Abstract
Objective
The objective of this study was to test the association between calpain-10 (CAPN10), a diabetes susceptibility gene, with risk of pancreatic cancer (PC).
Methods
DNA samples from 83 incident exocrine PC cases and 166 controls, all of whom were smokers, were genotyped for four markers of CAPN10 in a nested case–control study based on the Beta-Carotene and Retinol Efficacy Trial (CARET), a randomized chemoprevention trial of subjects at high risk of lung cancer. Controls were matched on sex, race, age, CARET intervention arm, duration of exposure to asbestos, and smoking history. Conditional logistic regression was used for statistical analyses.
Results
The minor allele of SNP-43 (rs3792267) in intron 3 was associated with increased risk of PC with an odds ratio of 1.57 (95%CI 1.03–2.38, p = 0.035) per allele. The three markers of the highest risk haplotype had an odds ratio of 1.98 (95%CI 1.12–3.49, p=0.019) for risk of PC compared to the most common haplotype. There was no evidence of interaction between either of these associations by diabetes status.
Conclusion
These results suggest that variation in CAPN10 may be associated with increased risk of PC among smokers. Thus, studies of genes associated with diabetes risk in PC are warranted in a larger population.
Keywords: pancreatic cancer, diabetes, calpain-10, CAPN10
Introduction
Pancreatic cancer (PC) is the fourth leading cause of cancer death in the USA [1]. It is estimated that PC will be diagnosed in over 37,000 people in 2008 resulting in over 34,000 deaths [1]. The majority of PC are exocrine tumors, and 95% of these are pancreatic ductal adenocarcinomas. Exocrine PC has a short survival time, with an average of only 4 months [2]. Smoking is the most established risk factor for PC with population attributable fraction of 22% [3].
A recent meta-analysis has shown that a family history of PC increases risk nearly twofold [4], suggesting genetic susceptibility to PC. Linkage and gene expression studies reported a mutation in Palladin (PALLD) gene linked with PC in a single family [5, 6]. Variants in the methionine synthase reductase (MTRR) gene, the mouse double minute 2 homologue (MDM2) gene, and the apoptotic genes, Fas-Fas ligand (FasL) and caspase-8 (CASP8), have been associated with PC in hospital-based case–control studies [7–9]. However, the presence of common susceptibility alleles for PC that could lead to earlier diagnosis and even cancer prevention in the general population remains to be confirmed. Although somatic mutations in oncogenes and tumor suppressor genes, including K-ras, p16, and p53 [10], have been identified in pancreatic adenocarcinomas, it is not known whether germline mutations in these genes contribute to the familial aggregation of PC.
A relationship between PC and type 2 diabetes (T2D) is also well established. A meta-analysis of 17 case–control studies and 19 cohort studies concluded that there is an approximately 80% greater risk of PC among individuals with type 2 diabetes, with the strongest association among those with <5 years duration of diabetes [11]. At least two additional case–control studies support the hypothesis that diabetes may be an early manifestation of PC rather than a risk factor for its development [12, 13]. Other studies have suggested that factors associated with abnormal glucose metabolism, a hallmark of diabetes, might have an important role in the etiology of PC [14], though a causal effect remains to be established. Based on these data, we postulated that variation in genes associated with diabetes risk may also be involved in the development of PC.
Calpain-10 (CAPN10) was identified as a candidate gene for T2D by positional cloning [15], and the CAPN10 protein product is a regulator of insulin secretion in pancreatic β-cells [16]. Single nucleotide polymorphisms (SNPs) within CAPN10 were associated with diabetes in a case–control study with over 6,000 white subjects [17] and in a meta-analysis in Europeans [18]. Because of this relationship to diabetes and its functional role in the pancreas, we evaluated the association between four variants in the CAPN10 gene and risk of incident PC.
Materials and Methods
Cases and Matched Controls
Study subjects have been described previously [19]. Briefly, subjects were selected from the Beta-Carotene and Retinol Efficacy Trial (CARET), a randomized chemoprevention trial of heavy smokers or asbestos-exposed workers at high risk of lung cancer [20]. As of September 1, 2004, 83 confirmed incident exocrine PC cases were available for analysis from this cohort, excluding cases diagnosed with lung cancer or those who had no history of smoking. For each eligible case, two controls without cancer were selected. Controls were matched to cases on age (5-year intervals), sex, race, CARET intervention arm, asbestos exposure, and smoking history using a two-step process. First, cases and controls were matched on smoking status (former or current). Current smokers were further matched on number of cigarettes per day (±10), while former smokers were matched on years elapsed between quitting and study enrollment, resulting in 166 controls used in the analysis. Race and diabetes status were self-reported, and body mass index (BMI), defined as weight per height squared (kg/m2), was based on measurements performed at baseline (study enrollment).
DNA Extraction and Genotyping
DNA was obtained from whole blood (N=216) or serum (N=33) and was extracted using QIAamp DNA Blood Mini Kit/protocol (Qiagen, Inc, Valencia, CA). DNA from serum was whole genome amplified by QIAGEN REPLI-g Services (Hilden, Germany).
SNP-44 (rs2975760) and SNP-43 (rs3792267) in intron-3, SNP-63 (rs5030952) in intron-6, and Indel-19 (rs3842570) in intron-13 of CAPN10 were selected based on previous evidence of association with T2D [17, 18]. Using TaqMan™ assays, genotypes were analyzed using an ABI Prism™ 7900HT Fast Real-Time PCR System. Sequential genotyping of the DNA samples was performed until unambiguous genotypes were ascertained, and thus, there were no missing genotypes in the analysis.
Statistical Analysis
All statistical analyses were performed using either R Gui, version 2.5.0 (www.r-project.org) or STATA 9.0 (Stata Corp. College Station, TX). The Fisher exact test was performed to test for deviations from Hardy–Weinberg equilibrium (HWE) in controls. Conditional logistic regression assuming additive genetic models was used to obtain odds ratios (OR), 95% confidence intervals (CI), and p values, and multiple comparisons were evaluated using the Bonferroni correction. We estimated haplotypes using the expectation-maximization algorithm implemented in Haplo. stats module in R. We then tested for association with reference to the most common haplotype. To test for interaction of the association between CAPN10 variants and PC by diabetes status, we compared models with and without an interaction term between each marker and diabetes status using a likelihood ratio test.
Linkage disequilibrium (LD) of the CAPN10 genomic region was evaluated using genotype data for Caucasian samples from the HapMap project (http://www.hapmap.org) and graphically displayed using HaploView [21]. Pairwise R2 and D′ values among controls were also calculated by HaploView.
Results
The matching procedure resulted in similar distributions for gender, race, and age between cases and controls (Table 1). Among all subjects, 75% were male and 94% were white, and the mean baseline age was approximately 61 years. Mean BMI for cases and controls were similar (28.0 versus 26.8, respectively). Sixteen percent of cases, compared with 13% of controls, self-reported diabetes at baseline. All subjects were current or former smokers at baseline.
Table 1.
Characteristics of pancreatic cancer cases and controls from the CARET cohort
| Characteristics | Cases (N=83) |
Controls (N=166) |
|||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Gender | Male | 62 | 74.7 | 124 | 74.7 |
| Female | 21 | 25.3 | 42 | 25.3 | |
| Race | White | 78 | 94.0 | 156 | 94.0 |
| Black | 3 | 3.6 | 6 | 3.6 | |
| Other | 2 | 2.4 | 4 | 2.4 | |
| Baseline age (years) | 45–49 | 2 | 2.4 | 4 | 2.4 |
| 50–54 | 12 | 14.5 | 25 | 15.1 | |
| 55–59 | 21 | 25.3 | 40 | 24.1 | |
| 60–64 | 21 | 25.3 | 42 | 25.3 | |
| 65–70 | 27 | 32.5 | 55 | 33.1 | |
| Mean age (SD) | 61.3 (5.5) | 61.1 (5.9) | |||
| CARET participant group | Asbestos-exposed | 16 | 19.3 | 32 | 19.3 |
| Heavy smoker | 67 | 80.7 | 134 | 80.7 | |
| Smoking status at baseline | Current smoker | 54 | 65.1 | 108 | 65.1 |
| Cigarettes per day (±10) | |||||
| <10 | 9 | 16.7 | 17 | 15.7 | |
| 10–20 | 20 | 37.0 | 40 | 37.0 | |
| 20–30 | 15 | 27.8 | 29 | 26.9 | |
| 30–40 | 7 | 13.0 | 15 | 13.9 | |
| >40 | 3 | 5.5 | 7 | 6.5 | |
| Former smoker | 29 | 34.9 | 58 | 35.0 | |
| Years since quit smoking | |||||
| <1 | 3 | 10.3 | 6 | 10.3 | |
| 1–5 | 13 | 44.8 | 26 | 44.8 | |
| 5–10 | 12 | 41.4 | 23 | 39.7 | |
| >10 | 1 | 3.5 | 3 | 5.2 | |
| Baseline BMI | <18.5 | 0 | 0.0 | 2 | 1.2 |
| 18.5–24.9 | 22 | 26.5 | 60 | 36.1 | |
| 25–29.9 | 38 | 45.8 | 67 | 40.4 | |
| 30–34.9 | 17 | 20.5 | 28 | 16.9 | |
| 35–39.9 | 5 | 6.0 | 7 | 4.2 | |
| >40 | 1 | 1.2 | 2 | 1.2 | |
| Mean (SD) | 28.0 (4.5) | 26.8 (4.7) | |||
| Self-reported baseline diabetes | Yes | 13 | 15.7 | 21 | 12.7 |
| No | 70 | 84.3 | 145 | 87.3 | |
Each of the four CAPN10 genetic markers had a minor allele frequency (MAF) >10% (Table 2), and none showed a deviation from HWE in controls (data not shown). Table 2 presents the conditional logistic regression results for the association of each marker with PC, assuming an additive genetic model. SNP-43 (rs3792267) was associated with risk of PC (p=0.035), although this result was not significant after Bonferroni correction. For each additional copy of the minor allele “A” of this marker, there was a 57% increased risk of PC (OR=1.57, 95%CI 1.03–2.38). No significant association was found for the three remaining markers.
Table 2.
Association between markers in CAPN10 gene and pancreatic cancer using conditional logistic regression, assuming additive genetic models, based on cases (N=83) and matched controls (N=166) in the CARET cohort
| Individual SNPs and three-marker haplotype | ||||||
| SNP | Minor allele | MAF (case/control) | OR | 95%CIa | p value | |
| SNP-44 | C | (0.18/0.11) | 1.69 | (0.99-2.91) | 0.055 | |
| SNP-43 | A | (0.35/0.26) | 1.57 | (1.03–2.38) | 0.035 | |
| Indel-19 | del | (0.41/0.36) | 1.31 | (0.86–1.99) | 0.203 | |
| SNP-63 | T | (0.11/0.11) | 1.00 | (0.53–1.90) | 1.000 | |
| Three-marker haplotypea | Frequency | OR | 95% CI | p value | ||
| TG-ins | 0.58 | 1 | (reference) | |||
| CG-ins | 0.14 | 1.98 | (1.12–3.49) | 0.019 | ||
| TA-ins | 0.28 | 1.78 | (1.15–2.80) | 0.010 | ||
| Interaction by diabetes status | ||||||
| SNP | Minor allele | With diabetes | Without diabetes | p value for the interaction | ||
| OR | 95% CI | OR 95%CI | ||||
| SNP-44 | C | 1.77 | (0.25–12.69) | 1.59 | (0.89–2.85) | 0.878 |
| SNP-43 | A | 1.39 | (0.34–5.69) | 1.61 | (1.00–2.59) | 0.754 |
| Indel-19 | del | 1.83 | (0.36–9.32) | 1.20 | (0.77–1.86) | 0.481 |
| SNP-63 | T | 3.40 | (0.13–87.0) | 0.92 | (0.78–1.77) | 0.307 |
| Three-marker haplotypea | ||||||
| TG-ins | 1 | (reference) | 1 | (reference) | 0.906 | |
| CG-ins | 2.53 | (0.31–20.7) | 1.87 | (1.02–3.46) | ||
| TA-ins | 1.76 | (0.39–8.02) | 1.82 | (1.11–3.01) | ||
OR odds ratio, risk per copy of the minor allele, 95%CI 95% confidence interval, indel insertion/deletion, ins insertion, MAF minor allele frequency
Three-marker haplotypes formed by SNP-44, SNP-43, and Indel-19 (rs2975760, rs3792267, and rs3842570)
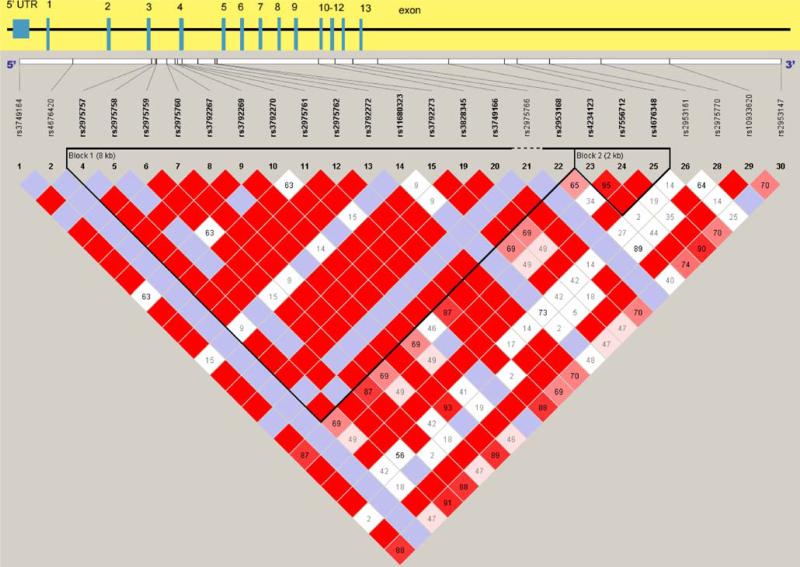
Pairwise correlations among the markers for the controls, measured by R2, were low, ranging from 0.04 to 0.23. However, all pairwise D′ values were 1, illustrating a lack of recombination events between the markers. These data imply that the marker alleles were inherited together on the same chromosome, but none of the markers alone capture all the variations for the region. Thus, haplotypes were constructed to capture as much variation as possible for the association analysis As shown in Fig. 1, the haplotype structure in CAPN10 using the Caucasian genotypes from the HapMap project, SNP-44 (intron-3), SNP-43 (intron-3), and Indel-19 (intron-6) are within a region of high LD. SNP-63 (intron-13) is 3 kb downstream of this region. Therefore, we performed three-marker haplotype analysis excluding SNP-63.
Fig. 1.
Linkage disequilibrium (LD)of CAPN10 gene region based on data from the HapMap Project, Caucasian population genotype data, visualized by HaploView [21]. Color coding: D′=1 (bright red);: D′<1 (pink/red);: D’ not statistically significant (blue/white). The black triangles frame regions of high LD. Only SNP-44(rs2975760) and SNP-43(rs3792267), located in intron-3, were available in the HapMap data, and they are outlined in yellow. Indel-19(rs3842570) is located in intron 6 within the same LD region. SNP-63(rs5030952) is located 3 kb downstream of this region
Table 2 shows the frequency of the resulting haplotypes, and their association with risk of PC, using the most common haplotype “TG-ins” as the reference group. Each of the two remaining haplotypes, “CG-ins” and “TA-ins,” approximately doubled the risk of PC (OR=1.98, p=0.019 and OR=1.78, p=0.0099, respectively). As shown in Table 2, there was no evidence of interactions between the SNPs and diabetes status with risk of PC.
Discussion
To our knowledge, this pilot study is the first to suggest an association between variants in the diabetes susceptibility gene CAPN10 with PC among smokers. The minor allele “A” of SNP-43 was associated with an increased risk of PC (OR=1.57, p=0.035). For α-level 0.05 with four markers, the expected number of false positives was less than our observed number of significant findings (expected, 4 × 0.05=0.2), suggesting that this finding was not observed by chance. However, this result was not statistically significant after Bonferroni correction.
Haplotype analysis showed that haplotype “TA-ins” increased risk of PC by 80% compared with the common haplotype “TG-ins” (OR=1.80, p=0.01). These two haplotypes differed only at the position for SNP-43, and the risk of having allele “A” in SNP-43 in this haplotype was consistent with the result for SNP-43 alone. Similarly, haplotype “CG-ins” nearly doubled the risk of PC compared with the reference haplotype (OR=1.98, p=0.019). These results suggest that haplotypes formed by multiple markers were more informative than analysis based on individual markers and may reflect the combined effects of multiple marker variants.
A number of studies have reported that an association between “homozygous G” genotype at SNP-43 of the CAPN10 gene was associated with increased risk of diabetes [15, 17, 18]. In the present study, however, increased risk of PC was associated with the “A” allele of SNP-43, suggesting that the CAPN10 risk for PC may not be attributable to its association with risk of diabetes. However, the CAPN10 variant could have pleiotropic effects on diabetes and PC through different alleles. Alternatively, this difference could be due to a “flip-flop” effect that can occur when different LD patterns are present in the populations being sampled due to cryptic population structure [22]. Because SNP-43 is intronic, and thus less likely to be the causal variant than a coding variant, such an effect could be present.
Because CAPN10 is a diabetes candidate gene, we examined a possible interaction between baseline diabetes on the association of CAPN10 markers with PC. There was no evidence of interaction from these data, but power was limited due to the small sample size of this pilot study.
The CAPN10 protein is expressed ubiquitously, with the highest levels of expression in the heart, pancreas, brain, liver, and kidney [15]. An immunohistochemical study identified CAPN10 specifically in the insulin-containing cells and the islet cells of the pancreas [16]. Inhibition of CAPN10 activity in mouse islets has been shown to increase glucose-induced insulin secretion [23], implying a relationship between CAPN10 and glucose homeostasis in pancreatic islets. However, these pathologic processes may or may not be relevant because exocrine PC develops in pancreatic ductal cells, not islet cells.
A study showing that overexpression of human CAPN10 in beta cells of transgenic mice islets enhanced ryanodine-induced apoptosis [24] implies that the association of CAPN10 with PC seen in this study might be mediated through apoptosis, the programmed cell death process that may be blocked in cancer cells, resulting in abnormal growth of tissue. Recent case–control studies showed that the apoptotic genes, FasL and CASP8, were associated with PC [9]. Whether or not CAPN10 acts through the same apoptotic pathway warrants further investigation.
A major strength of this study is its prospective design and the availability of incident PC cases, avoiding survival bias in case–control studies due to the rapid fatality of pancreatic cancer. The use of cases and controls who were all smokers from the CARET cohort [19] reduces the possibility of confounding due to smoking, the most established risk factor for PC. However, the smoking history in this cohort is also a study limitation since such a high-risk sample may not be representative of the general population. The power to detect association was also low due to the small sample size. Other limitations include the fact that diabetes status was self-reported and that only four markers in the CAPN10 gene were used, possibly not capturing all of the important variation in the CAPN10 gene.
In conclusion, we found that the intronic SNP-43 and its haplotype in CAPN10, a diabetes susceptibility gene, were associated with PC among smokers from the CARET cohort. Building on these results, future studies of diabetes candidate genes with greater statistical power are likely to increase our understanding of genetic susceptibility to PC.
Acknowledgments
This work was supported by NIH grants R21 CA115878-01, R25 CA115878-01, U01-CA063673, and P30ES07033. Specimens were from CARET Serum Bank maintained at Fred Hutchinson Cancer Research Center (FHCRC).
Contributor Information
Pui-yee Fong, Institute for Public Health Genetics, University of Washington, Box 357236, Seattle, WA 98195, USA.
Megan D. Fesinmeyer, Institute for Public Health Genetics, University of Washington, Box 357236, Seattle, WA 98195, USA Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Emily White, Department of Epidemiology, School of Public Health, University of Washington, Seattle, WA, USA; Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Federico M. Farin, Department of Environmental and Occupational Health Sciences, School of Public Health, University of Washington, Seattle, WA, USA
Sengkeo Srinouanprachanh, Department of Environmental and Occupational Health Sciences, School of Public Health, University of Washington, Seattle, WA, USA.
Zahra Afsharinejad, Department of Environmental and Occupational Health Sciences, School of Public Health, University of Washington, Seattle, WA, USA.
Margaret T. Mandelson, Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA GH Center for Health Studies, Group Health Cooperative, Seattle, WA, USA.
Teresa A. Brentnall, Department of Medicine, School of Medicine, University of Washington, Seattle, WA, USA
Matt J. Barnett, Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
Gary E. Goodman, Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
Melissa A. Austin, Institute for Public Health Genetics, University of Washington, Box 357236, Seattle, WA 98195, USA Department of Epidemiology, School of Public Health, University of Washington, Seattle, WA, USA; Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Fesinmeyer M, Austin M, Li C, De Roos A, Bowen D. Differences in survival by histologic type of pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1766–73. doi: 10.1158/1055-9965.EPI-05-0120. [DOI] [PubMed] [Google Scholar]
- 3.Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M. Comparative Risk Assessment collaborating g. Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366:1784–93. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- 4.Permuth-Wey J, Egan KM. Family history is a significant risk factor for pancreatic cancer: results from a systematic review and meta-analysis. Fam Cancer. 2009;8:109–17. doi: 10.1007/s10689-008-9214-8. [DOI] [PubMed] [Google Scholar]
- 5.Eberle M, Pfützer R, Pogue-Geile K, et al. A new susceptibility locus for autosomal dominant pancreatic cancer maps to chromo-some 4q32-34. Am J Hum Genet. 2002;70:1044–8. doi: 10.1086/339692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pogue-Geile K, Chen R, Bronner M, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohnami S, Sato Y, Yoshimura K, et al. His595Tyr polymorphism in the methionine synthase reductase (MTRR) gene is associated with pancreatic cancer risk. Gastroenterology. 2008;135:477–88. doi: 10.1053/j.gastro.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 8.Asomaning K, Reid AE, Zhou W, et al. MDM2 promoter polymorphism and pancreatic cancer risk and prognosis. Clin Cancer Res. 2008;14:4010–5. doi: 10.1158/1078-0432.CCR-07-4187. [DOI] [PubMed] [Google Scholar]
- 9.Yang M, Sun T, Wang L, et al. Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res. 2008;14:3230–6. doi: 10.1158/1078-0432.CCR-08-0177. [DOI] [PubMed] [Google Scholar]
- 10.Cowgill S, Muscarella P. The genetics of pancreatic cancer. Am J Surg. 2003;186:279–86. doi: 10.1016/s0002-9610(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 11.Huxley R, Ansary-Moghaddam A, de Berrington González A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005;92:2076–83. doi: 10.1038/sj.bjc.6602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Gupta S, Holly E. Diabetes mellitus and pancreatic cancer in a population-based case–control study in the San Francisco Bay Area, California. Cancer Epidemiol Biomarkers Prev. 2006;15:1458–63. doi: 10.1158/1055-9965.EPI-06-0188. [DOI] [PubMed] [Google Scholar]
- 13.Chari S, Leibson C, Rabe K, et al. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology. 2008;134:95–101. doi: 10.1053/j.gastro.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gapstur S, Gann P, Lowe W, Liu K, Colangelo L, Dyer A. Abnormal glucose metabolism and pancreatic cancer mortality. JAMA. 2000;283:2552–8. doi: 10.1001/jama.283.19.2552. [DOI] [PubMed] [Google Scholar]
- 15.Horikawa Y, Oda N, Cox N, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–75. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 16.Marshall C, Hitman G, Partridge C, et al. Evidence that an isoform of calpain-10 is a regulator of exocytosis in pancreatic beta-cells. Mol Endocrinol. 2005;19:213–24. doi: 10.1210/me.2004-0064. [DOI] [PubMed] [Google Scholar]
- 17.Jensen D, Urhammer S, Eiberg H, et al. Variation in CAPN10 in relation to type 2 diabetes, obesity and quantitative metabolic traits: studies in 6018 whites. Mol Genet Metab. 2006;89:360–7. doi: 10.1016/j.ymgme.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Tsuchiya T, Schwarz P, Bosque-Plata L, et al. Association of the calpain-10 gene with type 2 diabetes in Europeans: results of pooled and meta-analyses. Mol Genet Metab. 2006;89:174–84. doi: 10.1016/j.ymgme.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Fesinmeyer MD, Standford JL, Brentnall TA, et al. Association between the Peroxisome proliferator-activated receptor-gamma Pro12Ala variant and haplotype and pancreatic cancer in a high-risk cohort of smokers: a pilot study. Pancreas. 2009;38(6):631–7. doi: 10.1097/MPA.0b013e3181a53ef9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omenn G, Goodman G, Thornquist M, et al. The beta-carotene and retinol efficacy trial (CARET) for chemoprevention of lung cancer in high risk populations: smokers and asbestos-exposed workers. Cancer Res. 1994;54:2038s–43. [PubMed] [Google Scholar]
- 21.Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Lin P, Vance J, Pericak-Vance M, Martin E. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–8. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Sreenan S, Pan C, et al. A 48-hour exposure of pancreatic islets to calpain inhibitors impairs mitochondrial fuel metabolism and the exocytosis of insulin. Metabolism. 2003;52:528–34. doi: 10.1053/meta.2003.50091. [DOI] [PubMed] [Google Scholar]
- 24.Johnson J, Han Z, Otani K, et al. RyR2 and calpain-10 delineate a novel apoptosis pathway in pancreatic islets. J Biol Chem. 2004;279:24794–802. doi: 10.1074/jbc.M401216200. [DOI] [PubMed] [Google Scholar]