems to correlate with tumor presence and type. The overall ROC performance of all spectroscopic techniques confirms this observation and indicates that even single spectrum measurements contain significant diagnostic information. A particularly interesting result is that the ratio of dye fluorescence to autofluorescence calculated for a single measurement through the head reported perfect diagnosis of the U251 tumors. The ability of this method to distinguish 9L tumors from controls was also better than most diffusion-modeling techniques as determined by ROC analysis. This method is particularly attractive because it is derived from a single measurement and does not require a separate excitation measurement, much less MRI scanning, FEM mesh generation, or light propagation modeling. Also, the numerator and denominator of the ratio are extracted from the same spectrum and therefore are perfectly calibrated to one another, making the data processing particularly robust.
Fluorescence spectroscopy measurements acquired in this study likely benefited from the MRI-guided alignment procedures used to ensure that the fiber plane intersected at least a portion of the tumor bulk. However, in many cases, the tumor was not visible in the pre-Gd images used for alignment, and thus these images were used only to confirm that the fiber plane intersected as close to the axial midpoint of the brain as possible. Thus, it is unclear as to whether a nonguided measurement system designed to ensure repeatable fiber positioning would provide similar performance, although we suspect this is the case. If fiber positioning in an unguided system has a major impact on tissue diagnosis, protocols that sample several locations through the head and either use the maximum ratio values to identify an appropriate measurement location or use the maximum ratio value itself may readily be implemented. These protocols would likely promote repeatable and reliable tissue diagnosis.
The data suggest that bulk fluorescence spectroscopy provides diagnostic performance similar to that of MRI-FMT imaging. Most of these simple techniques showed perfect discriminating power between tumor lines and between the U251 and control groups. The diagnostic power of the spectroscopic data for these tumor lines was first suggested in a study by Gibbs-Strauss et al.,27 which used the fluorescence-to-excitation ratios normalized to values of a control group using the same optical detection system as used herein. Even with the extra normalization procedure, the ROC performance reported in the previous study was lower than what is reported here. The discrepancy is likely attributable to a modification of the fiber positioning system between the two studies. The original interface did not ensure adequate fiber contact with the animal’s head and was not optimized to absorb stray light signals. The new interface, shown in Fig. 1b, eliminates the stray light issue and provides reliable contact between the fiber and tissue surface. This modification produced dramatic improvements in the imaging capabilities and seems to have also positively impacted the diagnostic power of the spectroscopic measurements.
The previously reported study and system modifications notwithstanding, these results are rather surprising and suggest that under the experimental conditions used here, FMT and MRI-FMT hold little advantage over simple bulk tissue spectroscopy for diagnosing EGFR status in these orthotopic brain tumors. Due to the practical and algorithmic complexities and uncertainties involved with model-based imaging, spectroscopy techniques may be preferred in applications with proven diagnostic performance. While diffusion-based algorithms are commonly used for small animal imaging, accurate modeling of light propagation through these small, heterogeneous volumes is challenging. More accurate models have been developed,19, 32 although they have not been widely used in animal imaging to date. The modeling problem is also subject to the effects of estimating the tissue optical properties used in the models, as most FMT systems do not recover these values explicitly. In the paradigm used in this study, literature values were assigned to two large tissue regions; however, the values used may be substantially off of true values, a reality that is difficult to test or validate in each mouse. Also, optical properties in tissue are far more heterogeneously distributed than the modeling used in this study. Incorrect estimation of optical properties is likely one of the largest sources of uncertainty in the imaging problem. Other contributions include uncertainty in the MR image segmentation and using a 2-D model rather than a full volumetric recovery procedure.
It is unlikely that the diagnostic performance reported here can be directly extrapolated to different tissue volumes and optical probes. The brain is unique in producing high levels of fluorescence contrast with very low autofluorescence in the relevant wavelength range. Also, the tissue volumes sampled were relatively small and most of the tumors occupied a relatively large region of the sampled volume. Diagnoses in larger tissue volumes should favor imaging techniques, since these volumes will generally adhere more closely to the diffusion model. Measuring through larger volumes also amplifies the effects of tissue scattering, making spectroscopic measurements less specific and therefore more difficult to interpret. The trade-offs between the tissue volume size and geometry, tumor size and depth, tissue optical properties, and drug contrast will affect whether imaging is necessary for a particular diagnostic test. Until these limits have been established, transmission spectroscopy based on data ratios should be considered for each application under study.
Conclusion
All imaging and fluorescence spectroscopy analysis techniques provide excellent potential for diagnostic classification of EGFR(+) tumors in vivo. That spectroscopic measurements alone provide equivalent classification to MRI-guided FMT suggests that MRI-FMT may not necessarily provide improvements over nonguided FMT or even bulk fluorescence spectroscopy, at least for diagnosing EGFR status in brain tumors. Hybrid imaging is still required when the location of the lesion is uncertain, as knowing the suspected volume is still a critical part of diagnosis. However, it is not apparent that spatial reconstruction of the fluorescence signal provides quantifiable improvement in detection beyond analysis of the bulk measurement parameters, as long as they are processed in a ratio that is robust. The speed and simplicity of bulk spectroscopy techniques make them particularly attractive for high-throughput screening of research animals, and image-guided spectroscopy may be the most robust approach for translation to human diagnostic testing.
Acknowledgments
We would like to thank Nathan Watson and Mark Israel for use of the U251 tumor cell line. The 9L cells were a gift from Alexei Bogdanov. This research was funded by the National Institutes of Health Grant Nos. RO1 CA109558 and RO1 CA069544 and Philips Research Hamburg, Department of Defense Breast Cancer predoctoral fellowship BC051058.
References
- Weissleder R., Tung C. H., Mahmood U., and A.Bogdanov, Jr., “In vivo imaging of tumors with protease-activated near-infrared fluorescent probes,” Nat. Biotechnol. 17, 375–378 (1999). 10.1038/7933 [DOI] [PubMed] [Google Scholar]
- Ntziachristos V., Tung C. H., Bremer C., and Weissleder R., “Fluorescence molecular tomography resolves protease activity in vivo,” Nat. Med. 8(7), 757–760 (2002). 10.1038/nm729 [DOI] [PubMed] [Google Scholar]
- McCann C. M., Waterman P., Figueiredo J. L., Aikawa E., Weissleder R., and Chen J. W., “Combined magnetic resonance and fluorescence imaging of the living mouse brain reveals glioma response to chemotherapy,” Neuroimage 45, 360–369 (2009). 10.1016/j.neuroimage.2008.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlu A., Choe R., Durduran T., Rosen M. A., Schweiger M., Arridge S. R., Schnall M. D., and Yodh A. G., “Three-dimensional in vivo fluorescence diffuse optical tomography of breast cancer in humans,” Opt. Express 15(11), 6696–6716 (2007). 10.1364/OE.15.006696 [DOI] [PubMed] [Google Scholar]
- Li C., Mitchell G. S., Dutta J., Ahn S., Leahy R. M., and Cherry S. R., “A three-dimensional multispectral fluorescence optical tomography imaging system for small animals based on a conical mirror design,” Opt. Express 17(9), 7571–7585 (2009). 10.1364/OE.17.007571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V., Schellenberger E. A., Ripoll J., Yessayan D., Graves E., A.BogdanovJr., Josephson L., and Weissleder R., “Visualization of antitumor treatment by means of fluorescence molecular tomography with an annexin V-Cy5.5 conjugate,” Proc. Natl. Acad. Sci. U.S.A. 101(33), 12294–12299 (2004). 10.1073/pnas.0401137101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. C., Samkoe K. S., O’Hara J. A., Gibbs-Strauss S. L., Payne H. L., Hoopes P. J., Paulsen K. D., and Pogue B. W., “MRI-coupled fluorescence tomography quantifies EGFR activity in brain tumors,” Acad. Radiol. 17(3), 271–276 (2010). 10.1016/j.acra.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves E. E., Culver J. P., Ripoll J., Weissleder R., and Ntziachristos V., “Singular-value analysis and optimization of experimental parameters in fluorescence molecular tomography,” J. Opt. Soc. Am. A 21(2), 231–241 (2004). 10.1364/JOSAA.21.000231 [DOI] [PubMed] [Google Scholar]
- Gao F., Zhao H., Tanikawa Y., and Yamada Y., “A linear, featured-data scheme for image reconstruction in time-domain fluorescence molecular tomography,” Opt. Express 14(16), 7109–7124 (2006). 10.1364/OE.14.007109 [DOI] [PubMed] [Google Scholar]
- Gao F., Zhao H., Zhang L., Tanikawa Y., Marjono A., and Yamada Y., “A self-normalized, full time-resolved method for fluorescence diffuse optical tomography,” Opt. Express 16(17), 13104–13121 (2008). 10.1364/OE.16.013104 [DOI] [PubMed] [Google Scholar]
- Jiang H. B., “Frequency-domain fluorescent diffusion tomography: a finite-element-based algorithm and simulations,” Appl. Opt. 37(22), 5337–5343 (1998). 10.1364/AO.37.005337 [DOI] [PubMed] [Google Scholar]
- Kepshire D., Mincu N., Hutchins M., Gruber J., Dehghani H., Hypnarowski J., Leblond F., Khayat M., and Pogue B. W., “A microcomputed tomography guided fluorescence tomography system for small animal molecular imaging,” Rev. Sci. Instrum. 80(4), 043701 (2009). 10.1063/1.3109903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. T., Raymond S. B., Boverman G., Boas D. A., and Bacskai B. J., “Time resolved fluorescence tomography of turbid media based on lifetime contrast,” Opt. Express 14(25), 12255–12270 (2006). 10.1364/OE.14.012255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S., Lesage F. and Intes X., “Time domain fluorescent diffuse optical tomography: analytical expressions,” Opt. Express 13(7), 2263–2275 (2005). 10.1364/OPEX.13.002263 [DOI] [PubMed] [Google Scholar]
- Niedre M. J., Turner G. M., and Ntziachristos V., “Time-resolved imaging of optical coefficients through murine chest cavities,” J. Biomed. Opt. 11(6), 064017 (2006). 10.1117/1.2400702 [DOI] [PubMed] [Google Scholar]
- Patwardhan S. V., Bloch S. R., Achilefu S., and Culver J. P., “Time-dependent whole-body fluorescence tomography of probe bio-distributions in mice,” Opt. Express 13(7), 2564–2577 (2005). 10.1364/OPEX.13.002564 [DOI] [PubMed] [Google Scholar]
- Davis S. C., Dehghani H., Wang J., Jiang S., Pogue B. W., and Paulsen K. D., “Image-guided diffuse optical fluorescence tomography implemented with Laplacian-type regularization,” Opt. Express 15(7), 4066–4082 (2007). 10.1364/OE.15.004066 [DOI] [PubMed] [Google Scholar]
- Davis S. C., Pogue B. W., Springett R., Leussler C., Mazurkewitz P., Tuttle S. B., Gibbs-Strauss S. L., Jiang S. S., Dehghani H., and Paulsen K. D., “Magnetic resonance-coupled fluorescence tomography scanner for molecular imaging of tissue,” Rev. Sci. Instrum. 79(6), 064302 (2008). 10.1063/1.2919131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose A. D. and Hielscher A. H., “Fluorescence tomography with simulated data based on the equation of radiative transfer,” Opt. Lett. 28(12), 1019–1021 (2003). 10.1364/OL.28.001019 [DOI] [PubMed] [Google Scholar]
- Hyde D., Kleine R. D., Maclaurin S. A., Miller E., Brooks D. H., Kruckerb T., and Ntziachristos V., “Hybrid FMT–CT imaging of amyloid-ß plaques in a murine Alzheimer’s disease model,” Camera 44, 1304–1311 (2009). [DOI] [PubMed] [Google Scholar]
- Lin Y., Gao H., Nalcioglu O., and Gulsen G., “Fluorescence diffuse optical tomography with functional and anatomical a priori information: feasibility study,” Phys. Med. Biol. 52(18), 5569–5585 (2007). 10.1088/0031-9155/52/18/007 [DOI] [PubMed] [Google Scholar]
- Brooksby B., Jiang S., Kogel C., Doyley M., Dehghani H., Weaver J. B., Poplack S. P., Pogue B. W., and Paulsen K. D., “Magnetic resonance-guided near-infrared tomography of the breast,” Rev. Sci. Instrum. 75(12), 5262–5270 (2004). 10.1063/1.1819634 [DOI] [Google Scholar]
- Gulsen G., Yu H., Wang J., Nalcioglu O., Merritt S., Bevilacqua F., Durkin A. J., Cuccia D. J., Lanning R., and Tromberg B. J., “Congruent MRI and near-infrared spectroscopy for functional and structural imaging of tumors,” Technol. Cancer Res. Treat. 1(6), 497–505 (2002). [DOI] [PubMed] [Google Scholar]
- Guven M., Yazici B., Intes X., and Chance B., “Diffuse optical tomography with a priori anatomical information,” Phys. Med. Biol. 50, 2837–2858 (2005). 10.1088/0031-9155/50/12/008 [DOI] [PubMed] [Google Scholar]
- Intes X., Maloux C., Guven M., Yazici B., and Chance B., “Diffuse optical tomography with physiological and spatial a priori constraints,” Phys. Med. Biol. 49, N155–N163 (2004). 10.1088/0031-9155/49/12/N01 [DOI] [PubMed] [Google Scholar]
- Schweiger M. and Arridge S. R., “Optical tomographic reconstruction in a complex head model using a priori boundary information,” Phys. Med. Biol. 44(11), 2703–2722 (1999). 10.1088/0031-9155/44/11/302 [DOI] [PubMed] [Google Scholar]
- Gibbs-Strauss S. L., Samkoe K. S., O’Hara J. A., Davis S. C., Hoopes J., Hasan T., and Pogue B. W., “Detecting epidermal growth factor receptor tumor activity in vivo during Cetuximab therapy of murine gliomas,” Acad. Radiol. 17(1), 7–17 (2010). 10.1016/j.acra.2009.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliolanis N., Lasser T., Hyde D., Soubret A., Ripoll J., and Ntziachristos V., “Free-space fluorescence molecular tomography utilizing 360 degree geometry prejections,” Opt. Lett. 32(4), 382–384 (2007). 10.1364/OL.32.000382 [DOI] [PubMed] [Google Scholar]
- Lasser T. and Ntziachristos V., “Optimization of 360 degree projection fluorescence molecular tomography,” Med. Image Anal. 11(4), 389–399 (2007). 10.1016/j.media.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Brooksby B. A., Dehghani H., Pogue B. W., and Paulsen K. D., “Near infrared (NIR) tomography breast image reconstruction with a priori structural information from MRI: algorithm development for reconstructing heterogeneities,” IEEE J. Sel. Top. Quantum Electron. 9, 199–209 (2003). 10.1109/JSTQE.2003.813304 [DOI] [Google Scholar]
- van der Zee P., “Measurement and modeling of the optical properties of human tissue in the near infrared,” PhD Thesis, Department of Medical Physics and Bioengineering, University College London (1992). [Google Scholar]
- Klose A. D., Ntziachristos V., and Hielscher A. H., “The inverse source problem based on the radiative transfer equation in optical molecular imaging,” J. Comput. Phys. 202(1), 323–345 (2005). 10.1016/j.jcp.2004.07.008 [DOI] [Google Scholar]