Abstract
Bacteria use small molecule signals to access their local population densities in a process called quorum sensing (QS). Once a threshold signal concentration is reached, and therefore a certain number of bacteria have assembled, bacteria use QS to change gene expression levels and initiate behaviors that benefit the group. These group processes play central roles in both bacterial virulence and symbiosis, and can have significant impacts on human health, agriculture, and the environment. The dependence of QS on small molecule signals has inspired organic chemists to design non-native molecules that can intercept these signals and thereby perturb bacterial group behaviors. The opportunistic pathogen Pseudomonas aeruginosa has been the target of many of these efforts due to its prevalence in human infections. P. aeruginosa uses at least two N-acyl L-homoserine lactone signals and three homologous LuxR-type receptors to initiate a range of pathogenic behaviors at high cell densities, including biofilm formation and the production of an arsenal of virulence factors. This review highlights recent chemical efforts to modulate LuxR-type receptor activity in P. aeruginosa, and offers insight into the development of receptor-specific ligands as potential anti-virulence strategies.
Introduction to quorum sensing
Quorum sensing (QS) is an intercellular communication process driven by small molecules.1–3 Bacteria use a set of QS signals termed autoinducers to monitor the size of their growing population. This social activity can confer advantages unavailable to a singular bacterium.4–6 As bacterial populations reach a critical density, or quorum, the bacteria alter gene expression levels to coordinate a diverse range of behaviors, including virulence factor production, biofilm formation, bioluminescence, sporulation, and conjugation.7–9 In the case of pathogenic bacteria, QS allows the bacteria to amass in sufficiently high densities before launching a coordinated attack on the host and overwhelming its defenses.10–12
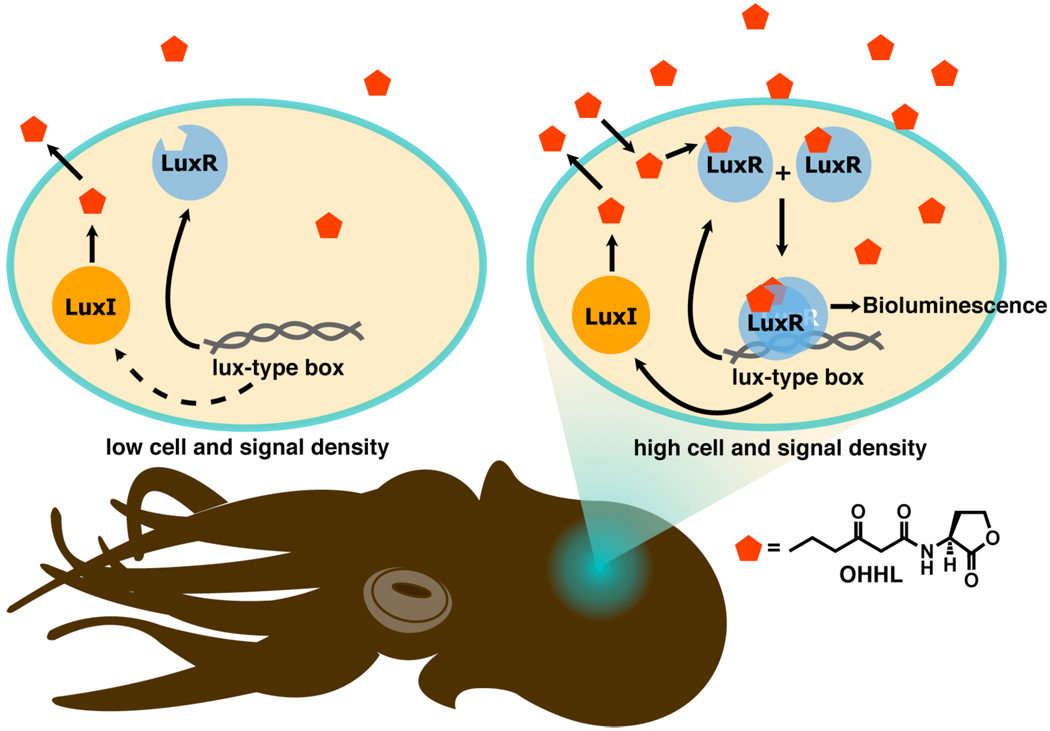
Gram-negative bacteria use N-acyl L-homoserine lactones (AHLs) as their primary autoinducers.13–15 The ~20 known AHLs differ only in the number of carbons (4–18) and substitution on their acyl chain (mostly oxidation at the 3-position and/or a cis-alkene). QS in Gram-negative bacteria was first described in the bioluminescent marine symbiont Vibrio fischeri, which resides within the light organs of certain squid and fish.16,17 V. fischeri uses a three-part QS circuit to coordinate symbiosis, made up of an autoinducer signal, its synthase, and its receptor (Figure 1). The synthase enzyme, LuxI, produces the 3-oxo-hexanoyl homoserine lactone (OHHL) autoinducer, which readily diffuses into the local environment from the organism. The concentration of signal grows with the size of the bacterial population. Once an intracellular threshold concentration of OHHL is reached, OHHL binds to its cognate cytoplasmic receptor protein, the LuxR transcription factor. The OHHL:LuxR complex then dimerizes and activates transcription of genes (via binding to QS promoters) involved in symbiosis, including those that induce bioluminescence.18 The elucidation of this canonical QS system in V. fischeri demonstrated that a single chemical signal could initiate a set of complicated binding events that controlled important functions for a bacterial group. All subsequently characterized AHL-type QS circuits contain homologues of the LuxI and LuxR regulatory proteins.6,13
Figure 1.
Schematic of QS and its role in the symbiotic relationship between V. fischeri and the Hawaiian bobtail squid (Euprymna scolopes). V. fischeri inhabits the light organ of the squid, and uses QS to bioluminesce at high cell densities. In turn, the squid uses this bioluminescence for camouflage and other processes. The lux-type-box is a short, palindromic sequence of DNA recognized by the [AHL:LuxR]2 complex.
QS in the opportunistic pathogen Pseudomonas aeruginosa has been the focus of considerable attention, due in part to this bacterium’s rapidly growing resistance to traditional antibiotics and prevalence in lung infections associated with cystic fibrosis.10–12,19,20 P. aeruginosa produces myriad virulence factors (e.g., extracellular proteases and toxins) and grows into sessile, drug-resistant biofilms at high cell densities, which make P. aeruginosa infections particularly problematic to treat.21 These phenotypes are controlled by a relatively complex QS system that contains at least three LuxR-type receptors and a pair of AHL signals (see below). Each of these ligand:receptor pairs represents an attractive target for the interruption of QS, and thereby virulence, in this pathogen. Indeed, developing non-native ligands capable of intercepting AHL:LuxR-type receptor binding has emerged as a valuable strategy to perturb QS signaling in Gram-negative bacteria,4 and this approach has been applied with increasing frequency in P. aeruginosa.22–24 Non-native QS modulators represent not only useful chemical tools to study the mechanisms of QS in P. aeruginosa, but could, with further development, provide a pathway for the generation of new therapies to treat P. aeruginosa infections. Notably, inhibition of QS should only temper virulence, as opposed to growth, and thus the development of resistance might be averted. Such antivirulence strategies are of interest for both fundamental and applied research, and represent a paradigm shift for the treatment of bacterial infection.25–28
QS is a rapidly growing field of research, and in view of its reliance on chemical signals, provides many opportunities for chemists to contribute. Several comprehensive reviews of small molecule approaches to targeting QS in Gram-negative bacteria have been reported elsewhere.4,29–31 For brevity, we have focused this Perspective on a selection of efforts that showcase chemical methods for controlling virulence in P. aeruginosa by targeting its three LuxR-type receptors.
The P. aeruginosa LuxR-type receptor triumvirate
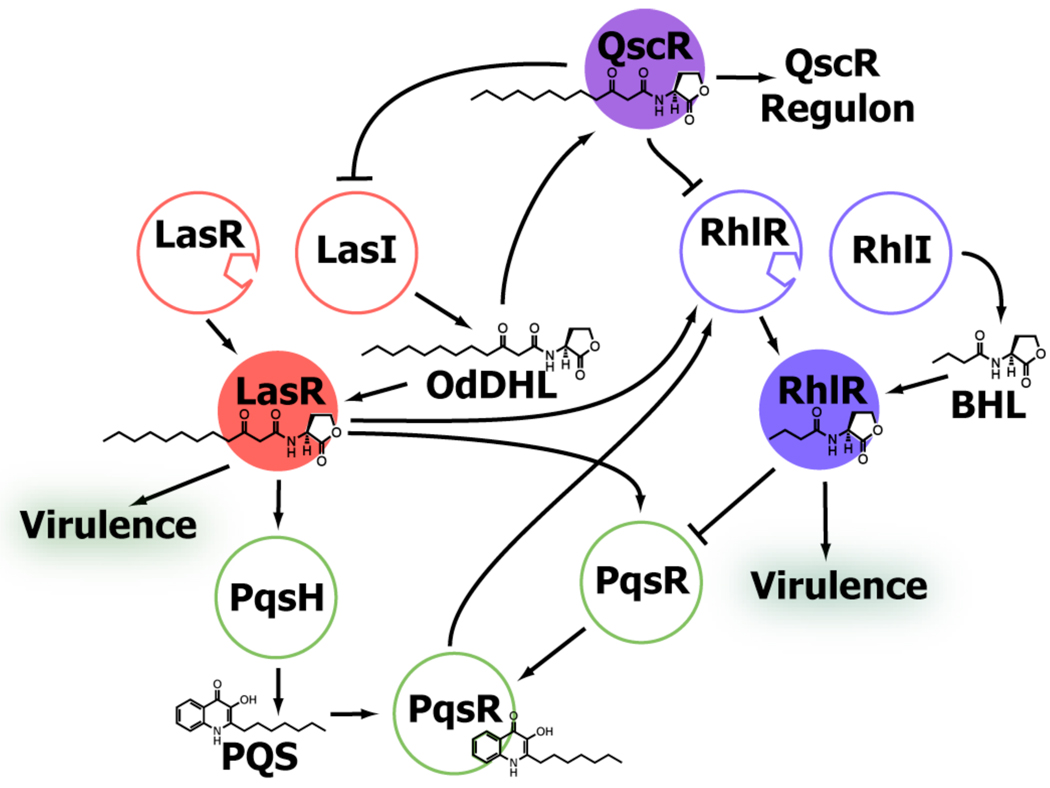
P. aeruginosa uses a set of receptors and signals to control QS, many of which have overlapping and poorly understood roles. Three of these receptors are LuxR homologs: LasR, RhlR, and QscR (Figure 2). A fourth receptor, PqsR, is a LysR-like receptor that recognizes the Pseudomonas quinolone signal PQS, and is intimately connected with the other three.32–34 The LasR system positively regulates the RhlR system, and together these two systems are responsible for regulating PqsR. In turn, QscR represses the LasR and RhlR systems.35–37
Figure 2.
Simplified schematic of QS signaling in P. aeruginosa. Pointed arrows indicate positive regulation, while flattened arrows indicate negative regulation. Ligand:receptor complexes highlighted in this review are shown as filled circles.
The LasR system was first characterized in the early 1990s, with the identification of the LasR receptor and its cognate signal synthase enzyme, LasI.17 Similar to LuxR in V. fischeri, LasR appeared to control a discrete set of genes in P. aeruginosa, including the gene for its namesake, the extracellular protease elastase B, and it was suggested that LasR functioned as a global regulator of virulence in this pathogen.38–40 The subsequent identification of 3-oxo-dodecanoyl homoserine lactone (OdDHL, Figure 2) as LasR’s cognate autoinducer drew further attention to the LasR system and its potential as a possible anti-virulence target.41 Indeed, the majority of efforts to modulate QS and thereby regulate virulence in P. aeruginosa to date have focused on LasR.4,42–45 The recently reported structure of the LasR AHL-binding domain,46 a protein notoriously difficult to handle in vitro, has further enabled the development of non-native ligands that target LasR, and we return to these efforts later in this Perspective.
Shortly after the LasR circuit was delineated, another LuxR receptor/LuxI synthase system was identified in P. aeruginosa – RhlR/RhlI. The RhlI synthase generates the butanoyl homoserine lactone autoinducer (BHL, Figure 2), which is recognized by the RhlR receptor. The BHL:RhlR complex induces a variety of genes in P. aeruginosa, including those for rhamnolipid biosurfactant synthesis (and hence its name).47,48 Early studies suggested that the RhlR system was subordinate to the LasR system at both the transcriptional and posttranslational levels.49–51 Together, these two systems were shown to control ~6% of the P. aeruginosa genome, highlighting the significance of QS-controlled gene expression in this organism.52 Later studies, however, revealed that the hierarchal relationship between LasR and RhlR in P. aeruginosa is largely dependent on empirical conditions, which can be manipulated to produce virulent phenotypes independent of a functional QS system53 or to activate different levels of gene expression in the LasR and RhlR systems at various points in its growth.54–56 These studies indicate that RhlR can elicit its effects independently on virulence, and therefore represents a relevant target for modulating QS in P. aeruginosa, comparable to LasR.
The third LuxR-type receptor identified in P. aeruginosa was QscR, and it diverges from LasR and RhlR in a number of ways. We highlight three here. First, QscR is an “orphan” receptor that lacks an associated LuxI-type enzyme.57 Without a cognate synthase, QscR appears to rely on the OdDHL signal produced by LasI for activity. Second, early studies showed that a P. aeruginosa mutant lacking qscR was hypervirulent in vivo,57 suggesting that QscR acts as a repressor of QS. This mechanism of action is the origin of QscR’s name as the quorum sensing control repressor protein (QscR). (We note that several other negative QS regulators have recently been identified in P. aeruginosa.57–61) The specific mechanism of QscR’s negative regulation remains elusive; however, QscR’s shared affinity for OdDHL with LasR has led to speculation that QscR sequesters OdDHL from LasR and/or forms inactive heterodimers with the LasR and RhlR monomers to negatively regulate QS.61 Even so, QscR appears to control another set of genes distinct from those involved in QS repression.57,61–63 Third, in contrast to LasR and RhlR, QscR can bind numerous AHLs with a range of acyl tail lengths and oxidation states at the 3-position, which has lead to speculation that QscR may play a role in not only intraspecies, but also interspecies bacterial communication.62 As an orphan receptor with relatively high ligand promiscuity, QscR makes a particularly attractive candidate for modulation with non-native small molecules, and may provide novel access to enhanced virulence control in P. aeruginosa (see below).
To develop ligands for interception of the LuxR-type receptor triumvirate in P. aeruginosa, researchers have either focused on ligands that are similar in structure to the native AHLs, or examined structures entirely different from AHLs by screening sizeable libraries of small molecules or natural product isolates. Both approaches have been successful. Below, we review several unbiased screening efforts first, and then return to the more expansive research efforts focused on the development of AHL analogs as probes of LasR, RhlR, and QscR. Thereafter, we close with a short discussion of structural studies and associated computational design efforts for the development of LuxR-type receptor modulators in P. aeruginosa.
Screening efforts for the identification of QS modulators in P. aeruginosa
Screening small molecule libraries for their ability to modulate LuxR-type proteins requires a robust biological assay, regardless if these are high-throughput screens (HTS) of large, structurally unbiased libraries or the targeted analysis of ~5 compounds. The most typical assays have relied on reporter gene read-outs, and are performed either in mutant P. aeruginosa strains that lack functional LuxI-type synthase enzymes, or in heterologous E. coli strains that produce the receptor of interest. These strains contain gene fusions that allow for expression of a colorimetric, fluorescent, or enzymatic reporter (such as green fluorescent protein (GFP) or β-galactosidase) upon activation of the LuxR-type receptor, and provide a straightforward read-out of receptor activity. Phenotypic assays in P. aeruginosa strains have also been utilized for the screening of LuxR-type protein modulators. These assays involve quantification of a native phenotype, most commonly the production of a virulence factor (e.g., an enzyme or colored metabolite) or biofilm formation, as a read-out of QS activity. Either reporter gene or phenotypic assays were utilized for all of the studies of small molecules reported herein.
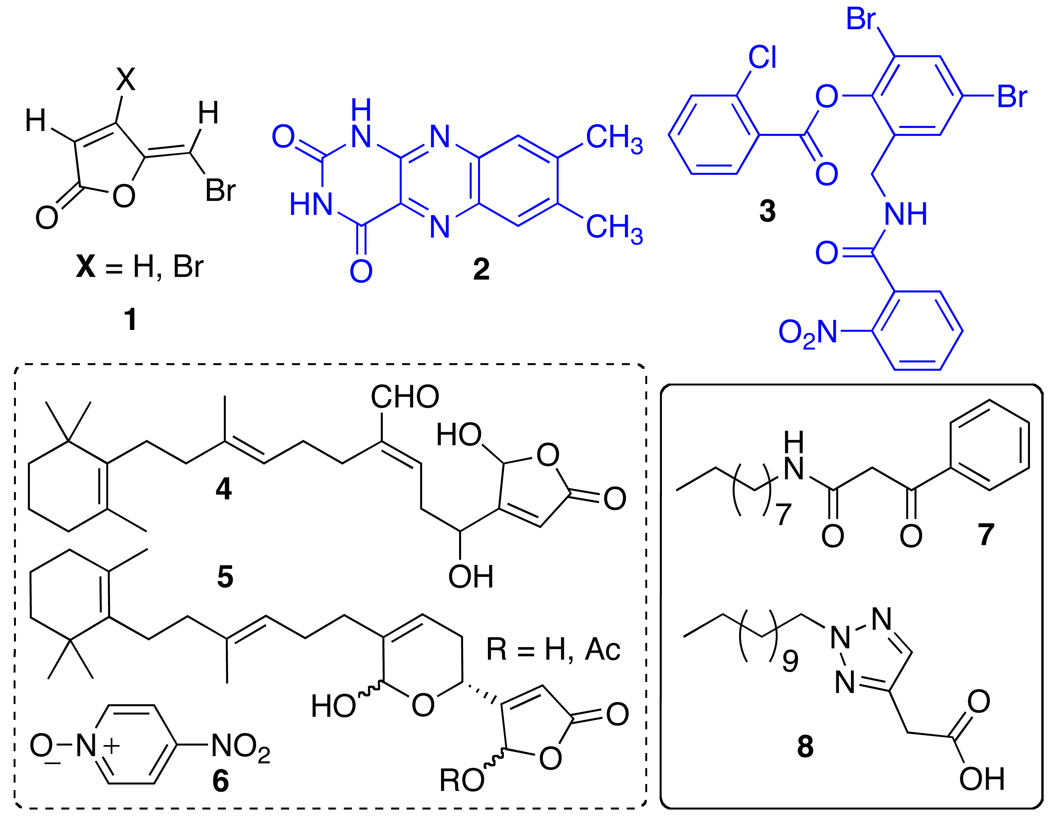
HTS efforts have revealed a diverse set of structures that can modulate LuxR-type receptors and QS phenotypes in P. aeruginosa (Figure 3). For example, small molecules that are structurally unrelated to natural QS signals have been successful at disrupting QS either by generally antagonizing genes associated with the QS circuit (7 and 8),64 or by activating LasR specifically (3).65 In addition, several known antibiotics were identified via screens to affect QS in P. aeruginosa. Azithromycin, ciprofloxacin, and ceftazidime (at sub-antibacterial concentrations) attenuate virulence and the transcription of genes in the QS pathway, although the specific targets for these drugs remain unknown.66,67 In related work, Givskov and coworkers developed a collection of reporter-type strains called QS inhibitor selectors (QSIS) to screen a set of nontoxic compounds (including pure chemicals, food isolates, and herbal medicines) for QS activity in P. aeruginosa.68 These studies revealed 4-nitro-pyridine-N-oxide (4-NPO, 6) as a potent QS inhibitor. Notably, administration of 6 to the model nematode Caenorhabditis elegans provided protection against P. aeruginosa infection to 95% of the infected worms (over 2 hours), while the untreated worms all succumbed to the infection.
Figure 3.
Non-AHL derived modulators of LasR. Blue: Agonists; Black: Antagonists. Contributors: Givskov and Høiby (1), Phillips (2), Greenberg (3, 7, and 8), and Givskov (4–6). Greenberg agonist 3 is shown as the corrected structure as determined by later X-ray studies (see text).
Several natural products have been shown to attenuate virulence in P. aeruginosa in vitro and in vivo, and suggest the tantalizing possibility for not only interspecies, but also interkingdom communication via these molecules. Plant extracts, for example, are a particularly rich source of such compounds. Rajamani et al. isolated and purified extracts from the green algae Chlamydomonas reinhardtii in order to study potential QS active components produced by this eukaryote. QS-active extracts were discovered and shown to contain lumichrome (2, Figure 3), which could modestly agonize LasR in an E. coli reporter strain. Follow-up studies indicated that 2 promoted LasR binding to DNA, suggesting a mechanism of agonsim.69 Recently, garlic and garlic extracts have been established to have broad effects on the expression of QS-related genes in P. aeruginosa, and have been shown to attenuate virulence and bacterial population growth in vivo in mouse model studies.68,70 Most notably, these agents are being evaluated in a human clinical trial for the treatment of lung infection in cystic fibrosis patients.71
Natural products containing furanones and their derivatives have consistently demonstrated strong effects on QS phenotypes through specific interaction with LuxR-type receptors.72–76 These compounds have been studied extensively as virulence inhibitors in P. aeruginosa by Givskov and coworkers, and continue to be a particularly interesting source of compounds with potent QS inhibitory activity in general. For example, the bromo-halogenated furanones (1) derived from a metabolite produced by the red seaweed Delisea pulchra can disrupt P. aeruginosa biofilms and promote clearance of the bacteria in vivo in the presence of exogenous antibacterials or a functional host immune system (e.g., in mice).73,77 Givskov and coworkers also identified the potent anti-inflammatory metabolites 4 and 5 from Luffariella variabilis as modest inhibitors of LasR activity.78
Relative to the unbiased screening efforts outlined above, the design and study of non-native AHL signals represents a much more expansive area of investigation into QS modulators. We continue this Perspective by highlighting a selection of studies that have utilized AHL analogs to control the activity of P. aeruginosa LuxR-type receptors. As the first LuxR-type receptor identified in P. aeruginosa, LasR has been the major target of these studies. Generally, these studies can be divided into two approaches: maintaining the natural ligand acyl tail while varying the lactone group, or maintaining the natural lactone group while exploring diversity in the acyl tail. We discuss each in turn below.
Modulation of LasR activity: Varying the AHL head group
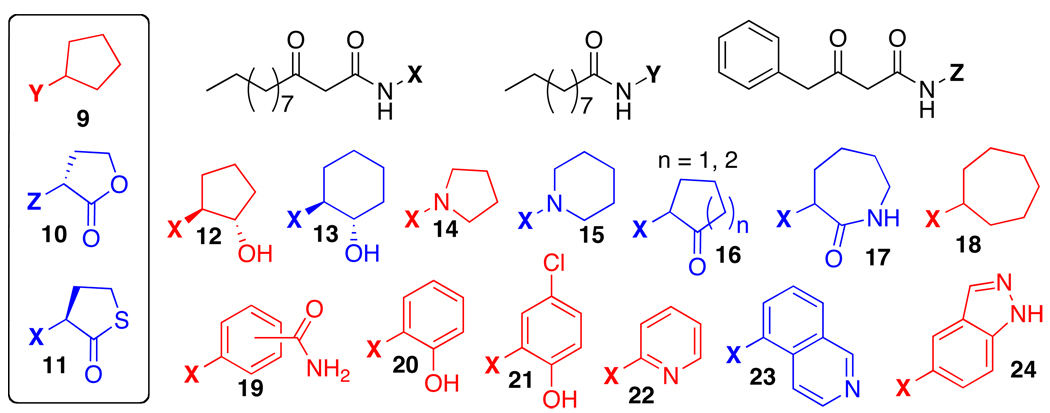
In order to identify the components of AHL structure that were essential for activity in LasR, early studies of AHL analogs included modest diversification of the lactone head group and limited library sizes. Iglewski and coworkers studied variations to the AHL acyl tail length, while replacing the lactone with a thiolactone, a lactam, or Meldrum’s acid ester. While most non-lactone derivatives showed no or limited activity, the thiolactone analog of OdDHL (11, Figure 4) activated LasR equally as well as OdDHL in an E. coli LasR reporter strain.79 Kato and coworkers replaced the lactone with a simple cyclopentyl group. By shortening the acyl tail length to 10 carbons (i.e., in 9), the researchers generated a compound that could significantly inhibit LasR activity in a P. aeruginosa reporter strain at high micromolar concentrations.80 In 2007, our laboratory examined a small set of d-lactone AHL analogs in LasR, and found that 10 could modestly agonize LasR in an E. coli LasR reporter strain at mid-micromolar concentrations.81 This result was surprising, as the native (l) stereochemistry has most commonly been invoked as essential for LuxR-type protein modulation by AHLs.
Figure 4.
Selected heterocyclic AHL analogs active in LasR. Blue: Agonists. Red: Antagonists. Contributors: Kato (9), Blackwell (10), Iglewski (11), and Suga (12–24).
Expanding beyond these relatively limited alterations to the natural lactone, Suga and coworkers performed an extensive search for heterocyclic AHL mimics capable of LasR modulation.82,83 These compounds maintained the native 3-oxo-C12 acyl group of OdDHL, yet had a variety of non-native heterocyclic head groups in place of the lactone. Suga’s team identified several compounds with significant activity as agonists or antagonists of LasR (12–24). In addition to modulating the activity of LasR in a P. aeruginosa GFP reporter system, the most active of these compounds could also attenuate QS phenotypic responses in the wild-type organism. For example, high concentrations (100 micromolar) of 16 (n = 2 in Figure 4) could nearly abolish pyocyanin expression in P. aeruginosa, while low micromolar concentrations of antagonist 20 were shown to affect biofilm development and to decrease elastase production by nearly 50%.82,83 However, with the few exceptions highlighted here, the study of AHL mimics with non-native head groups has yielded relatively few active modulators of LuxR-type proteins in P. aeruginosa. This could be due to the strength and apparent necessity of multiple hydrogen-bonding contacts between the AHL lactone and the ligand-binding site of the targeted receptor, which were unknown at the time of these prior studies (see structural section below). In general, more efforts have been focused on modifying other regions of the AHL, and we continue with a discussion of this approach below.
Modulation of LasR activity: Varying the AHL acyl tail
Numerous studies of AHL analogs in LasR have focused on modifications to the aliphatic acyl tail of OdDHL. These acyl group modifications vary from modest alterations to drastic changes. Early explorations into LasR’s tolerance for acyl diversity by Iglewski and coworkers focused on simply changing the length or degree of saturation of the AHL alkyl chain.79 The most active agonists in this set of AHLs, with nanomolar activity in an E. coli reporter system, had acyl tails close to the native 3-oxo-C12 carbon tail of OdDHL, ranging from 10 to 14 carbons (25–27, Figure 5). AHLs with acyl tail lengths outside of this range had dramatically reduced agonistic activities or were altogether inactive, indicating a high level of selectivity for the native 12-carbon AHL acyl tail by LasR. The aliphatic chains of active AHLs also require some degree of flexibility, as shown in a later study of conformationally constrained acyl chain analogs by Passador and coworkers.84 While none of their constrained analogs were found to be active as LasR modulators, non-enolizable perfluorinated analogs 28 and 29 were found to be weak LasR agonists in a P. aeruginosa reporter system.84 These data suggest that, while enolizable β-keto functionality is important for AHL activity in LasR, it is not as critical as flexibility in the acyl tail.
Figure 5.
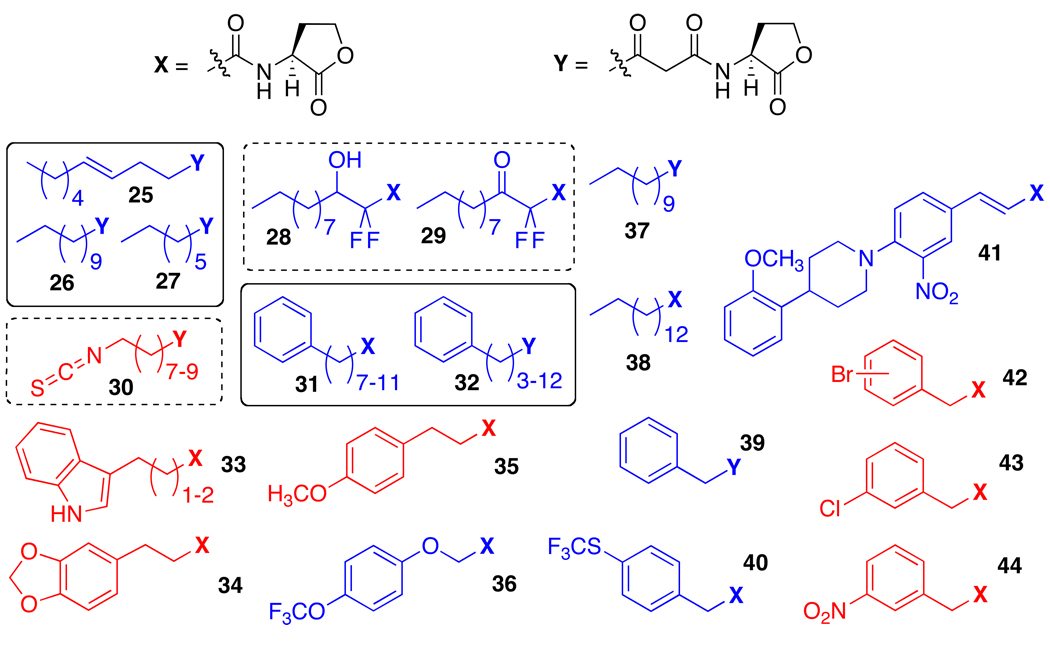
Selected AHL analogs with non-native acyl tails active in LasR. Blue: Agonists; Red: Antagonists. Contributors: Iglewski (25–27), Passador (28 and 29), Meijler (30), Sufrin (31 and 32), and Blackwell (33–44). Note, derivatives from Meijler are irreversible inhibitors of LasR.
In the recent years, our laboratory has performed several expansive studies into modification of AHL acyl tail functionality. Libraries totaling ~100 compounds were designed, synthesized, and systematically screened for agonistic and antagonistic activities in a set of reporter systems for LasR, LuxR in V. fischeri, and TraR in the plant pathogen Agrobacterium tumefaciens.81,85–87 These libraries yielded several compounds capable of agonizing or antagonizing the LasR receptor in both P. aeruginosa and E. coli reporter systems (e.g., 33–44). A second-generation library based on these initial hits yielded some of the most active modulators of LasR known at that time.85 Several of these compounds are especially notable in light of their selectivities for LasR over other LuxR-type reporters, such as TraR, LuxR, and QscR. For example, meta-substituted phenylacetanoyl HLs 42–44 show low micromolar to high nanomolar IC50 values and are among the most potent antagonists in the libraries against LasR, but are only moderate antagonists of TraR and LuxR. Likewise, indole propanoyl HL 33 (n = 1 in Figure 5) is a potent inhibitor of LasR with a high nanomolar IC50 value, but only shows minimal activity in QscR, even though these two receptors appear to share the same native signal molecule, OdDHL.81,88 Propanoyl HL 34 is particularly interesting in view of its moderate selectivity as an inhibitor of LasR over TraR or LuxR (<20% activity in the other receptors),85 and its simultaneous ability to activate QscR (EC50 = 72 nM; unpublished results, Mattmann and Blackwell). Indeed, this “dual activity” of 34 in LasR and QscR might make it a particularly potent inhibitor of QS in P. aeruginosa, as the tandem inhibition of LasR and activation of QscR should be synergistic.
Alternative methods for LasR inhibition with small molecules
Recently, Meijler and coworkers developed a set of probes capable of inhibiting LasR via covalent modification of the receptor.89 These irreversible inhibitors contain isothiocyanate functionality (30, Figure 5) and take advantage of a fortuitous cysteine residue present in the AHL binding site of LasR (as determined via prior structural studies, see below).46 In addition to providing insight into the mode of action of small molecule LasR inhibitors, the most active of these compounds (n = 8 in Figure 5) was also capable of significantly reducing biofilm formation and pyocyanin production in wild-type P. aeruginosa.
In addition to its properties as a QS signaling molecule, OdDHL also has novel anticancer properties; however, its agonistic effect on P. aeruginosa virulence is problematic if it is to be used as a therapeutic. By screening a small library of non-native AHLs in a LasR reporter strain, Sufrin and coworkers identified a set of AHLs that were both potent cancer inhibitors (31 and 32, Figure 5) and LasR agonists.90 Further studies revealed that the extended chain analogs of 31 and 32 (12 alkyl carbons and greater) had reduced LasR agonistic activities, yet still retained their anticancer activities. By taking advantage of LasR’s apparent high selectivity for AHL acyl tail length, these researchers were able to pursue novel cancer therapeutics and avoid a potentially harmful side effect.
Modulation of RhlR activity with small molecules
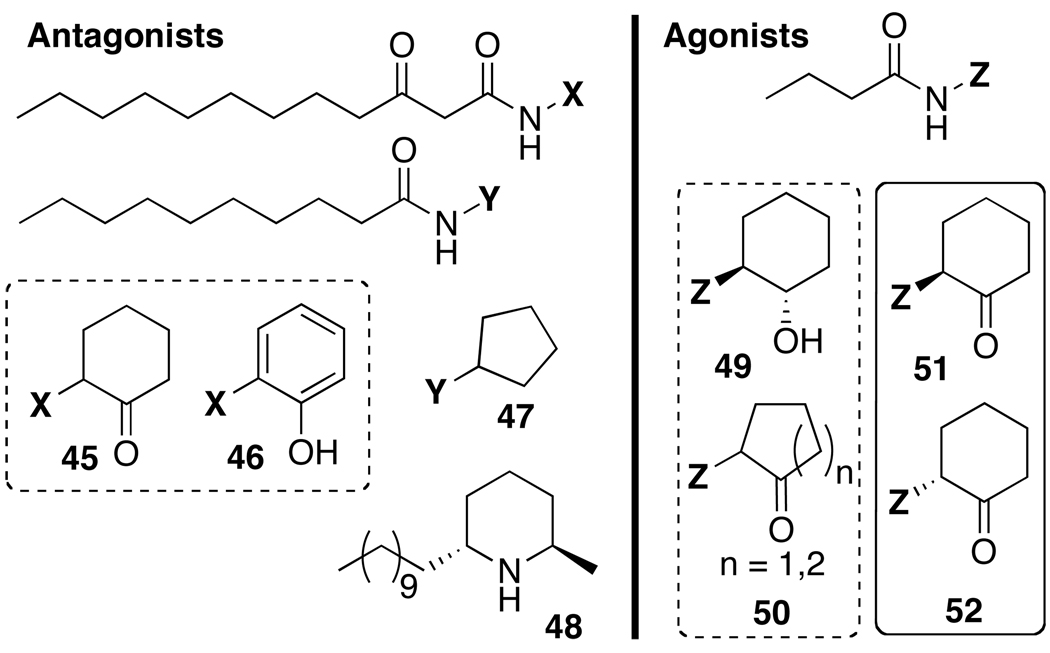
As RhlR has emerged as an important component of the P. aeruginosa QS circuit, increasing efforts have been devoted to the development of non-native, RhlR-specific modulators (Figure 6). Approaches in which previously identified modulators of LasR have been readapted to target RhlR instead have yielded a number of ligands active in RhlR. For example, simultaneous with their work in LasR (see above), Suga and coworkers also affixed their library of non-native heterocyclic head groups with a butanoyl tail, rather than a 3-oxo-C12 acyl tail, in order to potentially alter these ligands to target RhlR. This method was successful and yielded compounds (e.g., 49 and 50) that, at relatively high concentrations (>100 µM) in a P. aeruginosa GFP reporter, were able to activate RhlR to levels analogous to BHL.82 Interestingly, one compound retaining its 3-oxo-C12 tail and originally designed to modulate LasR (45) was found to moderately antagonize RhlR (at 50 µM). Note, this observed inhibitory activity against RhlR could occur through indirect inhibition of the LasR circuit, direct competitive inhibition of RhlR, or a combination of these effects.82 In a follow-up study by Suga and coworkers, another compound initially designed for LasR inhibition was also found to strongly inhibit RhlR at concentrations as low as 10 µM. This compound (46) also had a modest inhibitory effect on elastase production in P. aeruginosa, and unexpectedly appeared to enhance biofilm production. Biofilm morphology was altered, however, and this observation suggested that 46 had some negative effect on biofilm formation.83
Figure 6.
Selected compounds active in RhlR. Left: Antagonists; Right: Agonists. Contributors: Suga (45, 46, 49, and 47), Kato (47), Janda (48), and Spring (51 and 52).
Spring and coworkers continued the approach of tailoring LasR active compounds in order to target RhlR. Previously, Suga and coworkers had determined that the racemic, butanoyl cyclohexanone derivative (i.e., 51 + 52) could agonize LasR in a P. aeruginosa reporter at high micromolar concentrations.82 In a nice follow up study, Spring’s team synthesized the enantiopure cyclohexanones 51 and 52 (Figure 6) and tested them in P. aeruginosa QS phenotypic assays.91 High nM concentrations of 51 were found to restore pyocyanin and pyoverdin production levels to that of wild-type in a P. aeruginosa QS mutant. The R enantiomer 52 showed an order of magnitude less activity in this experiment, indicating that the S enantiomer 51 is the more active compound.
In addition to the 3-oxo-C12 analogs mentioned above, other extended alkyl chain AHL analogs have also been found to inhibit RhlR. The C10-cyclopentyl derivative 47, which is a potent LasR inhibitor, was also shown by Kato and coworkers to inhibit RhlR in P. aeruginosa.80 This dual-receptor activity could account for its observed potent inhibitory effect on biofilm formation and elastase, pyocyanin, and rhamnolipid production. The natural product solenopsin A (48), or fire ant venom, was recently shown to compete with BHL for RhlR binding, thus affecting the expression of QS controlled genes and phenotypes in P. aeruginosa.92 Its moderate structural similarity to several of the heterocyclic RhlR modulators introduced in this section could be a reason for this activity profile.
Modulation of QscR activity with small molecules
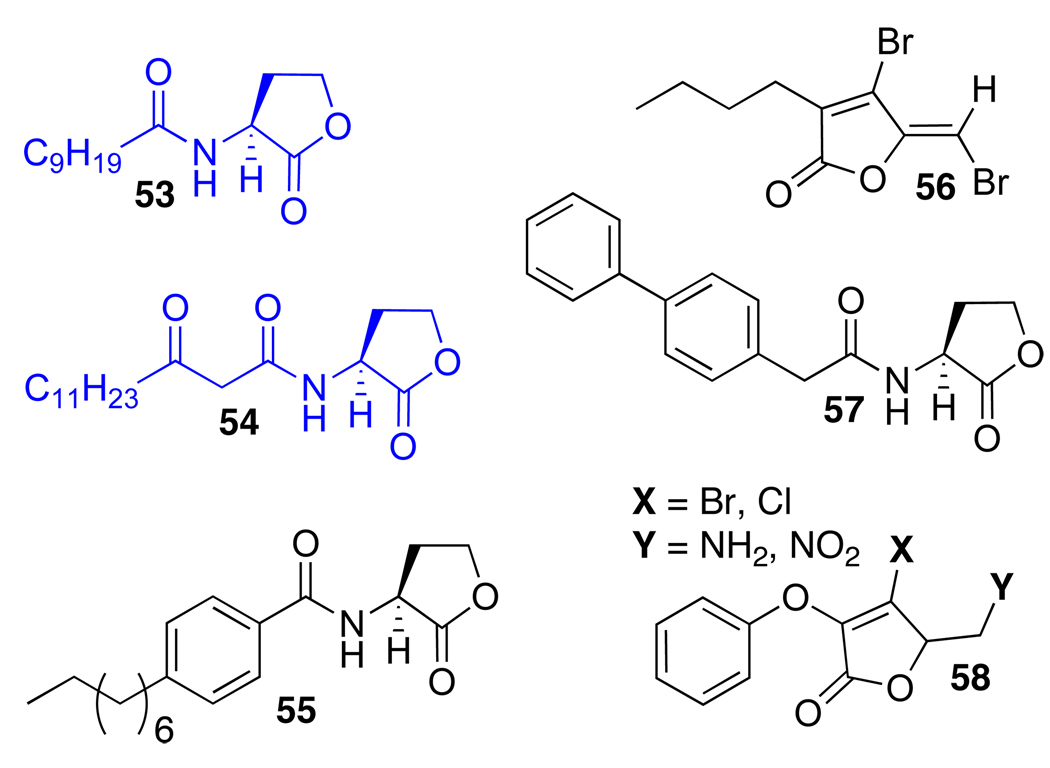
As highlighted above, QscR was determined to be a component of the P. aeruginosa QS circuit later than LasR and RhlR. As a result, studies of chemical modulators of this novel receptor in P. aeruginosa have been limited (Figure 7). We describe the most pertinent examples here. Working with multiple Pseudomonas strains, Wood and coworkers found that furanone 56 stimulated less siderophore biosynthesis in a mutant lacking the qscR gene relative wild-type. This led to the proposal that 56 might interact with the qscR gene or its product, QscR.76 In 2008, our laboratory screened the AHL analog libraries introduced above for ligands capable of agonizing or antagonizing QscR using an E. coli reporter strain.88 These studies revealed a set of hit compounds with modest activities in QscR. In terms of agonists, QscR was significantly more tolerant of AHLs bearing unbranched, aliphatic acyl groups relative to LasR. Our studies indicated that AHLs with acyl chains ranging from 10–14 carbons (e.g., 53 and 54) could strongly activate QscR, which corroborated with Greenberg and coworkers’ earlier report of QscR’s promiscuity in terms of its preferred “native” ligand.62 In terms of antagonists, AHLs containing steric bulk near the AHL amide bond position and throughout the acyl chain (e.g., 55 and 57) strongly inhibited QscR, which is in direct contrast to the most potent AHL-derived inhibitors of LasR. Several of our QscR inhibitors were capable of modulating the association of purified QscR with its cognate DNA sequence, as demonstrated by electromobility shift assays (EMSAs). The EMSAs showed differences in the binding of QscR to a short DNA sequence containing the QscR-specific promoter (PA1897) in the presence of compound, which suggested that these compounds elicit their antagonistic effects, at least in part, by inhibiting QscR:DNA binding.
Figure 7.
Selected compounds active in QscR. Blue: Agonists; Black: Antagonists. Contributors: Blackwell (53–55 and 57), Wood (56), and Park (58).
Recently, Liu et al. revisited the furanone substructure in search of QscR modulators, and uncovered a set of analogs (58) that inhibited QscR in both E. coli and P. aeruginosa reporter strains. Similar to our work, the inhibitory activities of these ligands were also confirmed by EMSAs with QscR and its target DNA.74 Overall, these initial studies define QscR as having requirements for small molecule control that are distinct from LasR and RhlR. This presents a particularly interesting opportunity to develop ligands capable of differentiating between LasR and QscR, despite their common native ligand OdDHL. Such studies are ongoing in our laboratory.
Structure-guided design of QS regulators in P. aeruginosa
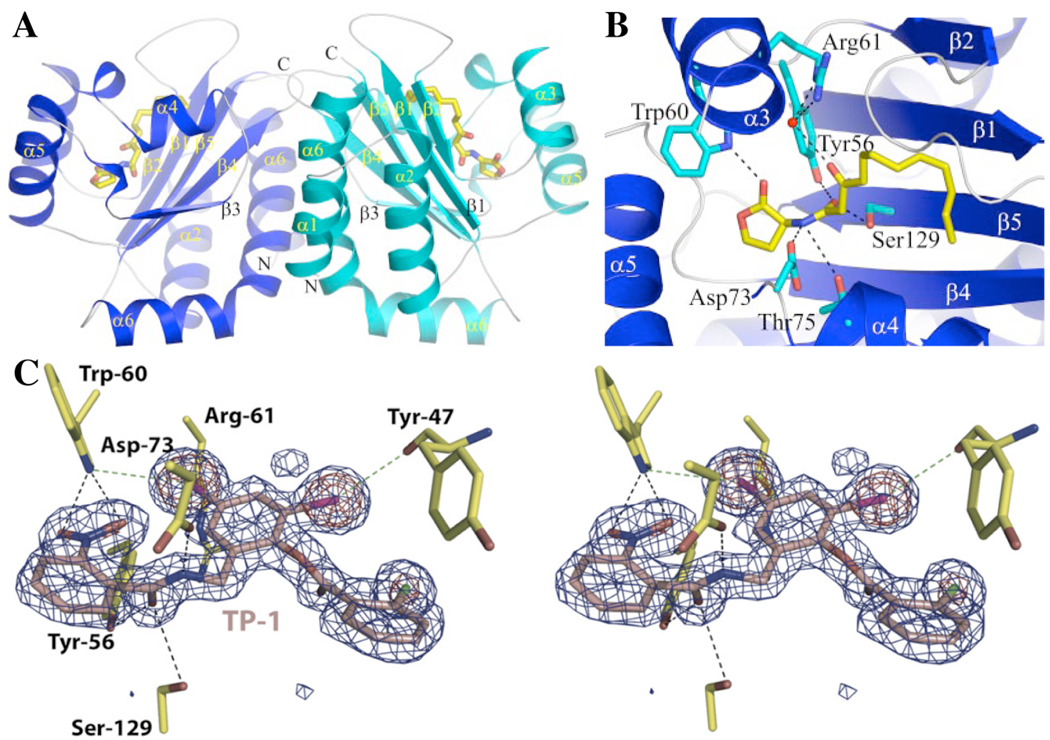
The lack of structural information for LuxR-type proteins has slowed computational design efforts for the generation of new QS modulators. LuxR-type proteins typically consist of two domains: an N-terminal ligand-binding domain, and a C-terminal DNA-binding domain. Most LuxR-type proteins require their native AHL ligand in order to adopt a stable conformation, and even as AHL complexes, these receptors have relatively short half lives.35 This instability has been problematic for the study of LuxR-type receptors in vitro, and has slowed biochemical and structural studies of these proteins. However, over the past eight years, several important structures of LuxR-type proteins have been reported. Two X-ray crystal structures of the TraR receptor from A. tumefaciens bound to its native ligand 3-oxo-octanoyl homoserine lactone (OOHL) and complexed to its target DNA sequence were reported in 2002.93,94 In 2006, Dyson and coworkers solved the structure of the N-terminal domain of SdiA from E. coli by NMR.95 SdiA is a LuxR-type homolog that, like QscR in P. aeruginosa, has no cognate synthase or native ligand, and is thought to play some part in interspecies communication. A year after the report of SdiA, the X-ray structure of the N-terminal domain for LasR bound to OdDHL was published, providing the first structural information about this important P. aeruginosa QS regulator (Figures 8A and 8B).46 Collectively, these structural data have been a major contribution to the QS field, and have corroborated considerable genetic and biochemical data about the LuxR-type receptor mechanism of action. In general, the structures for TraR, SdiA, and LasR were highly similar (in the N-terminal ligand-binding domain), suggesting that LuxR-type proteins can adopt like conformations when bound to native ligand.
Figure 8.
Image of the X-ray crystal structure of the LasR N-terminal ligand-binding domain (A),46 close-up views of OdDHL (B),46 and stereoviews of the non-native agonist 3 (C)96 bound in the LasR ligand-binding domain. Images reproduced with permission (see Acknowledgements).
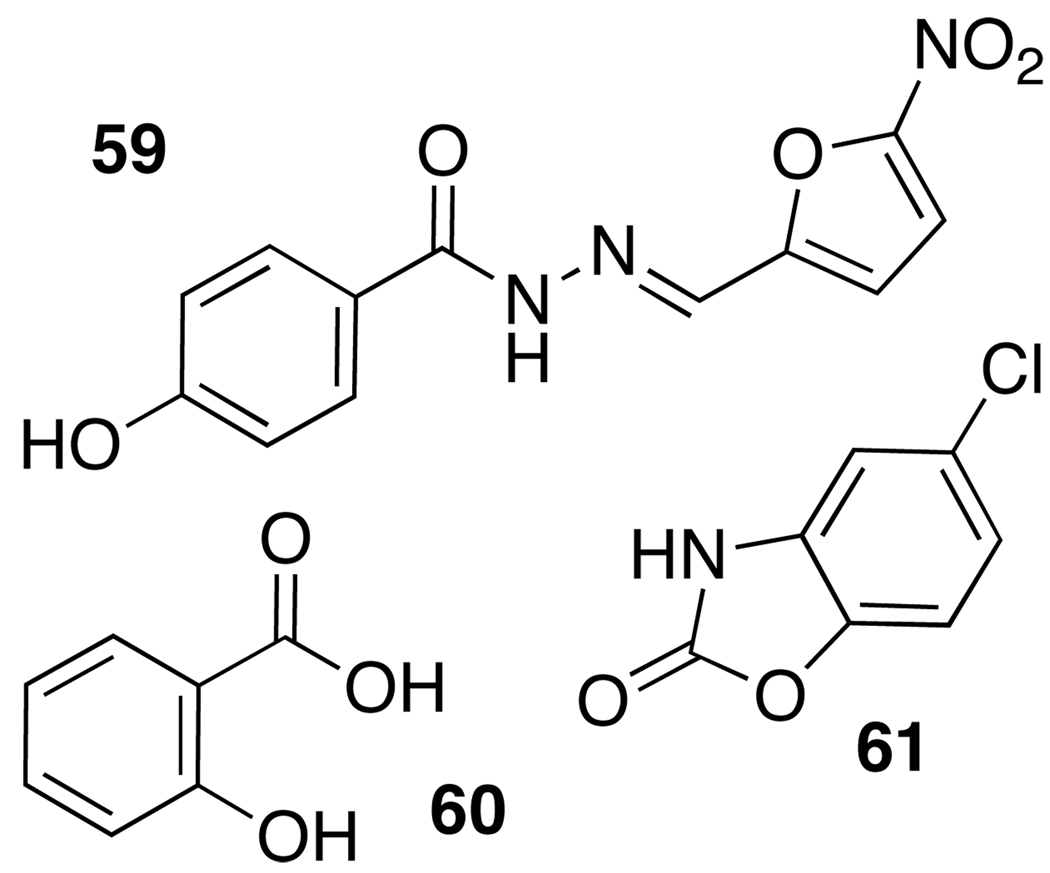
The structural data above have permitted the first computational and rational design approaches for the study and development of new QS modulators. “Virtual” high-throughput screening of natural product and known drug libraries against the OdDHL binding site in LasR, followed by cell-based reporter gene and phenotypic assays of virtual hits, revealed three active compounds (59–61, Figure 9). The furanone derivative nifuroxazide (59) was identified alongside chlorzoxazone (60) and salicylic acid (61), and each was shown to modestly inhibit LasR and RhlR activity and QS-related phenotypic responses in P. aeruginosa.97
Figure 9.
QS-active nifuroxazide (antimicrobial agent), chlorzoxazone (known drug: muscle relaxant), and salicylic acid (natural product) identified through virtual screening.97
Ahumedo et al. recently reported a comparative study of the LasR and TraR structures and ligands known to interact with these receptors.98 A series of active compounds, including the native AHLs and several previously identified non-native AHLs, were computationally docked into the ligand-binding sites of the receptors. In both LasR and TraR, the docking studies confirmed hydrogen-bonding interactions between ligand-binding site tryptophan, aspartic acid, and tyrosine residues and the lactone carbonyl, amide bond, and 3-oxo functionality of the AHL, respectively. In LasR, serine-129 neighbors tyrosine-56, and may either present an additional hydrogen bonding contact with the 3-oxo AHL functionality or be available for electronic interaction with the acyl tail (Figure 8B). Both ligand-binding sites, and especially that within LasR, revealed the AHL acyl tail buried within a hydrophobic pocket deep inside the protein, a feature that limits the accessibility of this site to ligands of inappropriate size. The latter feature in LasR perhaps explains its extreme selectivity for AHL tail length (see above). Overall, this study by Ahumedo et al. highlighted the potential utility of computational methods for the analysis of QS modulators, and established a foundation for further in silico design and screening of small molecule libraries in this area.
Very recently, the LasR ligand-binding domain was crystallized with the structurally distinct LasR inhibitors identified by Greenberg and coworkers, including triaryl agonist 3 (Figure 3), and studied using X-ray crystallography by Zou and Nair (Figure 8C).96 The unambiguous electron density in the X-ray structure of the LasR:3 complex suggested an important amendment to the chemical structure of 3, that is, the nitro and chlorine functional groups should be transposed (the corrected structure is depicted in Figure 3).64,96 Interestingly, despite its structural dissimilarity to OdDHL, compound 3 engages in interactions within the LasR binding site that are very similar to those of the natural ligand. One aromatic ring of 3 interacts with LasR residues that typically associate with the OdDHL lactone (i.e., tryptophan-60 and serine-129), while the other two aromatic rings occupy the space usually bound by the hydrophobic acyl tail of OdDHL (Figure 8C). Though some small structural adjustments are required for LasR to accommodate 3 relative to OdDHL (i.e., a shift in arginine-61), the halogens in 3 are postulated to contribute to favorable electronic interactions with polar residues in the LasR ligand-binding site (tyrosine-47 and tryptophan-60) and to participate in stronger hydrogen bonds than those possible with AHL aliphatic acyl groups.96
Conclusions and outlook
The three LuxR-type receptors in the pathogen P. aeruginosa form a powerful signaling triumvirate that directs bacterial group behavior. LasR, RhlR, and QscR are each important and collaborative components in a QS system designed for adaptability and persistence in unforgiving environments. Each of these receptors recognizes a specific AHL signal produced by P. aeruginosa in order to initiate a series of events that control QS. Inspiration for the chemical interception of AHL:LuxR-type receptor complexes in P. aeruginosa has come from different approaches, including HTS, rational design of small libraries based on the chemical structures of active AHLs, and computational methods. The majority of these efforts have been focused on methods to intercept LasR signaling, although in recent years attention has shifted to also include RhlR and QscR. Promising lead compounds have shown variable, and sometimes highly potent, activities in attenuating virulence using both reporter and wild-type strains of P. aeruginos a, and we have provided a summary of pertinent examples herein.
The current understanding of QS in Gram-negative bacteria is rapidly expanding since its discovery in V. fischeri over 30 years ago. As QS is identified in a growing number of bacterial species, the complexity of the known circuitries controlling group behavior is also increasing. Bacteria have been identified that utilize as many as seven QS signals to coordinate virulent behavior.6 P. aeruginosa, with only three LuxR-type proteins characterized so far (LasR, RhlR, and QscR), two AHL signals, and a pqs system driven by quinolones, controls QS via a highly sophisticated network. Additional components essential to this bacterium’s QS system are constantly being identified, and the relationships between already established factors remain to be fully characterized. What has become clear, however, is that no receptor single-handedly controls QS in P. aeruginosa, and therefore approaches to target a single receptor using a chemical approach will most likely be insufficient for broad QS control. It is therefore critical to further clarify the relationships that these receptors have with one another and the roles that they have in QS. Armed with this knowledge, we can then begin to exploit the relationships between LasR, RhlR, and QscR (and beyond) in tandem in order to establish an effective QS control strategy in P. aeruginosa and provide new pathways for therapeutic intervention.
Acknowledgements
We apologize to those authors that we could not cite in this review due to space limitations. The NIH (AI063326), NSF (CHE-0449959), ONR (N000140710255), and the Burroughs Welcome Fund are each acknowledged for their generous support of both fundamental and applied quorum sensing research in our laboratory. H.E.B. is an Alfred P. Sloan Foundation fellow. Figures 8A and 8B are reprinted from reference 46. Figure 8C is reprinted with permission from reference 96.
References
- 1.Ng W-L, Bassler BL. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler BL, Losick R. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Camilli A, Bassler BL. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geske GD, O'Neill JC, Blackwell HE. Chem. Soc. Rev. 2008;37:1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowery CA, Dickerson TJ, Janda KD. Chem. Soc. Rev. 2008;37:1337–1346. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 6.Boyer M, Wisniewski-Dyé F. FEMS Microbiol. Ecol. 2009;70:1–19. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 7.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 8.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 9.Marketon MM, Gronquist MR, Eberhard A, Gonzalez JE. J. Bacteriol. 2002;184:5686–5695. doi: 10.1128/JB.184.20.5686-5695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Nat. Rev. Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjarnsholt T, Givskov M. Anal. Bioanal. Chem. 2007;387:409–414. doi: 10.1007/s00216-006-0774-x. [DOI] [PubMed] [Google Scholar]
- 12.de Kievit TR, Iglewski BH. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua C, Greenberg EP. Nat. Rev. Mol. Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 14.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. FEMS Microbiol. Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 15.Welch M, Mikkelsen H, Swatton JE, Smith D, Thomas GL, Glansdorp FG, Spring DR. Mol. Biosyst. 2005;1:196–202. doi: 10.1039/b505796p. [DOI] [PubMed] [Google Scholar]
- 16.Eberhard A. J. Bacteriol. 1972;109:1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuqua WC, Winans SC, Greenberg EP. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uroz S, Dessaux Y, Oger P. ChemBioChem. 2009;10:205–216. doi: 10.1002/cbic.200800521. [DOI] [PubMed] [Google Scholar]
- 19.Wilder CN, Allada G, Schuster M. Infect. Immun. 2009;77:5631–5639. doi: 10.1128/IAI.00755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooley M, Chhabra SR, Williams P. Chem. Biol. 2008;15:1141–1147. doi: 10.1016/j.chembiol.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett John G, Edwards J. Clin. Infect. Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 22.Williams P, Camara M. Curr. Opin. Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Venturi V. FEMS Microbiol. Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 24.Suga H, Smith KM. Curr. Opin. Chem. Biol. 2003;7:586–591. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Rasko DA, Sperandio V. Nat. Rev. Drug. Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 26.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escaich S. Curr. Opin. Chem. Biol. 2008;12:400–408. doi: 10.1016/j.cbpa.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 28.Clatworthy AE, Pierson E, Hung DT. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 29.Janssens JCA, De Keersmaecker SCJ, De Vos DE, Vanderleyden J. Curr. Med. Chem. 2008;15:2144–2156. doi: 10.2174/092986708785747580. [DOI] [PubMed] [Google Scholar]
- 30.Ni N, Li M, Wang J, Wang B. Med. Res. Rev. 2009;29:65–124. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- 31.Pan J, Ren D. Expert Opin. Ther. Pat. 2009;19:1581–1601. doi: 10.1517/13543770903222293. [DOI] [PubMed] [Google Scholar]
- 32.Rasko DA, Sperandio V. Nat. Rev. Drug Discovery. 2010:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 33.Wade DS, Calfree MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. J. Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubern J-F, Diggle SP. Mol. BioSyst. 2008;4:882–888. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 35.Schuster M, Greenberg EP. In: Chemical Communication among Bacteria. Winans SC, Bassler BL, editors. Washington, DC: ASM Press; 2008. pp. 133–144. [Google Scholar]
- 36.Wagner VE, Li LL, Isabella VM, Iglewski BH. Anal. Bioanal. Chem. 2007;387:469–479. doi: 10.1007/s00216-006-0964-6. [DOI] [PubMed] [Google Scholar]
- 37.Schuster M, Greenberg EP. Zent. Bakteriol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 38.Gambello MJ, Iglewski BH. J. Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 40.Gambello MJ, Kaye S, Iglewski BH. Infect. Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Proc. Natl. Acad. Sci. U. S. A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen TB, Givskov M. Microbiology (Reading, Engl) 2006;152:895–904. doi: 10.1099/mic.0.28601-0. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Mol. Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stintzi A, Evans K, Meyer JM, Poole K. FEMS Microbiol. Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 45.Wagner VE, Iglewski BH. Clin. Rev. in Allergy and Immunol. 2008;35:124–134. doi: 10.1007/s12016-008-8079-9. [DOI] [PubMed] [Google Scholar]
- 46.Bottomley MJ, Muraglia E, Bazzo R, Carfì A. J. Biol. Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 47.Ochsner UA, Koch AK, Fiechter A, Reiser J. J. Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson JP, Passador L, Iglewski BH, Greenberg EP. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pesci EC, Pearson JP, Seed PC, Iglewski BH. J. Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson JP, Pesci EC, Iglewski BH. J. Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Kievit TR, Kakai Y, Register JK, Pesci EC, Iglewski BH. FEMS Microbiol. Lett. 2002;212:101–106. doi: 10.1016/s0378-1097(02)00735-8. [DOI] [PubMed] [Google Scholar]
- 52.Schuster M, Lostroh CP, Ogi T, Greenberg EP. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. Mol. Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 54.van Delden C, Pesci EC, Pearson JP, Iglewski BH. Infect. Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duan K, Surette MG. J. Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dekimpe V, Déziel E. Microbiology (Reading, Engl) 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 57.Chugani SA, Whiteley M, Lee KM, D’Argenio DA, Manoil C, Greenberg EP. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. Mol. Microbiol. 2007;66:1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- 59.Rampioni G, Schuster M, Greenberg EP, Zennaro E, Leoni L. FEMS Microbiol. Lett. 2009 doi: 10.1111/j.1574-6968.2009.01817.x. [DOI] [PubMed] [Google Scholar]
- 60.Siehnel R, Traxler B, An DD, Parsek MR, Schaefer AL, Singh PK. Proc. Natl. Acad. Sci. U. S. A. 2010 doi: 10.1073/pnas.0908511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ledgham F, Ventre I, Soscia C, Foglino M, Sturgis JN, Lazdunski A. Mol. Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- 62.Lee J-H, Lequette Y, Greenberg EP. Mol. Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 63.Lequette Y, Lee J-H, Ledgham F, Lazdunski A, Greenberg EP. J. Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Müh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Antimicrob. Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müh U, Hare B, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, Greenberg EP. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16948–16952. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skindersoe ME, Alhede M, Phipps RK, Yang L, Jensen PØ, Rasmussen TB, Bjarnsholt T, Tolker-Nielsen T, Høiby N, Givskov M. Antimicrob. Agents Chemother. 2008;52:3648–3663. doi: 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffmann N, Lee B, Hentzer M, Rasmussen TB, Song Z, Johansen HK, Givskov M, Høiby N. Antimicrob. Agents Chemother. 2007;51:3677–3687. doi: 10.1128/AAC.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Köte M, Nielsen J, Eberl L, Givskov M. J. Bacteriol. 2005;187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajamani S, Bauer WD, Robinson JB, Farrow JM, Pesci EC, Teplitski M, Gao M, Sayre RT, Phillips DA. Mol. Plant-Microbe Interact. 2008;21:1184–1192. doi: 10.1094/MPMI-21-9-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harjai K, Kumar R, Singh S. FEMS Immunol. Med. Microbiol. 2009 doi: 10.1111/j.1574-695X.2009.00614.x. [DOI] [PubMed] [Google Scholar]
- 71.Smyth AR, Cifelli PM, Ortori CA, Righetti K, Lewis S, Erskine P, Holland ED, Givskov M, Williams P, Camara M, Barrett DA, Knox A. Pediatr. Pulmonol. 2010;45:356–362. doi: 10.1002/ppul.21193. [DOI] [PubMed] [Google Scholar]
- 72.Defoirdt T, Miyamoto CM, Wood TK, Meighen EA, Sorgeloos P, Verstraete W, Bossier P. Environ. Microbiol. 2007;9:2486–2495. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- 73.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu H-B, Lee J-H, Kim JS, Park S. Biotechnol. Bioeng. 2010;106:119–126. doi: 10.1002/bit.22672. [DOI] [PubMed] [Google Scholar]
- 75.Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Microbiology. 1999;145(Pt 2):283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 76.Ren D, Zuo R, Wood TK. Appl. Microbiol. Biotechnol. 2005;66:689–695. doi: 10.1007/s00253-004-1691-6. [DOI] [PubMed] [Google Scholar]
- 77.Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, Høiby N. J. Antimicrob. Chemother. 2004;53:1054–1061. doi: 10.1093/jac/dkh223. [DOI] [PubMed] [Google Scholar]
- 78.Skindersoe ME, Ettinger-Epstein P, Rasmussen TB, Bjarnsholt T, de Nys R, Givskov M. Mar. Biotechnol. 2008;10:56–63. doi: 10.1007/s10126-007-9036-y. [DOI] [PubMed] [Google Scholar]
- 79.Passador L, Tucker KD, Guertin K, Journet M, Kende A, Iglewski BH. J. Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Appl. Environ. Microbiol. 2007;73:3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geske GD, O’Neill JC, Miller DM, Mattmann ME, Blackwell HE. J. Am. Chem. Soc. 2007;129:13613–13625. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith KM, Bu Y, Suga H. Chem. Biol. 2003;10:81–89. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 83.Smith KM, Bu Y, Suga H. Chem. Biol. 2003;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 84.Kline TR, Bowman J, Iglewski BH, De Kievit TR, Kakai Y, Passador L. Bioorg. Med. Chem. Lett. 1999;9:3447–3452. doi: 10.1016/s0960-894x(99)00626-5. [DOI] [PubMed] [Google Scholar]
- 85.Geske GD, Mattmann ME, Blackwell HE. Bioorg. Med. Chem. Lett. 2008;18:5978–5981. doi: 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geske GD, O’Neill JC, Miller DM, Wezeman RJ, Mattmann ME, Lin Q, Blackwell HE. ChemBioChem. 2008;9:389–400. doi: 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J. Am. Chem. Soc. 2005;127:12762–12763. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 88.Mattmann ME, Geske GD, Worzalla GA, Chandler JR, Sappington KJ, Greenberg EP, Blackwell HE. Bioorg. Med. Chem. Lett. 2008;18:3072–3075. doi: 10.1016/j.bmcl.2007.11.095. [DOI] [PubMed] [Google Scholar]
- 89.Amara N, Mashiach R, Amar D, Krief P, Spieser SAH, Bottomley MJ, Aharoni A, Meijler MM. J. Am. Chem. Soc. 2009;131:10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 90.Oliver CM, Schaefer AL, Greenberg EP, Sufrin JR. J. Med. Chem. 2009;52:1569–1575. doi: 10.1021/jm8015377. [DOI] [PubMed] [Google Scholar]
- 91.Glansdorp FG, Thomas GL, Lee JK, Dutton JM, Salmond GPC, Welch M, Spring DR. Org. Biomol. Chem. 2004;2:3329–3336. doi: 10.1039/B412802H. [DOI] [PubMed] [Google Scholar]
- 92.Park J, Kaufmann GF, Bowen JP, Arbiser Jack L, Janda KD. J. Infect. Dis. 2008;198:1198–1201. doi: 10.1086/591916. [DOI] [PubMed] [Google Scholar]
- 93.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Di Marco S. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang R-g, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 95.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. J. Mol. Biol. 2006;355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 96.Zou Y, Nair SK. Chem. Biol. 2009;16:961–970. doi: 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M, Tolker-Nielsen T. Antimicrob. Agents Chemother. 2009;53:2432–2443. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahumedo M, Diaz A, Vivas-Reyes R. Eur. J. Med. Chem. 2010;45:608–615. doi: 10.1016/j.ejmech.2009.11.004. [DOI] [PubMed] [Google Scholar]