Abstract
Maternal HIV-1-specific antibodies are efficiently transferred to newborns; their role in disease control is unknown. We administered non-sterilizing levels of neutralizing IgG, including the human neutralizing monoclonal IgG1b12, to six newborn macaques before oral challenge with SHIVSF612P3. All rapidly developed neutralizing antibodies and had significantly reduced plasma viremia for 6 months. These studies support the use of neutralizing antibodies in enhancing B cell responses and viral control in perinatal settings.
Neutralizing antibodies (NAbs) increase in titer and avidity during HIV-1 infection1 and in many subjects mature to neutralize divergent isolates, targeting diverse epitopes on Envelope (Env). A fraction of NAbs target conserved regions on Env including the CD4 binding site (CD4bs) 2,3. HIV-1-positive pregnant mothers transfer HIV-1-specific IgG across the placenta to the fetus in utero and mother-to-child transmission frequently occurs in the presence of NAbs, which can mediate selection of transmitted variants4. Passive administration of high levels of polyclonal neutralizing IgG or neutralizing mAbs (NMAbs) can fully protect against high dose intravenous5 or mucosal 6,7 SHIV challenge in nonhuman primates. Lower levels of NMAbs can prevent infection from high dose8 or serial low dose mucosal SHIV challenge in macaques when infused repeatedly9. Non-neutralizing antibodies, such as antibody-dependent cell-mediated viral inhibition (ADCVI)-mediating antibodies, are also found during HIV-1 and SIV infection and may reduce viral levels via natural killer or monocyte effector cells10. Passive infusion of non-neutralizing serum with potent ADCVI activity prevents infection in newborn rhesus macaques11.
Previously, we showed that early (d 1, 14) treatment of six SIV-infected juvenile macaques with SIV-specific neutralizing IgG (SIVIG) accelerated NAb development by 20 weeks12, accompanied by tight control of viremia for 5 years in the three macaques that seroconverted. Another group also demonstrated significant control of SIV plasma viremia afforded by passive neutralizing SIVIG (d 7 treatment)13. Defining the types and levels of antibodies that mediate beneficial effects is critical both for vaccine development and passive immunotherapy. To do so, we utilized a SHIV challenge model in newborn macaques to ask whether NAbs at physiologic, non-protective doses could limit viremia and CD4+ T cell destruction in newborns. Groups of six macaques were subcutaneously inoculated with Normal-IgG, Matched-IgG (SHIVIGSF162P3 plus IgG1-b12), or Mismatched-IgG (SHIVIG89.6P) 1 day prior to high dose (2 AID50) oral inoculation with SHIVSF162P3, which we previously showed is vertically transmitted and highly pathogenic in newborn macaques14. To ensure that the SHIVIG included CD4bs-directed NAb, IgG1b12, a potent, broadly reactive human NMAb directed to the CD4bs that neutralizes SHIVSF162P315, was incorporated in a ratio of 1:1,000 (w:w; ~2.5 µg ml−1 in vivo). The characteristics of the IgG preparations are summarized in Supplementary Table 1. At the doses delivered in vivo, Matched-IgG neutralized 93% of the challenge virus, while Mismatched-IgG neutralized 60% (Fig. 1a). Neutralization of SHIV89.6P was >90% in the Mismatched-IgG and 65% in the Matched-IgG (Fig. 1b).
Fig. 1. Matched-IgG, but not Mismatched-IgG, affects plasma viral load in SHIVSF162P3-infected infant macaques.
Neutralizing activity of Matched IgG, with IgG1-b12 (■) and without IgG1-b12 (□), and Mismatched IgG (●) against: the challenge virus SHIVSF162P3 (a) and heterologous virus SHIV89.6P (b). Vertical line at 2 × 103 denotes the estimated in vivo passive IgG concentration of 2 mg ml−1. (c) Mean PBMC proviral loads during the 24 weeks after initiation of infection as quantified by real-time PCR. (d) Mean plasma viral loads as quantified by real-time PCR during the 24 weeks after initiation of infection. (e) Differences in AUC comparing Normal-IgG, Mismatched-IgG, and Matched-IgG for the entire 24 weeks. (f) Differences in AUC calculations for post-acute viremia between all treatment groups (weeks 8–24). Horizontal bar indicates the median value for the group. P values are indicated.
Exposure to SHIVSF162P3 resulted in the infection of five of six Normal-IgG, all six Matched-IgG, and five of six Mismatched-IgG macaques, determined by DNA real-time PCR (Supplementary Table 1 and Supplementary Fig. 1). Peak peripheral blood mononuclear cell (PBMC) associated viremia was delayed by 1 week in both the Matched and Mismatched groups (Fig. 1c). At 1 wpi, proviral load was significantly lower in the Matched-IgG (P = 0.0159, Mann-Whitney U test) and Mismatched-IgG (P = 0.0159) groups but control was not sustained. Similar to cell-associated viremia, there was a 1 week delay in plasma viral load in both the Matched-IgG and Mismatched-IgG groups compared with Normal-IgG (Fig. 1d, and Supplementary Fig. 2). Differences in area under the curve minus baseline (AUC, Mann-Whitney U test) were significant in the Matched-IgG vs. Normal-IgG groups (P = 0.0173) but not the Mismatched-IgG group versus Normal-IgG (P = 0.6905) (Fig.1e). Steady state levels (weeks 8–24) were significantly different between Normal-IgG and the Matched-IgG group (P = 0.0303) but not the Mismatched-IgG group (P = 0.5556) (Fig.1f).
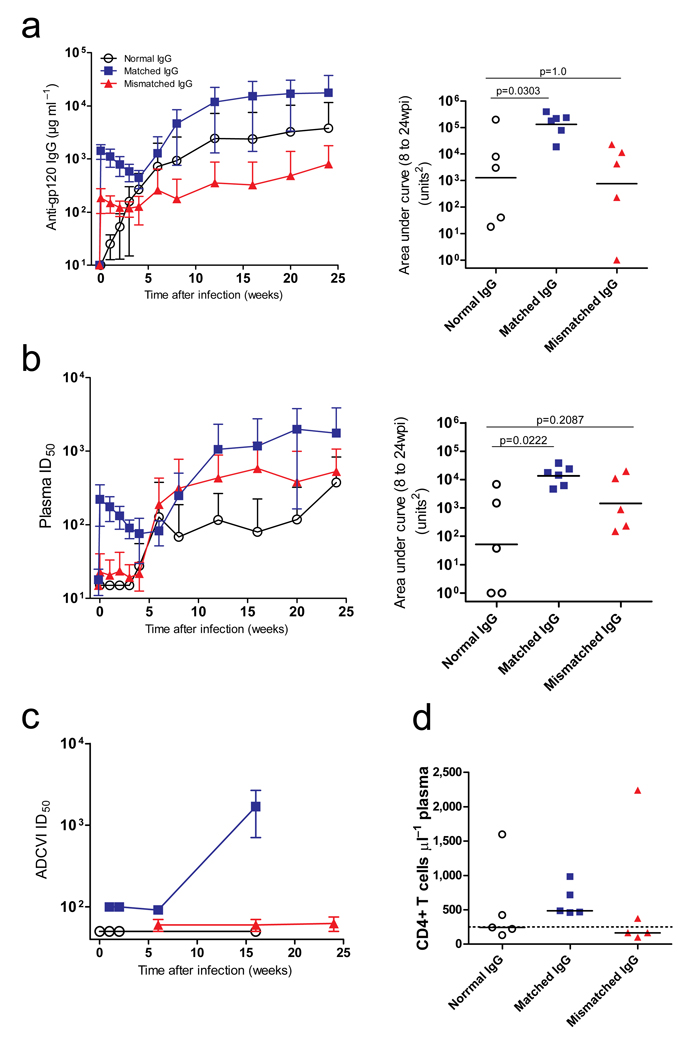
Prior to 24 wpi, only one macaque in the Normal-IgG group and two in the Mismatched-IgG group developed de novo gp120-specific antibodies. Matched-IgG was detected in the plasma 24 h post infusion, and by 6 wpi all six animals developed de novo antibodies above the passive transfer levels (Fig. 2a and Supplementary Fig. 3a–c). Total gp120-specific IgG (AUC, 8–24 weeks) of the Matched-IgG was significantly higher than the Normal-IgG group (P = 0.0303) but not for the Mismatched-IgG group (P = 1.0). By 24 wpi, mean titers in the Matched-IgG group were 19-fold higher than the Mismatched-IgG group and 10-fold higher than the combined control groups. All groups had similar kinetics and titers against the administered hepatitis B vaccine (P > 0.3; Supplementary Fig. 4), a control for non-HIV-1 B cell responses.
Fig. 2. Matched-IgG, but not Mismatched-IgG, improves humoral responses to 24 weeks post-infection.
(a) Left panel, mean HIV-1SF162 gp120-specific IgG concentration (±SD) as determined by kinetic ELISA for animals treated with Normal IgG, Matched IgG, and Mismatched IgG; right panel, analysis of the longitudinal effect of the SHIVIG pre-treatments on Env-specific IgG development (weeks 8–24) using AUC analysis (Mann-Whitney U test). (b) Left panel, neutralizing activity in the plasma of each group was evaluated against SHIVSF162P3 clone MC17, reported as the mean titers (±SD) for each group. ID50 is the plasma dilution necessary to inhibit infection by 50%; right panel, area under the curve from 8 wpi to 24 wpi, and the Matched-IgG group had significantly higher titers than the controls (AUC, Mann-Whitney U test) while the Mismatched-IgG group did not. (c) Mean ADCVI levels (±SD) were evaluated to examine sources of virus inhibition other than neutralization. (d) At 24 weeks post-infection, CD4 counts within the Matched group remained above 250 cells µl−1 blood (dotted line) while the Mismatched group had > 50% of infants at or below this level. * denotes P-value < 0.01.
To measure NAbs, we utilized a single pseudovirus cloned from the challenge stock in the TZM-bl assay14. Env clones in the challenge stock were 0.5% divergent (Supplementary Fig. 5) and exhibited a range of sensitivity to neutralization (Supplementary Fig. 6a). On the day of challenge, no NAbs were detected in the Normal-IgG group. Only one developed significant NAbs; two of five Normal-IgG infants had detectable NAbs only at 24 wpi (Supplementary Fig. 3d). On the day of challenge, two macaques in the Mismatched-IgG group had neutralizing activity just above the detection level and developed NAbs early, while the remaining macaques had detectable NAbs only at 24 wpi (Supplementary Fig. 3e). In contrast, all six macaques that received Matched-IgG had detectable NAbs on the day of challenge (Supplementary Fig. 3f) and developed de novo NAbs by 12 wpi, many detectable as early as 6 wpi, which increased and persisted until necropsy. Macaques that had NAbs by week 24 neutralized most or all variants from the inoculum (Supplementary Fig. 6b). Mean titers of NAbs (±SD) (Fig. 2b) and total NAb titer (AUC, 8–24 weeks) of the Matched-IgG group was significantly higher than the Normal-IgG group (P = 0.0069), while the Mismatched-IgG group was not (P = 0.0763).
Substantial de novo ADCVI activity was restricted to the Matched-IgG group in which five of six macaques developed 50% inhibitory titers ≥ 100 (P = 0.01, Fisher’s exact test; Fig. 2c). Several Mismatched-IgG animals had low-level ADCVI titers at the limit of detection (P = 0.4), and it is possible that titers increased after 6 months. Although ADCVI titers >100 were found only in plasma from animals with NAb responses, this degree of response was not present in all NAb-positive macaques (Supplementary Fig. 3g–i).
There were no significant differences in total peripheral CD4+ T cell levels (Supplementary Fig. 7a–d). However, by 24 wpi, none of the five surviving Matched animals had CD4 counts < 250 cells µl−1 plasma, while seven of ten animals in the Normal-IgG and Mismatched-IgG groups experienced early loss below 500; Six of the ten continued to < 250 (Fig. 2d). Macaques that developed NAbs, regardless of treatment, by week 8 maintained higher CD4 levels (P = 0.002, Mann-Whitney U test; Supplementary Fig. 7e). One of the ten macaques that developed NAbs had a CD4 count < 250 cells µl−1 at the end of 24 weeks, whereas those failing to develop NAbs had CD4 counts below that threshold (P = 0.019, Fisher’s exact test).
Matched-IgG with no ADCVI activity and neutralizing activity well below that used to provide sterilizing protection in other passive antibody studies5 and at a physiologic level for an SHIV-infected dam in vivo14 initiated rapid NAb and ADCVI development in neonatal macaques concurrent with improved control of plasma viremia. High levels of ADCVI developed only in those macaques that received the Matched-IgG, and an Fc effector function also may have contributed to reduced plasma viremia at later timepoints10,16. The ability to trigger strong NAbs and ADCVI responses provides indirect evidence that the early presence of NAbs protected B cells from the virus-induced destruction observed in HIV-1 infection17. Furthermore, NAb development was associated with higher CD4+ T cell count and suppressed viral replication. Mismatched-IgG that could only weakly neutralize the challenge virus failed to provide significant benefit, underscoring the importance of dosage. Although we did not observe increased CTL activity in SIV-infected passively treated juvenile macaques in a prior study12, others have observed preservation of central memory CD4+ T cells following passive treatment during SIV infection13. It is likely that improved B cell responses in this study were accompanied by enhanced T cell responses, which were not measured in this study due to sample size limitation.
Although we do not know the extent to which IgG1b12 within the Matched-IgG contributed to the early control of viremia and enhanced antibody production, the addition of IgG1b12 only increased the neutralization activity in the Matched IgG by 10%, and was less than 0.2 mg kg−1, five-fold lower than the amount shown to be protective against repeated low dose vaginal challenge with the same SHIV9. This idea is intriguing because it would signify that small fractions of NMAbs directed to the CD4bs or other key regions could be used to augment native immunity. The potential benefit is greatest for the prevention of Mother-to-child-transmission, where potent, broadly neutralizing NMAbs such as the recently discovered PG9/PG1618 and VRC0119 could be delivered to mothers prior to delivery or to newborns at parturition and during the period of breastfeeding. Future studies will be directed toward utilizing this nonhuman primate model to explore the protective and immunomodulatory effects of combination NMAb therapy and to understand the mechanisms underlying this beneficial effect.
Materials and Methods
IgG purification
Total IgG was purified from the plasma of adult Macaca nemestrina that were uninfected, or infected with SHIVSF162P3 or SHIV89.6P. Control IgG was purified from 1 L of normal plasma from a single simian retrovirus- and SIV-negative M. nemestrina screened for the absence of reactivity to SHIVSF162P3. Matched IgG was purified from approximately 100 ml of pooled plasma obtained from terminal bleeds of animals infected with SHIVSF162P3 that demonstrated narrow neutralizing activity (high neutralizing activity against the challenge virus and low neutralizing activity against SHIV89.6P) to mimic the typically narrow specificity of humoral responses in most HIV-1-patients. To ensure that the SHIVIG represented a mature NAb response, the monoclonal antibody IgG1b1215 specific for the CD4bs, was incorporated at a low concentration in a ratio of 1:1000 (w:w,140 µg ml−1). Lastly, Mismatched IgG was purified from 100 ml of pooled plasma obtained from terminal bleeds of M. nemestrina infected with SHIV89.6P that demonstrated high neutralizing activity against SHIV89.6P and low activity against the challenge virus. IgG purification was performed as previously described12. IgG preparations were tested for the presence of endotoxin via the limulus amoebocyte lysate test (Lonza Group Ltd) according to manufacturer’s instructions; all preparations contained < 5 EU ml−1. Activity based upon in vitro neutralization of SHIVSF162P3 and SHIV89.6P was monitored at each step. Purity was determined by quantifying scans of Coomassie-stained gels and was in the range of 85 to 90%.
Animal Care
This study used newborn 3–10 day old M. nemestrina (pigtailed macaques) obtained from the time-mated breeding colony and raised in the ABSL-3 infant nursery at the Washington National Primate Research Center in Seattle, WA (WaNPRC). All animal procedures were approved by University of Washington Institutional Animal Care and Use Committee (IACUC) in accordance with Office of Laboratory Animal Welfare (OLAW) and the National Institutes of Health Guide for Care of Laboratory Animals. The WaNPRC is an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) accredited facility.
IgG Administration
Eighteen M. nemestrina newborn macaques were treated with purified total M. nemestrina IgG from uninfected macaques (Control IgG, n = 6), SHIVIG from SHIVSF162P3-infected macaques (Matched IgG, n = 6), or SHIVIG from SHIV89.6P-infected macaques (Mismatched, n = 6). Purified IgG was delivered subcutaneously at multiple sites around the scruff of the neck at a dose of 170 mg kg−1 (Control IgG) or 200 mg kg−1 (Matched and Mismatched IgG) 24 h before challenge. No toxicities were observed with the subcutaneous administration. All 18 animals were challenged orally with two doses of 50% animal infectious doses (AID50) of a macaque cell-grown stock of SHIVSF162P3 administered 1 h apart. AID50 was determined in a titration experiment described previously14. Infants were monitored for 6 months for clinical signs of disease, including lymph node palpation and measurement, weight, appetite, etc. Blood samples were taken at weekly, bimonthly, or monthly intervals to determine lymphocyte subsets, antibody responses, and viral load in plasma and in PBMCs. As a positive control for functional humoral immunity, all macaques were vaccinated with recombinant hepatitis B vaccine (Engerix-B, GlaxoSmithKline Biologicals) at the time of IgG infusion (0 wpi) and at 4 wpi.
Quantitative PCR
PBMC DNA was extracted using QiaAmp DNA Blood Mini Kit (Qiagen) according to the manufacturer’s instructions. Proviral load was measured by quantitative PCR. PCR reactions for SHIV DNA contained TaqMan Universal PCR Master Mix (ABI, Norwalk, CT), 500 nM of forward and reverse primers (GAG5f: 5’–ACTTTCGGTCTTAGCTCCATTAGTG–3’; GAG3r: 5’–TTTTGCTTCCTCAGTGTGTTTCA–3’), and 200 nM of Taqman probe labeled with a 5’ FAM fluorescent reporter dye and a 3’ blackhole quencher (5’–FAM–TTCTCTTCTGCGTGAATGCACCAGATGA–BHQ TAMRA–3’). Real-time PCR was performed on a the ABI 7500 Fast machine (ABI) with the following cycling conditions: 2 min at 50 °C, 95 °C for 10 min, followed by 45 cycles at 95 °C for 15 s and at 60 °C for 1 min. Standards were dilutions of genomic DNA from Hut-78/E11S cells, which contain one copy SIV Gag per genome.
Quantitative RT-PCR
Viral stock RNA and plasma viral RNA samples were extracted using the QiaAmp viral RNA extraction kit (Qiagen) per manufacturer’s instructions. RNA copy number was measured by quantitative RT-PCR. RT reactions consisted of 500 µM dNTPs, 2.5 ng µl−1 random hexamers, 0.6 U µl−1 RnaseOut, 5.7 U µl−1 SuperScript III (Invitrogen) in a total volume of 14 µl. RT reaction conditions included 25 °C for 10 min, followed by 42 °C for 50 min, then 85 °C for 5 mins. To quantify cDNA, 2 µl of the RT reaction was used in the quantitative PCR described above in a total volume of 30 µl. Standard for RNA was an in vitro transcript of plasmid p239gag containing KpnI-BamHI SIV Gag fragment from SIVmac239 (gift of J. Lifson and M. Piatak). Ten-fold dilutions of this standard were made from 1 × 106 copies µl−1 to 10 copies µl−1. A final 2-fold dilution was made to obtain five copies µl−1. Plasma samples from the Titration group were only available for weeks 8–24.
Kinetic ELISA
Gp120-specific antibodies and antibodies to Hepatitis B Surface Antigen (HBsAg) were assessed by kinetic enzyme-linked immunoabsorbent assay (ELISA). Polystyrene 96-well flat-well plates (Maxisorp, Immulon; VWR) were coated with 1–2 µg ml−1 (100 µl per well) of SF162 gp120, 89.6P gp120, or HBsAg in carbonate-bicarbonate buffer (pH 9.5) and incubated 4 °C for 12 to 18 h. Wells were blocked with 1% normal goat serum in BLOTTO (5% skim dry milk in PBS) at 200 µl per well at 25 °C for 1 h. Plates were washed with 0.1% Triton–X 100 after which test plasma diluted to 1:60, 1:180, and 1:540 in 1% Triton–X 100 was added at 100 µl well−1 and incubated for 1 h at 25 °C. Antigen binding was detected with horseradish peroxidase conjugated goat anti-monkey antibody to whole IgG for 1 h at 25 °C and plates were developed with 3,3’,5,5’–tetramethylbenzadine (Sigma-Aldrich). Color development was monitored in real-time using an ELISA plate reader at a wavelength of 650 nm. To determine relative concentrations, the slope of the linear portion of the development curve was compared to those generated by a standard dilution series consisting of the purified Matched IgG used in the animals described above.
Cloning of gp160
Reverse-transcription, nested PCR was performed on RNA isolated from cell-free culture of SHIVSF162P3 using the SuperScript III First Strand Synthesis Kit (Invitrogen) per manufacturer’s instructions. Control amplifications with no reverse transcriptase were included to monitor for DNA contamination. The first round PCR was performed with 20 ρmol of primers, EO (5’–TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA–3’) and EO1 (5’–TCCAGTCCCCCCTTTTCTTTTAAAAA–3’), with 1.5 M MgCl2, 200 µM dNTP, and 3.5 U expand high fidelity Taq (Roche, Indianapolis, IN). The following cycling conditions were used: denature at 94 °C for 2 min, 35 cycles of 94 °C for 1 min, 61 °C for 1 min, and 72 °C for 3 min and 30 s, followed by a final extension at 72 °C for 10 min. Second round PCR was performed on 10 µl of first round PCR product using 20 ρmol of primers P3 5'gp160 NheI (5’–GCGGCGGCGGCTAGCGTAGAAAAATTGTGGGTCAC–3’) and P3 3' gp160 ClaI (5’–GCCGCCGCCATCGATTTATAGCAAAGCCCTTTC–3’) with 1.5 mM MgCl2, 200 µM dNTP, and 3.5 U expand high fidelity enzyme (Roche). The second round primers inserted an in-frame NheI restriction site 3’ of the Env leader sequence and a ClaI restriction site at the 3’ end of gp41. The second round cycling conditions included: 92 °C for 5 min, 40 °C for 1 min, 68 °C 5 min, 92 °C for 1 min, 45 °C for 1 min, 68 °C for 5 min, 2× (92 °C for 1 min, 50 °C for 1 min, 68 °C for 5 min), 2× (92 °C for 1 min, 55 °C for 1 min, 68 °C for 5 min), 25× (92 °C for 1 min, 60 °C for 1 min, 68 °C for 5 min), 68 °C for 10 min. The resulting PCR product was purified using the QIAquick PCR purification kit (Qiagen) according to the manufacturer’s instructions.
The purified nested PCR products were pooled and ligated into the 2.1 TOPO-TA vector (Invitrogen) using 4 µl of PCR product and 1 µl of vector. 2 µl of ligation mix were transformed into TOP10 chemically competent cells (Invitrogen). Transformed cells were selected for with ampicillin and X-gal. Individual colonies were inoculated into 3 ml of Luria-Bertani media with 10 µg/ml ampicillin and incubated in a shaking incubator at 30 °C for 12–16 h. Plasmid DNA was prepared using the Qiaprep Spin Miniprep Kit (Qiagen) according to the manufacturer’s instructions. Proper insertion of gp160 was confirmed by digestion with EcoRI. Clones that contained the proper length insertion of 2.4 kb were subsequently subcloned into the expression plasmid pEMC* (gift from Dr. Vicente Planelles). The insert was removed from the TOPO-TA vector by digestion with NheI (NEB) and ClaI (NEB) and the gp160 fragment was ligated in to pEMC* with rapid T4 DNA ligase (Roche). Approximately 8 ng of total plasmid DNA was transformed into 50 µl of MAX Efficiency DH10B Competent Cells (Invitrogen) as per manufacturer’s instructions and grown at 30 °C for 24 h. The plasmid was purified with a Qiaprep Spin Miniprep Kit as described above and sequenced for verification of the insert.
Fusion assay
Gp160 clones were tested for in vitro expression and functionality by direct transfection of TZM-bl cells. TZM-bl cells were plated to 70% confluency in a 96-well tissue culture plate. Each well was transfected with 0.1 µg plasmid DNA in 2.7 µl polyethylenimine (Polysciences, Inc, Indianapolis, IN) and 13 µl serum-free DMEM and incubated for 24 h at 37 °C. After 24 hours, cells were fixed with 1:1 Acetone: Methanol for 10 min at −20 °C. To stain for expression, the fixed cells were incubated with 25 µg ml−1 SHIVSF162P3 IgG in PBS/1% BSA/5% normal goat serum for 1 h at 37 °C, followed by incubation with goat anti-monkey fluorescein isothiocyanate (FITC) conjugated secondary antibody (in PBS/1% BSA/5% normal goat serum) at a dilution of 1:50 for 1 h at 37 °C. The plates were imaged on a Nikon eclipse E600 fluorescent scope.
Construction of pseudovirus
Individual Env clones contained within the pEMC* expression plasmid were co-transfected with the Env-deleted viral backbone plasmid Q23ΔEnv (kindly provided by Dr. Julie Overbaugh). 293T cells, plated to 80% confluency in 6-well plates, were transfected with 2.6 µg of total DNA (0.3 µg of Env plasmid and 2.3 µg of Q23ΔEnv plasmid) prepared in 10.5 µl of polyethylenimine (Polysciences, Inc) and 52 µl of serum-free DMEM. Pseudovirus was harvested 48 h later, spun at 2,000 RPM for 10 min, and stored at −80 °C until use. To determine the infectivity of prepared pseudovirus, each preparation was titered on TZM-bl cells. Four-fold serial dilutions of pseudovirus were added to complete media (DMEM, 10% FCS, 1% L-glutamine, 1% penicillin-streptomycin) in the presence of 7.5 µg ml−1 DEAE Dextran and incubated at 37 °C for 1 h. Each well received 100 µl of TZM-bl cells resuspended in media at 1×105 cells ml−1. 48 h later cells were lysed for 2 min directly on the plate using 100 µl of Bright-Glo Luciferase Assay Substrate (Promega) and immediately analyzed for luciferase activity on a luminometer. For use in neutralization assays, the 200 TCID50 was calculated according to the Reed-Muench method.
Sequencing
Cloned gp160 was sequenced using two forward primers (5’–GCACAGTACAATGTACACATGGAA–3’; 5’–ATGGGATCAAAGTCTAGAGCCATGTG–3’), two reverse primers (5’–CTTGCCCACTTATCCAATTC–3’; 5’–CACAATCCTCGCTGCAATCAAG–3’), and the vector primers provided in the TOPO TA kit (M13(–21)F: 5’–TGTAAAACGACGGCCAGT–3’; M13R: 5’–AACAGCTATGACCATG–3’). Sequencing was performed with Big Dye version three on the Applied Biosystems 3100 Genetic Analyzer. Sequences were assembled and edited using Sequencher and aligned with ClustalX.
TZM-bl neutralization assay
Plasma samples from each animal were tested at all available timepoints for neutralizing activity using the 96-well TZM-bl neutralization assay described previously14. Briefly, plasma samples were heat inactivated at 56 °C for 1 h. Serial dilutions of heat-inactivated plasma received 200 TCID50 of virus in the presence of 7.5 µg ml−1 DEAE Dextran and incubated in a total volume of 150 µl media (DMEM, 10% FCS, 1% L-glutamine, 1% penicillin-streptomycin) for 1 h at 37 °C. After incubation, each well received 1×104 TZM-bl cells suspended in 100 µl of media. 48 h later cells were lysed for 2 min directly on the neutralization plate using 100 µl of Bright-Glo Luciferase Assay Substrate (Promega, Madison, WI) and immediately analyzed for luciferase activity on a luminometer. The reciprocal dilution of plasma necessary to achieve 50% neutralization is reported. Any sample that did not neutralize at 1:30 is reported as 15. All values are calculated with respect to virus only [(virus-cell only)-(plasma-cell only)]/ (virus – cell only).
ADCVI assay
The ADCVI assay was performed as previously described20. Target cells for the ADCVI assay were prepared by infecting polybrene-treated CEM.NKR-CCR5 cells (National Institutes of Health AIDS Research Reference Reagent Program) with SHIVSF162p3 at a multiplicity of infection of ~0.05. After 48 h, target cells were washed to remove cell-free virus. Effector cells (PBMCs from healthy human donors) were added to target cells at an E:T ratio of 10:1. Test sera were added to target and effector cells to achieve a final dilution of 1/100. Seven days later, supernatant fluid was collected, and p24 was measured by ELISA (Zeptometrix). The percent inhibition due to ADCVI was calculated relative to pooled HIV-1-negative serum as follows: percent inhibition = 100(1 − [(p24p)/ (p24n)]), where (p24p) and (p24n) are concentrations of p24 in supernatant fluid from wells containing a source of SHIV-positive or -negative Ab, respectively.
Statistical analyses
To gain statistical power, the data from the titration animals and the Normal-IgG animals were combined after testing for differences for all comparisons except for plasma virus loads. No data for this assay were available for the titration macaques prior to week 4. To measure the median change in virus load and antibody levels during a timespan of interest, the area under the curve minus baseline was calculated after replacing values at the lower limit of detection with the midpoint value between zero and the lower limit. Statistical significance was then determined using the Mann-Whitney U test using nontransformed data. The Mann-Whitney U test was also used to calculate differences in continuous outcomes at specific timepoints. For differences in binary outcomes, we used Fisher’s exact tests to determine statistical significance. Significance level was set at P < 0.05. All uninfected animals were excluded from statistical analyses.
Supplementary Material
Acknowledgements
We would like to thank Z. Brower for technical support, D. Morris and K. Filer for manuscript preparation, M.L. Marthas and K.K.A. Van Rompay for advice on subcutaneous dosing of IgG and oral inoculation of infants, J. Overbaugh (Fred Hutchinson Cancer Research Center) for the env-deleted viral backbone plasmid Q23ΔEnv, S. Barnett and I. Srinivasan (Novartis) for recombinant gp120-SF162, and V. Planelles (University of Utah) for the plasmid pEMC*. M. Piatak and J.D. Lifson (SAIC) kindly provided RNA standards and advice on real time PCR. SHIVSF162P3 was supplied by N .Miller and R. Pal. Reagents were supplied by the NIH AIDS Research Reference Reagent Program. This study was supported by grants from NIH R01HD038653 (N.L.H.), R01AI33292 (D.R.B.) and R01AI052039 (D.N.F.) and from the Elizabeth Glaser Pediatric AIDS Foundation (N.L.H.).
Footnotes
Contributions of authors:
C. Ng, J.P Jaworski, W.F. Sutton and G. Landucci were responsible for experimental work, analyses, and preparation of figures; P. Delio and L, Kuller provided animal care and clinical and laboratory assessments; D. Anderson oversaw the study at the Washington NPRC; C. Ng, D. Forthal, D. Burton, D. Anderson, and P. Jayaraman contributed to the writing of the manuscript; B. Richardson performed the statistical analyses; D. Burton provided IgG1b12; N.L.Haigwood was responsible for study design and coordination, and writing the manuscript.
References
- 1.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steimer KS, Scandella CJ, Skiles PV, Haigwood NL. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to gp120. Science. 1991;254:105–108. doi: 10.1126/science.1718036. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, et al. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nature Medicine. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 6.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 7.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 8.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000433. e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forthal DN, Landucci G, Phan TB, Becerra J. Interactions between natural killer cells and antibody Fc result in enhanced antibody neutralization of human immunodeficiency virus type 1. J Virol. 2005;79:2042–2049. doi: 10.1128/JVI.79.4.2042-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forthal DN, et al. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J Virol. 2006;80:9217–9225. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haigwood NL, et al. Passive immunotherapy in simian immunodeficiency virus-infected macaques accelerates the development of neutralizing antibodies. J Virol. 2004;78:5983–5995. doi: 10.1128/JVI.78.11.5983-5995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto H, Kawada M, Takeda A, Igarashi H, Matano T. Post-infection immunodeficiency virus control by neutralizing antibodies. PLoS One. 2007;2:e540. doi: 10.1371/journal.pone.0000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayaraman P, et al. Evidence for persistent, occult infection in neonatal macaques following perinatal transmission of simian-human immunodeficiency virus SF162P3. J Virol. 2007;81:822–834. doi: 10.1128/JVI.01759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 16.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 17.Moir S, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forthal DN, Gilbert PB, Landucci G, Phan T. Recombinant gp120 vaccine-induced antibodies inhibit clinical strains of HIV-1 in the presence of Fc receptor-bearing effector cells and correlate inversely with HIV infection rate. J Immunol. 2007;178:6596–6603. doi: 10.4049/jimmunol.178.10.6596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.