Abstract
FcγRIII (CD16) is a receptor expressed on immune cells that selectively binds immmunoglobulin G (IgG) molecules, IgG binding results in cellular activation and cytokine release. IgG is an important factor in arthritis and can be found in arthritic temporomandibular joints (TMJ). We hypothesize that a reduction in FcγRIII expression in the TMJ tissues will reduce the nociceptive and inflammatory response in an inflamed joint. To test this hypothesis siRNA, either naked or complexed with linear polyethylenimine (PEI) was injected into the superior joint space of the TMJ. After administration of siRNA the joint was injected with saline or with complete Freund’s adjuvant (CFA) to induce arthritis. Nociceptive responses were quantitated in the rat by measuring the animal’s meal duration. FcγRIII expression in the TMJ tissue was assayed by immunocytochemistry or western. Cleavage of FcγRIII transcript was then assayed by 5′ rapid amplification of cDNA ends method (5′ RACE). Interleukin-1β (IL-1β) and IgG content was measured in the TMJ tissue by ELISA. The results indicate that injection of FcγRIII siRNA reduced the amount of FcγRIII in the TMJ tissues and that the transcript was cleaved in a manner consistent with a RNA interference mechanism. Moreover, injection of FcγRIII siRNA reduced the nociceptive response of rats with an arthritic TMJ and reduced the amount of pro-inflammatory cytokine IL-1β. We conclude that FcγRIII contributes to the pain resulting from inflammatory arthritis of the TMJ and that siRNA has the potential to be an effective treatment for this disorder.
Introduction
FcγRIII is a member of the Fc receptor family and a cellular component of both innate and adaptive immunity. FcγRIII will bind the Fc portion of antibodies activating or inhibiting a series of inflammatory responses (1–4). Binding to an Fc receptor can cause activation or inhibition of inflammation depending on the whether the receptor contains an intracellular immunoreceptor tyrosine-based activation motif (ITAM) or a immunoreceptor tyrosine-based inhibitory motif (ITIM). FcγRIII binding is preferential for small IgG dimer or trimer complexes, such as IgG anti-IgG antibody complexes that make up self antigens (5;6). Self antigens are potential triggers for onset or maintenance of arthritis (7–9). IgG antibodies bind Fc receptors on several types of leukocytes including neutrophils, macrophages, natural killer and mast cells activating arthritic mechanisms in both humans and rats (1–4;10). Notably, IgG levels are higher in humans that have TMJ arthritis (11), suggesting a potential role for FcγRIII.
FcγRIII is a valid therapeutic target first, because a significant sub-set of TMJ patients present with some level of inflammation (12;13) and deleting FcγRIII expression has been shown to decrease inflammatory arthritis (14). Second, FcγRIII is a receptor restricted to leukocytes that are in synovial tissues impacted by arthritis (15), including TMJ tissues (10) and third, because IgG, a ligand for FcγRIII, was significantly higher in the joint of humans that have arthritic TMJ disorders (11). Together these results suggest FcγRIII has a role in inflammatory TMJ arthritis and we hypothesize that a reduction in FcγRIII expression in the TMJ tissues will reduce the nociceptive response in an inflamed joint.
A viable method for knockdown of FcγRIII expression would be an intra-articular injection of siRNA having homology to the FcγRIII transcript (16). Administration of siRNA is often a challenging, but complexing siRNA with liner PEI polymer [H2N-(CH2CH2N-CH2CH2NH2)x-(CH2CH2NH)y-] increases the transfection efficiency of siRNA (17). PEI is a cationic polymer that forms nano-sized complexes with anionic nucleic acids mainly by attractive electrostatic interactions. When mixing PEI and nucleic acids, one adds a higher ratio of cationic PEI amines (N) than anionic nucleic acid phosphates (P); (called an N/P ratio). A high N/P ratio keeps the resulting complexes cationic causing electrostatic attraction between the cationic complex and the anionic phospholipid bilayer of cellular membranes. In this report we tested PEI complexed siRNA and in the event that siRNA would be used in future clinical applications we also tested naked siRNA, because the linear PEI used in these studies can have toxic effects, reducing cell viability (18). Moreover, injecting naked siRNA would eliminate the potential of activating the immune system as a result of PEI being present. After siRNA enters the cell it assembles with several proteins to form the siRNA-induced silencing complex (siRISC)(19–21). siRISC will bind a specific mRNA as a result of sequence complementarity to the siRNA loaded into the siRISC and silence gene expression, in part, by initiating cleavage of the bound mRNA (22;23). Activated RISC cleaves its target mRNA precisely between the nucleotides complementary to positions 10 and 11 of the siRNA anti-sense strand, generating a specific size mRNA cleavage product. This specific product can be detected by 5′ RACE (24).
To test our hypothesis we measured nociceptive responses, i.e., meal duration (25–29), in rats given a TMJ injection of FcγRIII siRNA and then a injection of saline or an arthritic adjuvant. Breakdown of FcγRIII protein and transcript in the TMJ tissue after siRNA treatment was determined by immunocytochemistry, western and 5′ RACE. In addition to these measurements we analyzed the effect of FcγRIII treatment on IL-1β expression in the inflamed joint.
Materials and Methods
These studies were approved by the Baylor College of Dentistry Institutional Animal Care and Use Committee in accordance of the guidelines of the USDA and National Institutes of Health Guide for Care and Use of Laboratory Animals. Male Sprague Dawley rats (220–250 grams) were purchased from Harlan Industries (Houston, TX). Upon arrival, animals were housed individually in a temperature-controlled room (23°C) under a 12:12 hour light:dark cycle with lights on at 0600 hour. The rats were given chow (Teklad 6% M/R Diet #7002, Harlan Industries, Houston, TX) and water ad libitum.
Treatment groups and experiments
Five treatment groups were used in this study. In Treatment Group 1 the superior joint space of the rat TMJ was injected with 11 μg/joint of carboxyfluorescein (FAM) conjugated FcγRIII siRNA [5′-ccuuauaauguuagcuacuccaucu-3′, 5′-ggaauauuacaaucgaugagguaga-3′] (Invitrogen, Carlsbad, CA). One day after injecting siRNA, the TMJ was injected with CFA. In Treatment Group 2 each TMJ was injected with a combination of 5.5 μg of FcγRIII siRNA #1 (5′-ccagcucucuagugugguutt-3′, 5′-aaccacacuagagagcuggtg-3′) and 5.5 μg of FcγRIII siRNA #2 (same sequence as the FAM conjugated FcγRIII siRNA) or 11 μg of a Silencer Negative Control #1 siRNA (5′-aguacugcuuacgauacggtt-3′, 5′-ccguaucguaagcaguacutt-3′) complexed to PEI. The Silencer Negative Control #1 siRNA has a random sequence that has no homology to any known gene and was expected to not cause degradation of any transcript. One day after injecting the siRNA, the TMJ was injected with saline or CFA. In Treatment Group 3 the siRNA from Treatment Group 2 without PEI complexing was injected into both TMJs, followed by a second TMJ injection of saline or CFA one day later. Treatment Group 4 includes injecting both TMJs with 10 μg of FcγRIII siRNA #1 and 10 μg of FcγRIII siRNA #2 or 20 μg of Silencer Negative Control #1 siRNA, followed by an injection of saline or CFA one day later. Treatment Group 5 was not injected with siRNA but did receive a TMJ injection of saline or CFA.
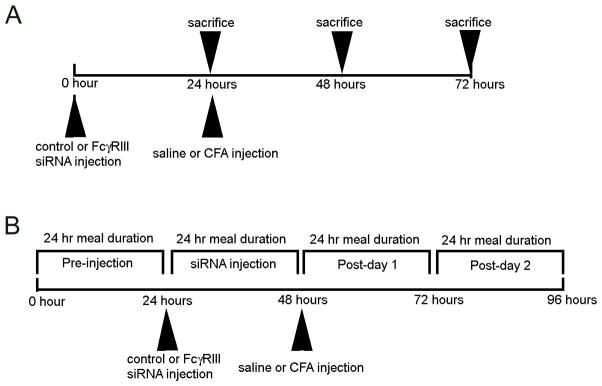
Five experiments were performed using one or more of the treatment groups described above. The first experiment included Treatment Group 1. 24 hours after CFA injection the rats were sacrificed and the tissue harvested for immunocytochemistry (Fig. 1A). Three rats were included in this experiment. For the second experiment the TMJ tissue was isolated 24, 48 or 72 hours after injecting rats in Treatment Group 2 with siRNA (Fig. 1A). Immunocytochemistry and 5′ RACE studies were completed on these tissues. Three rats were included per treatment. In the third experiment the nociceptive response was measured in rats from Treatment Groups 2 and 5 (Fig 1B). Four rats were included per treatment group. In the fourth experiment the nociceptive response was measured in rats from Treatment Groups 3 and 5 (Fig. 1B). Six rats were included per treatment. Finally, the fifth experiment included a western blot for measuring FcγRIII protein levels and an ELISA for quantitation of IL-1β and IgG in the TMJ, 48 hours after injecting rats from Treatment Groups 4 and 5 with saline or CFA (Fig. 1A). Four rats were included per treatment.
Figure 1. Experimental timeline for the tissue collection and meal duration studies.
A) Timeline for experiments that included harvesting tissue after no injection or after injection of siRNA followed by an injection of saline or complete Freund’s adjuvant (CFA) into the superior joint space of the rat temporomandibular joint (TMJ). The tissue was analyzed by fluorescent imaging, 5′ rapid amplification of cDNA ends method (5′ RACE), western and ELISA. B) Timeline for studies that included injecting siRNA complexed with polyethylenimine (PEI) or injecting naked siRNA into the upper joint space of the TMJ. siRNA injections were followed by injection of saline or CFA. Before and after injecting the TMJ meal duration measurements were completed in the feeding modules.
TMJ injection of siRNA, CFA and saline
To complete the TMJ injections Sprague Dawley male rats were anesthetized with isoflurane (5% flow) between 9:00–11:00 am. The injections were made using a 29-gauge, one-half-inch needle (Becton Dickinson, Franklin Lakes, NJ). The TMJ injections were completed by inserting the needle tip posterior to the zygomatic process of the temporal bone. The needle tip was then directed medioanteriorly along the roof of the mandibular fossa, where it entered the superior joint space of the TMJ (30) and the solution was expressed within 5 seconds. Injections included 15 μg of CFA (Mycobacterium tuberculosis, Chondrex, Redmond, WA) per joint or 0.9% saline or siRNA in a 15–30 μl volume. We have previously used CFA for the induction of nociceptive responses/inflammation in the TMJ (10;28–33). siRNA was either naked (in 0.9% saline) or was complexed with linear PEI polymer according to the manufactures directions (in vivo-jetPEI, N/P=6, Polyplus transfection Inc). Following injections and removal from anesthesia the rats are freely moving within two minutes. The animals were returned to their feeding modules to measure meal duration as described below.
Meal duration measurement to quantify nociception
Meal duration was characterized using data acquired from 32 feeding modules that were situated within sound-attenuated chambers equipped with photobeam computer-activated pellet feeders (Med Assoc. Inc., East Fairfield, VT). The rats were given 45 mg rodent chow pellets (Bioserv, Frenchtown, NJ). When the animal removed a pellet from the feeder trough, a photobeam placed at the bottom of the trough is no longer blocked, signaling the computer to drop another pellet, record the date and time, and keep a running tally of the total daily food consumption. The record of pellets dropped over time was computer-analyzed with a proprietary computer program to establish the meal duration (34) which is a continuous non-invasive biological marker of TMJ nociception (surface and deep) in the undisturbed animal (26;28;29;35). In the meal duration calculation for the rat, the end of a meal was defined as when no pellets were removed from the feeder for ten minutes (36). The minimum meal size needed to be at least three pellets.
Sample and tissue preparation
On the day tissue was collected animals were removed from their individual cages, taken to an adjacent room and sacrificed within 20 seconds by decapitation to minimize stress. Removal of TMJ tissue was performed. The soft tissue included the synovial membrane, joint capsule, retrodiscal tissue, articular disc, and a small amount of the lateral pterygoid muscle. After dissection, the tissues were placed in liquid nitrogen and stored in liquid nitrogen until RNA or protein was isolated. Alternatively, 0.5 cm TMJ tissue blocks centered on the TMJ condylar head were removed, fixed in 4% paraformaldyhyde for 48 hours. The tissue was demineralized in a 0.5 M EDTA solution and then to increase the demineralization rate the tissue was microwaved in a Pelco Biowave (Redding, CA). After demineralization the tissue was placed in 25% sucrose for 24 hours and sectioned on a cryostat (Damon International Equipment Company, Needham Heights, MA). The tissues were processed into serial 20 μm sections on superfrost plus slides (StatLab, Lewisville, TX).
Immunocytochemistry
Slides containing TMJ sections were rinsed twice in PBS for a total of 10 min, blocked with 2% BSA and 0.3% Triton-X 100 in PBS for 1 h at room temperature. Following three rinses in PBS, the slides were incubated in the primary antibody solution overnight at 4°C. Primary antibodies consisted of CD14 (T-19) (Goat polyclonal; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) diluted 1:10 and CD16 (H-80) (Rabbit polyclonal; Santa Cruz Biotechnology, Inc.) diluted 1:20. Primary antibodies were diluted with PBS and 0.3% Triton X-100. After incubation in primary antibody the slides were rinsed three times in PBS for a total of 15 min and placed for 1 hour in a 1:500 dilution of secondary antibody in PBS and 0.3% Triton X-100. Secondary antibodies included goat anti-rabbit-biotin or rabbit anti-goat-biotin (Invitrogen, Carlsbad, CA). After rinsing the slides three times in PBS for a total of 10 min, the slides were then placed in streptavidin-568 (Invitrogen) for 30 minutes at room temperature. Following three rinses in PBS the slides were counterstained with DAPI (Vector Labs, Burlingame, CA) and mounted with Fluoromount-G mounting medium (Electron Microscopy Sciences, Hatfield, PA). The fluorescent signal was imaged using a Nikon fluorescent microscope, MetaMorph Imaging System software (Molecular Devices Corporation, Sunnyvale, CA) and a Photometrics CoolSnap K4 CCD camera (Roper Scientific, Inc, Duluth, GA). Images were captured for DAPI using a filter with excitation between 395–410 nm and an emission between 450–470 nm. Images were captured for FAM using a filter with excitation between 490–505 nm and an emission between 515–545 nm and images for 568 nm were captured using a filter with an excitation filter between 520–570 nm and an emission filter was between 570–610 nm was utilized. Confocal images were collected with a Leica TCS SP2 microscope (Bannockburn, IL) using slides that were counterstained with Topro-3 (Invitrogen).
5′ RACE for the FcγRIII transcript
Total RNA was isolated from TMJ tissue using UltraSpec RNA total RNA isolation kit (Biotecx, Houston, TX). The total RNA was quantitated and the quality determined using an Agilent 2100 Bioanalyzer (Germany) following the manufacturer’s directions. 5′ RACE was completed using 10 μg of total RNA, as outlined in the manufactures directions (FirstChoice RLM-RACE, Applied Biosystems, Austin, TX). Exceptions to the protocol were that we did not remove the 5-methyl guanosine or dephosphorylate the RNA strands. First, the RACE adapter (5′-gcugauggcgaugaaugaacacugcguuugcuggcuuugaugaaa-3′) was ligated to the degraded RNA strands. The 5-methyl cap prevents ligation of the adaptor to full length transcripts. Second, reverse transcription of the FcγRIII mRNA was completed using the reverse transcription RACE primer (5′-ccgctgtttagccatacgat-3′). The reverse transcription product was added to a PCR reaction that included the RACE PCR primer for FcγRIII (5′-tgtggagccttgtactttccgact-3′) and the RACE PCR outer primer (5′-gctgatggcgatgaatgaacactg-3′) which hybridizes to the RACE adaptor. PCR reactions used a 3 min denaturation step 95°C, followed by 35 cycles of 94°C for 30 seconds, 63°C for 30 seconds, and 72°C for 1 min, followed by a 7 min 72°C extension. Electrophoresis of the PCR product was completed with a 2% agarose gel and the gel was stained with ethidium bromide. siRNA was purchased from Invitrogen (Carlsbad, CA). The adapter and primers were purchased from Integrated DNA Technologies (Coralville, Iowa).
Western and ELISA
At the time of the analysis, the TMJ tissue was placed in T-per lysis reagent (Thermo Scientific, Rockford, IL) and ground with a tissue homogenizer (Ultra-Turrax, Jankel & Kunkel, Germany). The total protein in the sample was determined using the BCA Protein Assay (Thermo Scientific) following the manufacturer’s directions. IL-1β or IgG concentration in the TMJ tissue was evaluated by ELISA following the manufacturer’s directions (R & D Systems, Minneapolis, MN and Alpha Diagnostics International Inc., San Antonio, TX, respectively). The concentrations were expressed as the amount of IL-1β or IgG per mg of total protein. For the western, 20 μg of total protein was loaded on a 8% tris-gylcine acrylamide gel, electrophoresis was performed and the protein in the gel was transferred onto a PVDF membrane in 25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3 (150 mA for 5 hrs, room temp). The membrane was blocked 1 hr in TBST buffer (100 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20, pH 7.4) plus 5% BSA (w/v) and then probed with the FcγRIII antibody clone H-80 (Santa Cruz, Santa Cruz, CA) at 1:500, 4°C overnight. The next day the membranes were washed in TBST buffer, incubated for 1 hour in goat anti rabbit secondary antibody (1:1000) conjugated to horseradish peroxidase (BioRad, Hercules, CA), washed in TBST buffer, reacted with ECL plus reagent (Thermo Scientific) and exposed to film.
Statistical Methods
Two-way ANOVA with repeated measures was used to analyze the rat meal duration, and cytokine data. The independent variables were treatment (siRNA, saline, CFA) and time. The dependent variable was either meal duration or the amount of cytokine. IgG values were analyzed by a t-test. Power for the meal duration experiments was 60% and power for the molecular studies was 80% with an α=0.05. P values less than 0.05 were deemed to be statistically significant.
Results
FcγRIII siRNA was inside the cells
A confocal image of the TMJ retrodiscal tissue taken 24 hours post-siRNA injection shows siRNA (green) was in the cytoplasm surrounding the blue nuclei (Fig. 2, bottom and right panel) indicating that siRNA was internalized as early as 24 hours post-injection.
Figure 2. siRNA was inside cells after injection of siRNA in the TMJ.
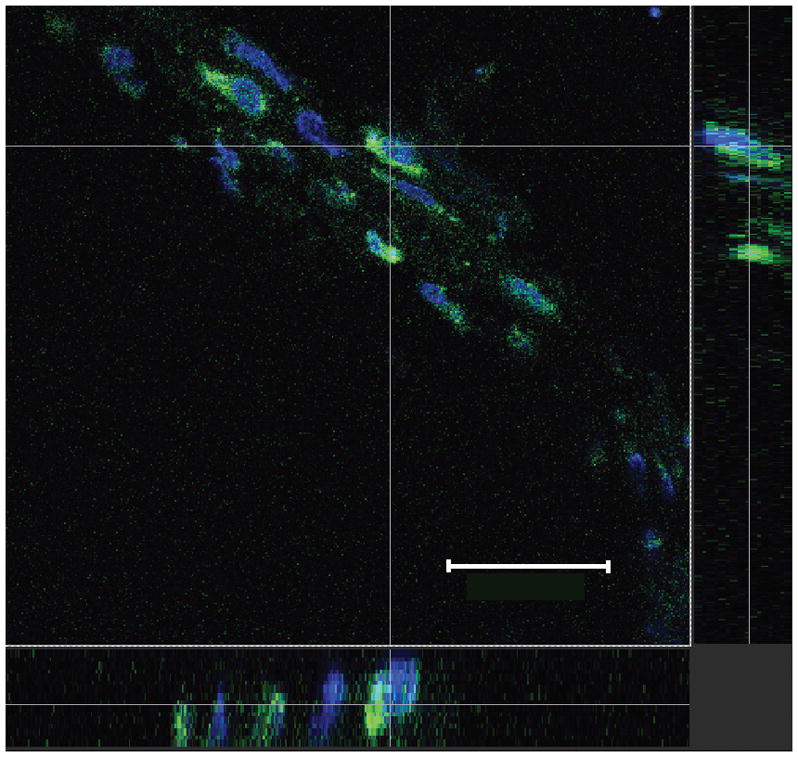
Carboxyfluorescein (FAM) conjugated FcγRIII siRNA complexed with linear PEI was injected into upper joint space of the male rat TMJ. Tissue was collected 24 hours post-siRNA injection, fixed and sectioned. FAM conjugated siRNA (green) surrounds nuclei stained with Topro-3 (blue) in the retrodiscal region. The main image, a view from above, is delineated by a dotted white line. A cross section image was produced in the horizontal and vertical plane, as indicated by thin, solid white lines. The cross section image for the X axis is on the bottom and the cross section image for the Y axis is to the right. See figure one legend for abbreviations. Bar= 50 μM. Representative image of 3 animals.
FcγRIII siRNA reduced FcγRIII expression
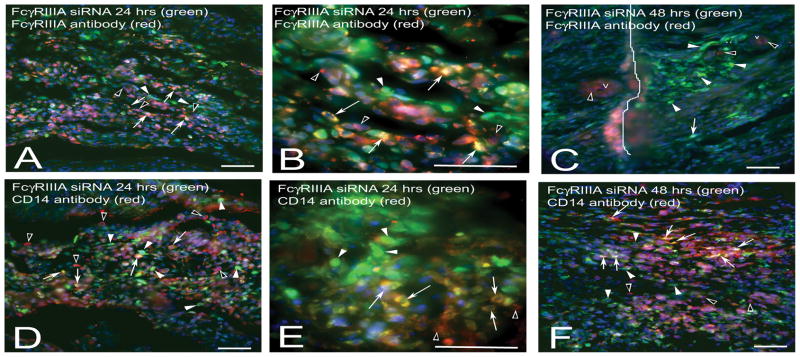
Injection of fluorescently labeled FcγRIII siRNA (green) into the TMJ showed that 24 (Fig. 3, A, B, D, E) and 48 hours (Fig. 3, C, F) after transfection many FcγRIII positive (red cells, Fig. 3, A, B) and CD14 positive (red cells, Fig. 3, D, E, F) cells were transfected with the fluorescently labeled FcγRIII siRNA (green). Images in panels A, B, D and E are before CFA injection indicating a number of FcγRIII positive (red cells, panels A and B) and CD14 positive cells (red cells, Panels D and E) are present in the joint before the onset of inflammation. Importantly, 24 hours after injecting CFA and 48 hours after injecting FcγRIII siRNA (green) a small number of red FcγRIII positive cells were present in the arthritic joint (Fig. 3, C) in comparison to the number of the number of FcγRIII positive cells in a non-inflamed joint 24 hours post-FcγRIII siRNA injection (Fig. 3A). Most of the remaining FcγRIII positive cells 48 hours post-FcγRIII siRNA treatment were localized near blood vessels (Fig. 3, C). In contrast to FcγRIII, the number of CD14 positive cells 48 hours post-injection (Fig. 3, F) was increased versus 24 hours post-injection (Fig. 3, D). Controls without primary antibody were negative and non-injected controls were negative (data not shown).
Figure 3. FcγRIII siRNA reduced the number of FcγRIII positive cells.
The upper joint space of the TMJ was injected with FAM conjugated FcγRIII siRNA complexed with PEI. Low (A) and high (B) magnification image of a section stained with FcγRIII antibody (red) 24 hours after injecting FAM conjugated FcγRIII siRNA (green). C) Tissue stained with FcγRIII antibody (red) 48 hours after injecting FcγRIII siRNA and 24 hours after injecting CFA. Low (D) and high (E) magnification images of a tissue section stained with CD14 antibody (red) 24 hours after injecting FcγRIII siRNA. F) Tissue stained with CD14 antibody (red) 48 hours after injecting FcγRIII siRNA and 24 hours after injecting CFA. Arrows point to double labeled cells (yellow) in the retrodiscal tissue containing FcγRIII siRNA and FcγRIII protein (A–C) or CD14 protein (D–F). Arrow heads point to cells containing only FcγRIII siRNA (green) and open arrowheads point to red cells having FcγRIII protein (A–C) or CD14 protein (D–F) with little co-labeling of siRNA. v= blood vessel on panel C. Line on panel C indicates a fold in the tissue section. See legends to figure one and two for abbreviations. Representative images are shown for 3 animals per treatment group. Size bars = 50 μm.
5′ RACE analysis detects cleavage of FcγRIII transcript by RNAi mechanism
Using an appropriately designed 5′ RACE protocol (24) the RISC cleavage product for the FcγRIII mRNA was 192 pb. The 192 bp 5′ RACE product was present in the TMJ tissue of rats treated with the FcγRIII siRNA and not in rats injected with the control siRNA (Fig. 4). Cleavage of the FcγRIII mRNA was observed 24 and 48 hours post-siRNA injection (Fig. 4). These results indicate the FcγRIII siRNA was degrading FcγRIII transcript through the RNA interference (RNAi) mechanism. Extraneous band are likely the result of non-specific cleavage of the FcγRIII transcript.
Figure 4. The FcγRIII gene was cleaved after injecting FcγRIII siRNA.
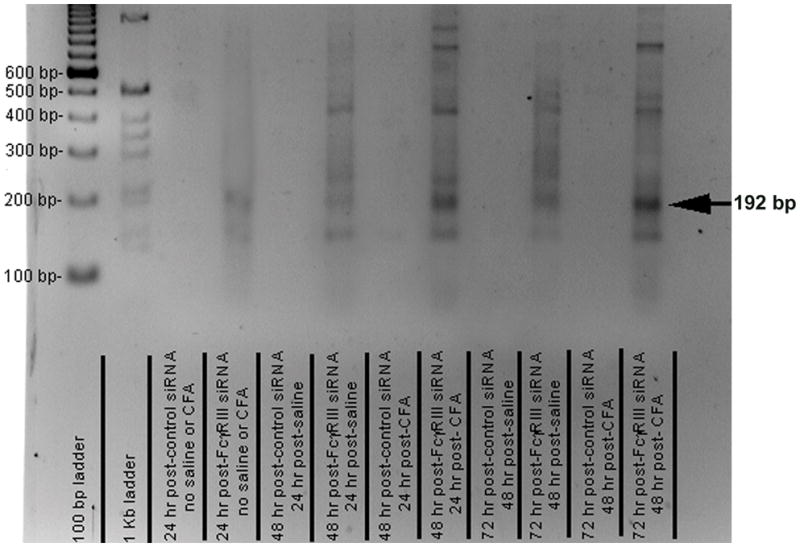
FcγRIII or control siRNA was complexed with PEI and injected into the upper joint space of the rat TMJ. 24 hours later the tissue was harvested from a portion of these rats. In the remaining portion of rats saline or CFA was injected. Tissue was isolated 48 and 72 hours post-siRNA injection; which was 24 and 48 hours following the saline or CFA injection, respectively. Using an appropriately designed oligo for reverse transcription and oligos for PCR primers the predicted 192-bp 5′ RACE product (arrow) was generated using RNA isolated from the TMJ tissue. Size markers were in the 2 left lanes. See figure one legend for abbreviations. Three rats were included for each treatment group, representative images are shown for each treatment group.
siRNA injected into the TMJ reduces nociceptive responses
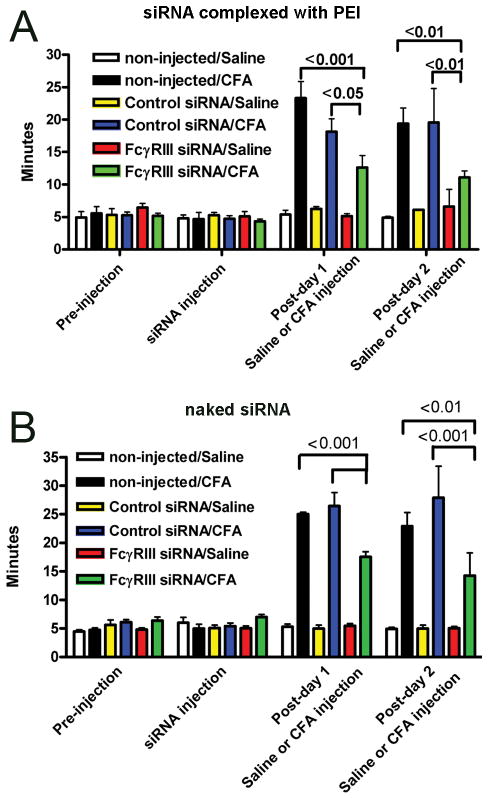
The nociceptive response was measured in rats before and after injection of the TMJ. On the day labeled “siRNA injection”, no change in the meal duration was observed when comparing the non-injected rats to the siRNA injected rats (Fig. 5A and B). CFA significantly increased the nociceptive response versus saline injected rats (Fig. 5A and B). One exception was on post-day 2 (Fig. 5, A), the meal duration of the FcγRIII siRNA/Saline group was not significantly different than the FcγRIII siRNA/CFA group. In CFA injected rats, administration of FcγRIII siRNA complexed with PEI caused a significant decrease in meal duration for two days; compare FcγRIII siRNA injected rats versus rats injected with control siRNA or the non-injected rats (Fig. 5, A). Similarly, injection of naked FcγRIII siRNA reduced the nociceptive response of the arthritic TMJ by 50% 2 days post-injection (Fig. 5, B). The reduction in nociception was concomitant with a reduction in FcγRIII expression in the TMJ tissues injected with FcγRIII siRNA (Fig. 6, A). No significant effect was detected after injection of control siRNA, compare non-injected rats with the control siRNA injected rats from the same treatment group (Fig. 5, A and B)
Figure 5. FcγRIII siRNA attenuated the nociceptive response in an arthritic TMJ.
A) Rats were injected with either control siRNA or FcγRIII siRNA complexed with PEI. Four animals were analyzed per treatment group. B) Rats were injected with naked control or FcγRIII siRNA. Six animals were analyzed per treatment group. Twenty-four hours after siRNA injection, saline or CFA was injected. “non-injected” rats did not receive a siRNA injection but did receive the saline or CFA injection. Daily meal duration was measured before injection (Pre-injection) after siRNA injection (siRNA injection) and one (Post-day 1) and two days (Post-day 2) after saline and CFA injection. See figure one legend for abbreviations. Values are the mean ± SEM for daily meal duration.
Figure 6. Naked FcγRIII siRNA reduced FcγRIII and IL-1β expression in an arthritic TMJ.
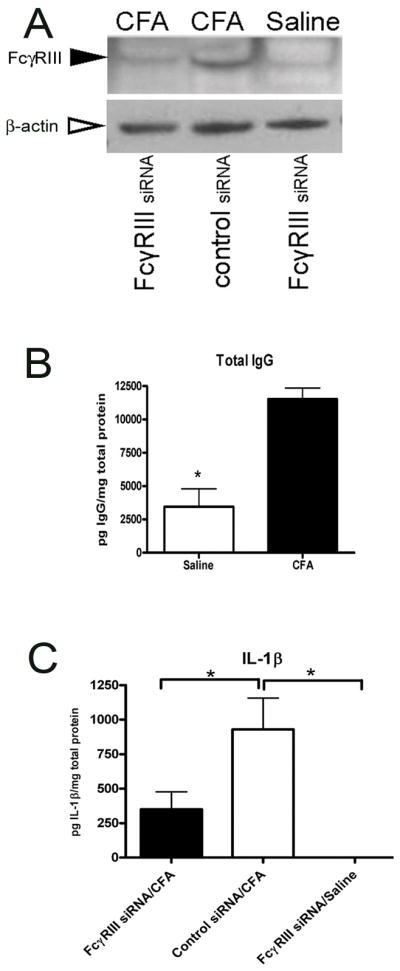
Male rats were first injected with control or FcγRIII siRNA. After 24 hours a second injection containing saline or CFA was given into the TMJ. Retrodiscal, disc and synovial TMJ tissues were collected 48 hours following saline or CFA injection. A) Image of a western blot using the TMJ tissues of the treated rats after being probed with the FcγRIII antibody. A single intense band with the correct size (~50 KDa) was detected (arrowhead). After stripping, the membrane was incubated with anti-β-actin antibody (open arrow). B) Rats were injected first with saline and then 24 hours later with saline or CFA the total IgG antibody levels were measured by ELISA. C) IL1-β, as measured by ELISA, in the TMJ tissues after administration of siRNA and CFA or saline injection. See figure one legend for abbreviations. Values are the mean ± SEM. *= P<0.05. Four animals were analyzed per treatment group.
Knockdown of FcγRIII reduced the immune response resulting from CFA injection
Injection of CFA increased the level of FcγRIII protein (Figure 6, A, middle lane) but injection of FcγRIII siRNA caused a decrease in FcγRIII protein (Figure 6, A, left lane), consistent with this result is that siRNA complexed with PEI decreased the number of FcγRIII positive cells (Fig. 3, C).
Injection of CFA significantly increased the amount of IgG in the TMJ tissue (Fig. 6, B). IgG binding to FcγRIII will activate immune cells causing release of cytokines such as IL-1β (1–4;10). CFA significantly increase the amount of IL-1β; compare the saline group (right bar, Fig 6, C) to the CFA group (middle bar, Fig. 6, C), consistent with previous results of our laboratory (29;37).
Discussion
To date, no study has reported the effect of injecting siRNA into an arthritic TMJ. In this study the arthritic response was induced by injecting CFA into the TMJ, CFA increased the amount of IgG in the TMJ tissue consistent with human TMJ arthritis (11). IgG binds the FcγRIII receptor, activating immune cells and causing release of cytokines such as IL-1β (1–4;10). Previous studies from our lab have demonstrated that IL-1β will increase as a result of IgG activating FcγRIII (38). A higher amount of IgG in the TMJ after CFA injection was expected to participate in the activation of FcγRIII positive immune cells, thus stimulating cytokine release. Consistent with this idea, IL-1β expression does increase in the TMJ after CFA injection (29;37;39). Treatment with FcγRIII siRNA would be expected to attenuate the number of receptors, decreasing the number of immune cells activated by IgG and reduce the amount of cytokine released. As expected, IL-1β levels decreased in the TMJ tissue after FcγRIII siRNA treatment. Because IL-1β participates in the arthritic nociceptive response of the TMJ (40) we suggest a reduction in IL-1β after FcγRIII siRNA injection would contribute to a decrease in nociception. Results from this study show siRNA, with homology to the FcγRIII gene, reduced the arthritic nociceptive response for two days after injection. The results also show that cleavage of the FcγRIII transcript was consistent with an RNAi mechanism. siRNA treatment reduced arthritic joint pain and inflammation suggesting this molecule has the potential as a pharmaceutical agent for the treatment of inflammatory TMJ arthritis.
A reduction in the nociceptive response was associated with a reduction in FcγRIII expression. As mentioned earlier, the number of CD14 positive cells increased in an FcγRIII siRNA/CFA treated joint, but the number of FcγRIII (CD16) positive cells decreased. Could the siRNA be reducing the nociceptive response through a pathway that does not involve FcγRIII? One alternative by which FcγRIII siRNA would reduce the nociceptive response is by binding and activating the Toll-like receptor 3 (TLR3) (41). TLR3 will bind double stranded RNA in a non-specific manner and if the nociceptive response was attenuated through a TLR3 mechanism then both FcγRIII and control siRNA should have decreased the nociceptive response. The activation of TLR3 does not appear likely because the control siRNA did not significantly modulate the nociceptive response in comparison to non-injected rats. Although, a decreasing trend in the nociceptive response was observed upon injection of control siRNA complexed with PEI. A second alterative by which the siRNA might regulate nociception, in a manner that does not involve FcγRIII, would be by regulating an off-target gene. A reduction in an off-target gene does occur by a sequence-dependent mechanism. Complementarity between the siRNA and a transcript can require as little as 11–14 base pairings within a typical 21–22 nucleotide siRNA (42;43). This base pairing results in binding and then breakdown of the off-target gene transcript through the RNAi pathway. Because the off-target mechanism requires sequence specificity the FcγRIII siRNA could reduce the nociceptive response while the control siRNA would not. We cannot exclude the possibility that FcγRIII siRNA reduced the nociceptive response by causing degradation of an off-target gene. Studies using large arrays to quantitate all the gene transcripts in the tissue would address this question. siRNA can also effect off-target protein synthesis without affecting expression at the transcript level, two examples are the TP53 (p53) and cyclin-dependent kinase inhibitor 1A (p21) gene (44). p53 and p21 are indicators of “off-target effects” that act upon the cell state. It is unclear if a change in cell status would affect the nociceptive response, but future studies will examine the levels of p53 and p21 in siRNA treated tissues. A third alternative by which siRNA can effect cellular physiology without actually degrading the target transcript is by causing cell death (45) or by inducing IFN-γ or IL-12 expression (41;46). IFN-γ and IL-12 expression can inhibit angiogenesis or activate the innate immune system, leading to stimulation of an inflammatory response (47–49). No sign of cell death was observed in tissue taken from TMJ tissue treated with siRNA. Also, the inflammatory response was not increased but decreased, as shown by a reduction in IL-1β, suggesting FcγRIII siRNA does not lead to cell death or an inflammatory response. In summary, a reduction in the nociceptive response can be due to a reduction in FcγRIII expression but further experiments are necessary to exclude siRNA modulating expression of an off-target gene that would result in the reduced nociceptive response.
Our initial assumption was that a higher amount of naked siRNA would be needed in comparison to siRNA complexed with PEI because previous work has shown PEI increases transfection efficiency (17). A higher amount of naked siRNA was injected into the rats TMJ and the tissue was harvested for western and ELISA studies. The assumption of needing more siRNA when PEI is absent appeared incorrect because inhibition of the nociceptive response was similar in rats injected with 11 μg of naked siRNA or 11 μg complexed siRNA. A simple explanation for this result would be that PEI did not improve the transfection efficiency and suggests future studies need to focus on the transfection efficiency protocol. In any event, both naked and PEI complexed siRNA decreased the nociceptive response suggesting siRNA can be used to treat diseased TMJ patients. In the clinic naked siRNA would be more desirable because PEI can be toxic (18) and may be deleterious when injected intra-articularly. It should be pointed out that our current experimental paradigm is limited because siRNA treatment was always given prior to onset of inflammation or pain. Since clinical treatment occurs after disease symptoms are diagnosed we will need to test siRNA treatment after onset of disease to assess the potential of using siRNA in a clinical setting
In conclusion, injection of FcγRIII siRNA reduced the amount of FcγRIII in the TMJ tissues and the transcript was cleaved in a manner consistent with an RNAi mechanism. Moreover, injection of FcγRIII siRNA reduced the nociceptive response of rats with an arthritic TMJ and reduced the amount of the pro-inflammatory cytokine IL-1β suggesting siRNA has the potential to be an effective treatment for this disorder.
Acknowledgments
This study was supported by R01 DE016059-01 to LLB and R01 DE015372 to PRK from the National Institute of Dental and Craniofacial Research (NIDCR) and the Office of Research on Women’s Health (ORWH).
Reference List
- 1.Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–90. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 2.Ravetch JV, Kinet JP. Fc receptors. Annu Rev Immunol. 1991;9:457–92. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 3.Raghavan M, Bjorkman PJ. Fc receptors and their interactions with immunoglobulins. Annu Rev Cell Dev Biol. 1996;12:181–220. doi: 10.1146/annurev.cellbio.12.1.181. [DOI] [PubMed] [Google Scholar]
- 4.Hulett MD, Hogarth PM. Molecular basis of Fc receptor function. Adv Immunol. 1994;57:1–127. doi: 10.1016/s0065-2776(08)60671-9. [DOI] [PubMed] [Google Scholar]
- 5.Klaassen RJ, Goldschmeding R, Tetteroo PA, Von dem Borne AE. The Fc valency of an immune complex is the decisive factor for binding to low-affinity Fc gamma receptors. Eur J Immunol. 1988;18(9):1373–7. doi: 10.1002/eji.1830180911. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams VM, Cambridge G, Lydyard PM, Edwards JC. Induction of tumor necrosis factor alpha production by adhered human monocytes: a key role for Fcgamma receptor type IIIa in rheumatoid arthritis. Arthritis Rheum. 2000;43(3):608–16. doi: 10.1002/1529-0131(200003)43:3<608::AID-ANR18>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Pope RM, Teller DC, Mannik M. The molecular basis of self-association of antibodies to IgG (rheumatoid factors) in rheumatoid arthritis. Proc Natl Acad Sci U S A. 1974;71(2):517–21. doi: 10.1073/pnas.71.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nardella FA, Teller DC, Mannik M. Studies on the antigenic determinants in the self-association of IgG rheumatoid factor. J Exp Med. 1981;154(1):112–25. doi: 10.1084/jem.154.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards JC, Cambridge G. Rheumatoid arthritis: the predictable effect of small immune complexes in which antibody is also antigen. Br J Rheumatol. 1998;37(2):126–30. doi: 10.1093/rheumatology/37.2.126. [DOI] [PubMed] [Google Scholar]
- 10.Guan G, Kerins CC, Bellinger LL, Kramer PR. Estrogenic effect on swelling and monocytic receptor expression in an arthritic temporomandibular joint model. J Steroid Biochem Mol Biol. 2005;97(3):241–50. doi: 10.1016/j.jsbmb.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Chang H, Israel H. Analysis of inflammatory mediators in temporomandibular joint synovial fluid lavage samples of symptomatic patients and asymptomatic controls. J Oral Maxillofac Surg. 2005;63(6):761–5. doi: 10.1016/j.joms.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Alstergren P. Cytokines in temporomandibular joint arthritis. Oral Dis. 2000;6(6):331–4. doi: 10.1111/j.1601-0825.2000.tb00125.x. [DOI] [PubMed] [Google Scholar]
- 13.Wadhwa S, Kapila S. TMJ disorders: future innovations in diagnostics and therapeutics. J Dent Educ. 2008;72(8):930–47. [PMC free article] [PubMed] [Google Scholar]
- 14.Hazenbos WL, Gessner JE, Hofhuis FM, Kuipers H, Meyer D, Heijnen IA, et al. Impaired IgG-dependent anaphylaxis and Arthus reaction in Fc gamma RIII (CD16) deficient mice. Immunity. 1996;5(2):181–8. doi: 10.1016/s1074-7613(00)80494-x. [DOI] [PubMed] [Google Scholar]
- 15.Bhatia A, Blades S, Cambridge G, Edwards JC. Differential distribution of Fc gamma RIIIa in normal human tissues and co-localization with DAF and fibrillin-1: implications for immunological microenvironments. Immunology. 1998;94(1):56–63. doi: 10.1046/j.1365-2567.1998.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin A, Gallou-Kabani C, Mathieu JR, Cabon F. Systemic delivery and quantification of unformulated interfering RNAs in vivo. Curr Top Med Chem. 2009;9(12):1117–29. doi: 10.2174/156802609789630820. [DOI] [PubMed] [Google Scholar]
- 17.Boussif O. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim MS, Kwon YJ. Acid-Responsive Linear Polyethylenimine for Efficient, Specific, and Biocompatible siRNA Delivery. Bioconjug Chem. 2009 doi: 10.1021/bc800436v. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301(5641):1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 20.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117(1):83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 21.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116(6):831–41. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 22.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 23.Mello CC, Conte DJ. Revealing the world of RNA interference. Nature. 2004;431:338–42. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 24.Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119(3):661–73. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerins C, Carlson D, McIntosh J, Bellinger L. A role for cyclooxygenase II inhibitors in modulating temporomandibular joint inflammation from a meal pattern analysis perspective. J Oral Maxillofac Surg. 2004;62(8):989–95. doi: 10.1016/j.joms.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Kramer PR, Bellinger LL. The effects of cycling levels of 17beta-estradiol and progesterone on the magnitude of temporomandibular joint-induced nociception. Endocrinology. 2009;150(8):3680–9. doi: 10.1210/en.2008-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thut PD, Hermanstyne TO, Flake NM, Gold MS. An operant conditioning model to assess changes in feeding behavior associated with temporomandibular joint inflammation in the rat. J Orofac Pain. 2007;21(1):7–18. [PubMed] [Google Scholar]
- 28.Kerins CA, Carlson DS, Hinton RJ, Grogan DM, Marr K, Kramer PR, et al. Specificity of meal pattern analysis as an animal model of dermining temporomandibular joint inflammation/pain. International Journal of Oral Maxiollofacial Surgery. 2005;34:425–31. doi: 10.1016/j.ijom.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Kerins CA, Carlson DS, McIntosh JE, Bellinger LL. Meal pattern changes associated with temporomandibular joint inflammation/pain in rats; analgesic effects. Pharmacol Biochem Behav. 2003;75(1):181–9. doi: 10.1016/s0091-3057(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 30.Hutchins B, Spears R, Hinton RJ, Harper RP. Calcitonin gene-related peptide and substance P immunoreactivity in rat trigeminal ganglia and brainstem following adjuvant-induced inflammation of the temporomandibular joint. Arch Oral Biol. 2000;45(4):335–45. doi: 10.1016/s0003-9969(99)00129-6. [DOI] [PubMed] [Google Scholar]
- 31.Harper RP, Kerins CA, McIntosh JE, Spears R, Bellinger LL. Modulation of the inflammatory response in the rat TMJ with increasing doses of complete Freund’s adjuvant. Osteoarthritis Cartilage. 2001;9(7):619–24. doi: 10.1053/joca.2001.0461. [DOI] [PubMed] [Google Scholar]
- 32.Harper RP, Kerins CA, Talwar R, Spears R, Hutchins B, Carlson DS, et al. Meal pattern analysis in response to temporomandibular joint inflammation in the rat. J Dent Res. 2000;79(9):1704–11. doi: 10.1177/00220345000790091101. [DOI] [PubMed] [Google Scholar]
- 33.Kerins CA, Spears R, Bellinger LL, Hutchins B. The prospective use of COX-2 inhibitors for the treatment of temporomandibular joint inflammatory disorders. Int J Immunopathol Pharmacol. 2003;16(2 Suppl):1–9. [PubMed] [Google Scholar]
- 34.Bellinger LL, Fabia R, Husberg BS. Meal patterns prior to and following liver transplantation in rats. Physiol Behav. 1997;62(3):525–9. doi: 10.1016/s0031-9384(97)80329-0. [DOI] [PubMed] [Google Scholar]
- 35.Kramer PR, Kerins CA, Schneiderman E, Bellinger LL. Measuring persistent temporomandibular joint nociception in rats and two mice strains. Physiol Behav. 2010;99(5):669–78. doi: 10.1016/j.physbeh.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castonguay TW, Kaiser LL, Stern JS. Meal pattern analysis: artifacts, assumptions and implications. Brain Res Bull. 1986;17(3):439–43. doi: 10.1016/0361-9230(86)90252-2. [DOI] [PubMed] [Google Scholar]
- 37.Kerins C, Carlson D, McIntosh J, Bellinger L. A role for cyclooxygenase II inhibitors in modulating temporomandibular joint inflammation from a meal pattern analysis perspective. J Oral Maxillofac Surg. 2004;62(8):989–95. doi: 10.1016/j.joms.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Kramer PR, Kramer SF, Guan G. 17 beta-estradiol regulates cytokine release through modulation of CD16 expression in monocytes and monocyte-derived macrophages. Arthritis Rheum. 2004;50(6):1967–75. doi: 10.1002/art.20309. [DOI] [PubMed] [Google Scholar]
- 39.Kramer PR, Winger V, Kramer SF. 17beta-Estradiol utilizes the estrogen receptor to regulate CD16 expression in monocytes. Mol Cell Endocrinol. 2007;279(1–2):16–25. doi: 10.1016/j.mce.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai YC, Shaftel SS, Miller JN, Tallents RH, Chang Y, Pinkert CA, et al. Intraarticular induction of interleukin-1beta expression in the adult mouse, with resultant temporomandibular joint pathologic changes, dysfunction, and pain. Arthritis Rheum. 2006;54(4):1184–97. doi: 10.1002/art.21771. [DOI] [PubMed] [Google Scholar]
- 41.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452(7187):591–7. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 43.Jackson AL. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 44.Scacheri PC. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1892–7. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao X, Tang S, Thrasher JB, Griebling TL, Li B. Small-interfering RNA-induced androgen receptor silencing leads to apoptotic cell death in prostate cancer. Mol Cancer Ther. 2005;4(4):505–15. doi: 10.1158/1535-7163.MCT-04-0313. [DOI] [PubMed] [Google Scholar]
- 46.Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, et al. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12(6):988–93. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, Maclachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–62. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 48.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–90. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–9. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]