Abstract
Mutation of the breast cancer susceptibility gene, BRCA2, leads to breast and ovarian cancers. Mechanistic insight into the functions of human BRCA2 has been limited by the difficulty of isolating this large protein (3,418 amino acids). Here we report purification of full length BRCA2 and show that it both binds RAD51 and potentiates recombinational DNA repair by promoting assembly of RAD51 onto single-stranded DNA (ssDNA). BRCA2 acts by: targeting RAD51 to ssDNA over double-stranded DNA; enabling RAD51 to displace Replication protein-A (RPA) from ssDNA; and stabilizing RAD51-ssDNA filaments by blocking ATP hydrolysis. BRCA2 does not anneal ssDNA complexed with RPA, implying it does not directly function in repair processes that involve ssDNA annealing. Our findings show that BRCA2 is a key mediator of homologous recombination, and they provide a molecular basis for understanding how this DNA repair process is disrupted by BRCA2 mutations, which lead to chromosomal instability and cancer.
One of the proposed driving forces behind the tumorigenic process is the onset of genomic instability that, when coupled to repeated rounds of cell division, promotes oncogenesis1. A hallmark of human and mouse cells that are mutant for BRCA2 is severe chromosomal instability marked by an accumulation of chromosomal breaks, translocations, exchanges, and other abnormal structures2. Accordingly, germline mutations in BRCA2 are associated with a highly penetrant incidence of breast and/or ovarian cancer as well as tumors in other tissues and organs3,4. BRCA2 possesses eight highly conserved repeated sequences, termed the BRC repeats, and a carboxy-terminal region that were shown to bind RAD515–7. RAD51 plays a central role in recombination, assembling onto single-stranded DNA (ssDNA) as a nucleoprotein filament, and catalyzing the invasion and exchange of homologous DNA sequences8,9.
At the cellular level, loss of BRCA2 function results in sensitivity to cross-linking agents, a decrease in homology-directed repair of double-stranded DNA breaks (DSB’s), and defects in replication and checkpoint control2,10–12. BRCA2 is also required for RAD51-induced focus formation after exposure to DNA damaging agents13,14. Prior studies utilizing fragments of BRCA2, fusions of the BRC repeats with the DNA-binding domain (DBD) of BRCA2, and analysis of the Ustilago maydis and Caenorhabditis elegans orthologues, Brh2 and BRC-2 respectively, have provided a framework for understanding the mediator role that BRCA2 plays in DSB repair by RAD51-driven homologous recombination15–23 (see Supplementary Fig. 1 for a model based on this report and the work referenced above). However, the large size of human BRCA2 (3,418 amino acids), difficulty in driving high level expression, insufficient solubility, and its propensity to degrade, have precluded isolation of the full length BRCA2 protein and have hampered fuller understanding of its functions. Here, we describe purification of the full-length protein from human cells, and report its biochemical functions with regard to recombinational DNA repair.
Purified BRCA2 binds RAD51 and DMC1
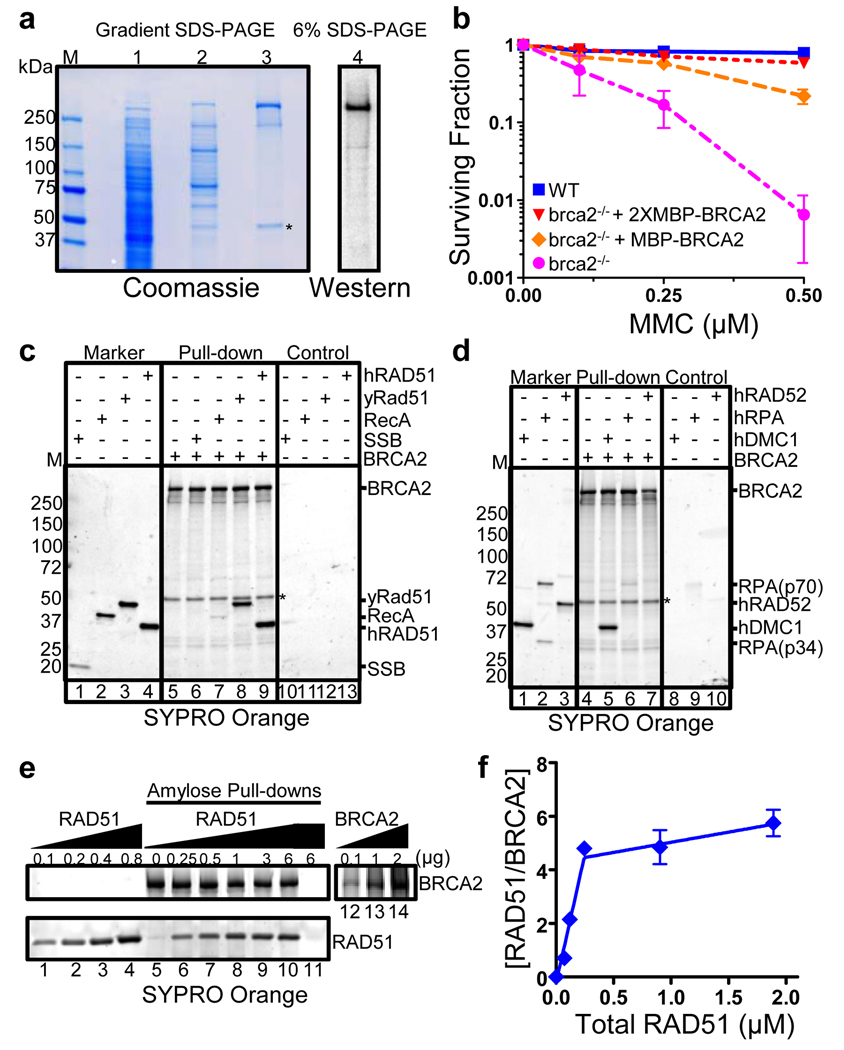
By utilizing a mammalian expression vector (phCMV1) driven by a CMV promoter and by adding two tandem repeats of the Maltose Binding Protein (designated 2XMBP) to the N-terminus of human BRCA2 (470 kDa including the two MBP tags), we expressed significant amounts of protein that could be purified to near homogeneity (Fig. 1a and Supplementary Fig. 2). The identity of full length BRCA2 was confirmed by western blotting using an antibody to the C-terminal region of BRCA2 (Fig. 1a, lane 4 and Supplementary Fig. 2b, c), an antibody to the N-terminal MBP tag, and by mass spectrometric analysis. Mass spectrometric analysis of a variable minor band directly below the full length protein confirmed the presence of a truncated BRCA2 species lacking the carboxy terminus. The band near the 50 kDa marker (Fig. 1a, asterisk) was confirmed by mass spectrometry to be β-tubulin. The presence of this contaminant appears not to interfere with any of our in vitro studies. We found that 2XMBP-BRCA2 fully complemented brca2 mutant (VC8) cells (Fig. 1b, Supplementary Fig. 3); therefore, the tag was not removed for the studies reported here. Hereafter, we refer to the N-terminal 2XMBP-tagged version of full length BRCA2 as BRCA2.
Figure 1. Protein interactions with purified full-length human BRCA2.
(a) Lane 1) 293T cell lysate, 2) amylose eluate, 3) purified BRCA2, 4) Western blot (Ab-2). “M”, standards. Asterisk, β-tubulin. (b) MMC survival. Wild type V79 ( ), brca2−/− + MBP-BRCA2 (
), brca2−/− + MBP-BRCA2 ( ), 2XMBP-BRCA2 (
), 2XMBP-BRCA2 ( ), and brca2−/− VC8 (
), and brca2−/− VC8 ( ) cells. Errors bars, s.d. (n=3). (c) and (d) Protein pull-downs. 2XMBP-BRCA2 with indicated proteins. “Marker”, protein input. ”Control”, proteins + amylose resin. (e) RAD51 titration of BRCA2: Lanes 1–4, RAD51 standards; 5–11, Pull-downs; 5, BRCA2 alone; 11, RAD51 alone; 12–14, BRCA2 standards. (f) Data from (e) fit to segmental linear regression. Errors bars, s.d. (n=2).
) cells. Errors bars, s.d. (n=3). (c) and (d) Protein pull-downs. 2XMBP-BRCA2 with indicated proteins. “Marker”, protein input. ”Control”, proteins + amylose resin. (e) RAD51 titration of BRCA2: Lanes 1–4, RAD51 standards; 5–11, Pull-downs; 5, BRCA2 alone; 11, RAD51 alone; 12–14, BRCA2 standards. (f) Data from (e) fit to segmental linear regression. Errors bars, s.d. (n=2).
Our initial criteria for determining that purified BRCA2 was properly folded and retained biochemical function was to test its ability to bind recombination proteins that were previously reported to interact. We incubated BRCA2 with several purified candidate proteins and used the MBP tag to capture the complexes on amylose beads, and analyze the complexes on SDS-PAGE gels stained with SyproOrange, (Fig. 1c, d). As expected from in vivo pull-down assays and interaction with a fusion protein construct5,15,24–26, human RAD51 bound to BRCA2 (Fig. 1c, lane 9). Also in agreement27, BRCA2 bound DMC1 (Fig. 1d, lane 5), the meiotic counterpart of RAD51. BRCA2 bound to yeast Rad51 (Fig. 1c, lane 8) but, given the high homology between the two orthologues (~67% identical; 83% homologous 28,29), this is not surprising. BRCA2 did not appreciably bind the E. coli homolog, RecA, (Fig. 1c, lane 7) showing that interaction did not extend to the evolutionary distant bacterial protein. We also did not detect any significant interaction with human RPA (Fig. 1d, lane 6), despite a report in the literature30, E. coli SSB (Fig. 1c, lane 6), or human RAD52 (Fig. 1d, lane 7).
The BRC repeat defines a RAD51 binding motif whose number varies widely amongst different organisms ranging from one BRC repeat in U. maydis to 15 repeats in Trypanosoma brucei31. Human BRCA2 contains eight BRC repeats. Various studies established that most of the human BRC repeats in isolation can bind RAD515,24–26; however, it remained unclear how many binding sites are occupied within the context of the full-length protein and whether the interaction affinities were comparable. Using known concentrations of purified RAD51 (Fig. 1e, lanes 1–4) and recombinant BRCA2 (Fig. 1e, lanes 12–14) as standards, and staining with SyproOrange, we quantified the binding; in the absence of BRCA2, RAD51 did not bind non-specifically to the amylose resin (Fig. 1e, lane 11). In the presence of BRCA2 (Fig. 1e, lanes 5–10), the amount of RAD51 bound increased linearly with concentration (indicative of tight binding in the nM range) until about 4.5 (± 0.9) RAD51 molecules were bound per BRCA2 (Fig. 1f); afterward, a weaker binding (in the µM range) was evident. At the maximum RAD51 concentration attainable, approximately 6 RAD51 proteins were bound to each BRCA2. The binding of RAD51 to BRCA2 was also examined under buffer conditions identical to those used for DNA strand exchange assays, and similar binding characteristics were found (Supplementary Fig. 4). The protein complexes formed between BRCA2 and RAD51 were not dependent on magnesium or calcium ions, nucleotide cofactors, or the presence of DNA (data not shown).
BRCA2 prefers to bind ssDNA over dsDNA
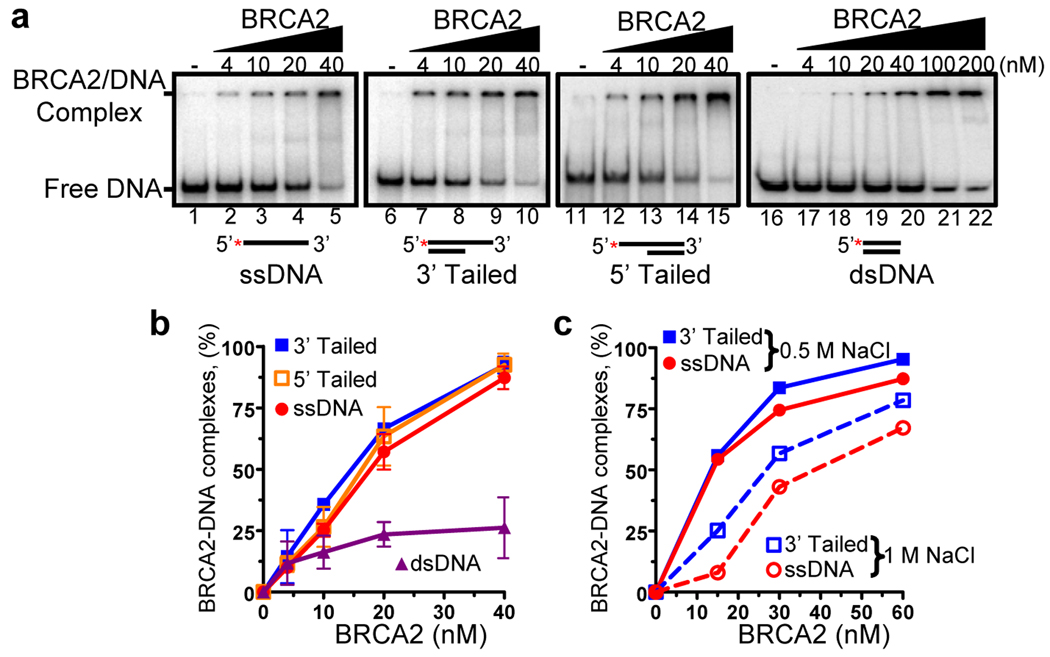
The DNA binding domain of BRCA2 contains both Oligonucleotide-Binding (OB) folds and a tower domain, which engender BRCA2 with potential sites for binding both ssDNA and dsDNA16. Indeed, both the carboxy-terminus of BRCA2 and a fusion protein containing BRC repeats 3 and 4 linked to the DNA binding domain of BRCA2 bind both ssDNA and dsDNA15,16. By using electrophoretic mobility shift assays (EMSA), we tested the ability of BRCA2 to bind ssDNA, dsDNA, and dsDNA with an ssDNA tail (3’ tailed or 5’ tailed). BRCA2 bound to all of these substrates; however, those containing ssDNA were strongly preferred over dsDNA (Fig. 2a, b). These results are consistent with previous reports on the DNA binding domain of BRCA215. A slight preference for tailed DNA over ssDNA was revealed at higher salt concentrations (Fig. 2c); however, the difference was modest. The binding specificity for the various DNA substrates was unaltered when BRCA2 was incubated with RAD51 prior to binding to the DNA (Supplementary Fig. 5).
Figure 2. BRCA2 displays a strong preference for binding tailed and ssDNA substrates over dsDNA.
(a) EMSA. BRCA2 binding: ssDNA, 3’ Tailed DNA, 5’ Tailed DNA, and dsDNA. (b) Quantification: 3’ Tailed DNA ( ), 5’ Tailed DNA (
), 5’ Tailed DNA ( ), ssDNA (
), ssDNA ( ), and dsDNA (
), and dsDNA ( ). (c) EMSA in 0.5 M or 1 M NaCl: 3’ tailed (squares) or ssDNA (circles). Error bars, s.d. (n=3).
). (c) EMSA in 0.5 M or 1 M NaCl: 3’ tailed (squares) or ssDNA (circles). Error bars, s.d. (n=3).
BRCA2 stimulates DNA strand exchange by RAD51
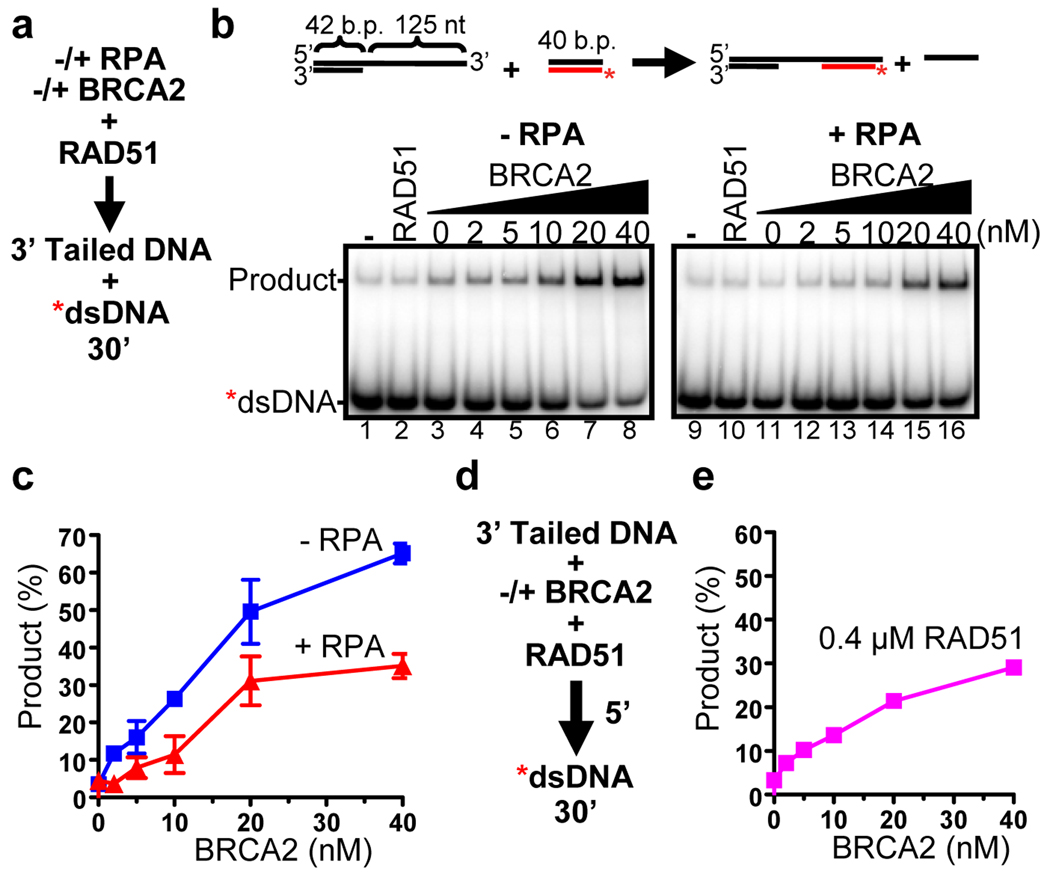
An essential function of RAD51 in recombinational DNA repair is its capacity to homologously pair and exchange DNA strands. To promote this process, RAD51 must assemble onto the 3’ ssDNA tails generated by resection of DNA breaks. To mimic the DNA intermediate generated after DSB resection in vivo, we utilized a tailed DNA substrate created by annealing a 42-mer oligonucleotide to a 167-mer to create a 42 base pair (bp) dsDNA region followed by a 125 nucleotide 3’ ssDNA overhang (Fig. 3b; termed 3’ tailed DNA). To validate the DNA substrate, we conducted DNA strand exchange assays as a function of RAD51 protein concentration using an optimized in vitro DNA strand exchange protocol (Supplementary Fig. 6a, c); optimal product formation was at a 1:3 (RAD51: nucleotide) ratio, consistent with the DNA binding stoichiometry of RAD5132. However, in vivo, filament assembly conditions are not optimal: RAD51 must compete with RPA for binding to the ssDNA33,34. Furthermore, RAD51 can bind to both ssDNA and dsDNA, but the binding to dsDNA is not productive and, in fact, blocks DNA strand exchange9,34. Thus, DNA strand exchange can be stimulated in at least two mechanistically distinct ways.
Figure 3. BRCA2 stimulates DNA strand exchange promoted by RAD51.
(a) DNA strand exchange reaction protocol for (b) and (c). (b) Reaction in the absence (left) or presence (right) of RPA. (c) Quantification: absence ( ); presence (
); presence (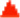 ) of RPA. Error bars, s.d. (n=3). (d) DNA strand exchange protocol for (e) in absence of RPA. (e) Quantification of (d).
) of RPA. Error bars, s.d. (n=3). (d) DNA strand exchange protocol for (e) in absence of RPA. (e) Quantification of (d).
Initially, to determine whether BRCA2 affects DNA strand exchange, reactions were performed by using an optimal amount of RAD51 but introducing it concurrently to a mixture of 3’ tailed DNA and dsDNA. Indeed, as demonstrated in Figures 3b and c, when RAD51 is permitted to assemble on both ssDNA and dsDNA, DNA strand exchange is reduced to background levels (Fig. 3b, lanes 2 & 10). However, if BRCA2 is incubated with RAD51 prior to mixing with DNA substrates, this inhibition is alleviated in a concentration-dependent manner (Fig. 3b, lanes 3–8; Fig 3c), suggesting that BRCA2 directs RAD51 to ssDNA or limits binding to dsDNA, or both. In the presence of RPA, stimulation by BRCA2 is maintained, although the magnitude is reduced (Fig. 3b, lanes 11–16; Fig. 3c). To confirm that the product did not result from “melting” of the donor duplex DNA and spontaneous annealing during the de-proteinization step35, we performed the same reaction with 10-fold excess unlabeled oligonucleotide complementary to the labeled pairing strand in the stop-mix, and the results were unchanged (Supplementary Fig. 7a, lane 9). The stimulation was ATP-dependent (Supplementary Fig. 7a, lanes 11 & 12) and did not occur with a heterologous template (Supplementary Fig. 7a, lane 10); furthermore, BRCA2 alone was unable to promote DNA strand exchange (Supplementary Fig. 7a, lane 3). These results support a role for BRCA2 in targeting RAD51 to ssDNA, limiting assembly onto the dsDNA partner, or both.
BRCA2 limits assembly of RAD51 on dsDNA
To determine whether BRCA2 slowed or prevented assembly on the dsDNA partner of DNA strand exchange, reactions were performed using a concentration of RAD51 (0.4 µM) sufficient to saturate both the ssDNA and dsDNA present. At such a concentration, DNA strand exchange is inhibited due to binding of excess RAD51 to the dsDNA target32,34 (see Supplementary Fig. 6a, lane 5 and Supplementary Fig. 6c). To optimize filament formation on the ssDNA, BRCA2 and RAD51 were incubated with the 3’ tailed ssDNA first, and then the dsDNA was added to initiate the reaction (Fig. 3d); however, the excess free RAD51 binds the dsDNA partner and inhibits the reaction (Supplementary Fig. 8b, lane 1). To eliminate complications from competition with RPA, these reactions were done in the absence of RPA. BRCA2 stimulated DNA strand exchange in a concentration dependent manner (Fig. 3e and Supplementary Fig. 8, lanes 2–6). To provide more direct evidence that BRCA2 prevents nucleation onto the dsDNA and targets RAD51 to the 3’ Tailed DNA, we preincubated BRCA2 and RAD51 and then analyzed RAD51-DNA complex formation by either EMSA or the partitioning of RAD51 onto biotinylated 3’ Tailed DNA in the presence of excess dsDNA (Supplementary Fig. 9); BRCA2 prevented binding of RAD51 to dsDNA (Supplementary Fig. 9b) and favored RAD51 binding to the ssDNA (Supplementary Fig. 9f). Taken together with the results of the previous section, these data support the idea that BRCA2 recruits RAD51 to ssDNA, likely by virtue of its affinity for ssDNA, and inhibits assembly of RAD51 onto dsDNA.
BRCA2 recruits RAD51 onto ssDNA complexed with RPA
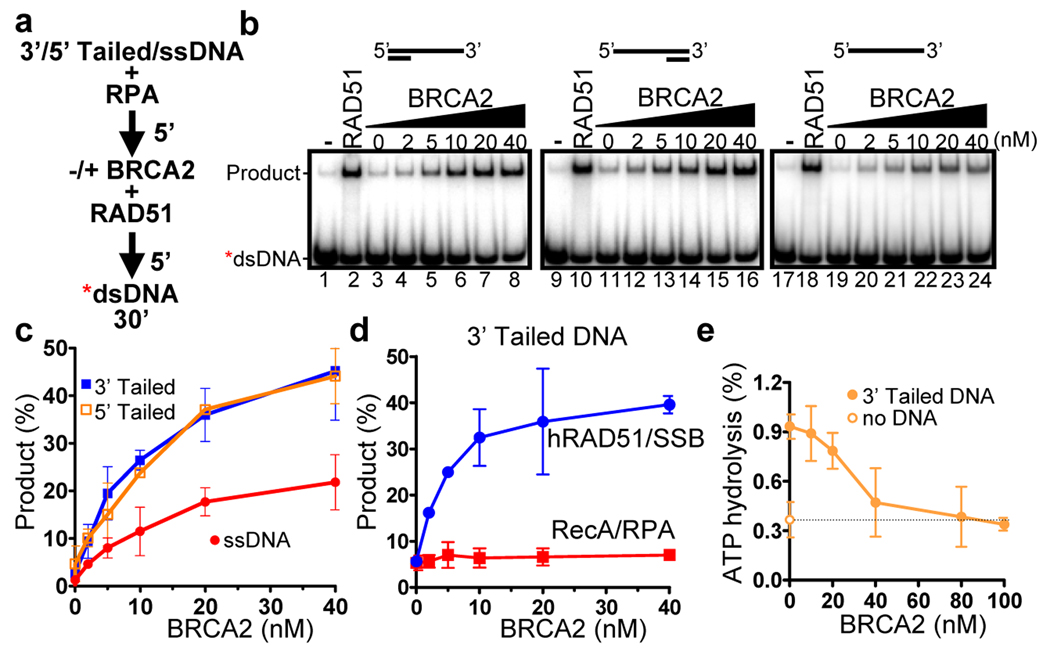
Prior work utilizing a human BRCA2 polypeptide fusion and U. maydis Brh2 demonstrated the ability of those proteins to promote RAD51 filament formation onto RPA-coated ssDNA15,17. To determine whether the full-length human BRCA2 protein provides a similar avenue for stimulation, we next performed DNA strand exchange assays using an optimal amount of RAD51 but, rather than permitting filament formation on naked ssDNA, the ssDNA was first complexed with RPA (Fig. 4a). RAD51 was subsequently introduced in the presence or absence of BRCA2, and finally, the labeled duplex DNA was added to start the reaction. As expected, incubation of the ssDNA with increasing concentrations of RPA prior to addition of RAD51 severely impaired DNA strand exchange (Supplementary Fig. 6b, lanes 4–7). As shown in Figure 4b & c, increasing amounts of BRCA2 stimulated DNA strand exchange as much as 20-fold, suggesting that BRCA2 accelerates formation of the RAD51 nucleoprotein filament at the presynaptic stage of recombination and alleviates the inhibition posed by RPA. Stimulation by BRCA2 occurred at concentrations as low as 2 nM (Fig. 4b, c), and that were sub-stoichiometric relative to the RAD51 concentrations (approximately 100-fold less than RAD51).
Figure 4. BRCA2 stimulates RAD51-mediated DNA strand exchange by promoting stable RAD51-ssDNA filament formation, overcoming inhibition by RPA.
(a) DNA strand exchange reaction protocol for (b) – (c). (b) Autoradiograms: 3’ Tailed, 5’ Tailed, and ssDNA substrates. (c) Quantification of (b). (d) Assays as in (a), except RecA replaced RAD51 (red squares) and SSB replaced RPA (blue circles). (e) Inhibition of RAD51 ssDNA-dependent ATP hydrolysis by BRCA2. Error bars, s.d. (n=3).
To confirm a presynaptic role for BRCA2, we also performed a kinetic analyses of the DNA strand exchange reactions by varying the time that BRCA2 was incubated with RAD51 and the RPA-ssDNA complex, either before (Supplementary Fig. 10a–c) or after (Supplementary Fig. 10d–f) addition of homologous dsDNA. BRCA2 imparts a significant stimulation (~20-fold) of DNA strand exchange in as little as one minute after incubation with RAD51 and the RPA-ssDNA complex (Supplementary Fig. 10c); only after 60 minutes does the yield without BRCA2 match that seen with BRCA2 at 1 minute.
In contrast to U. maydis Brh217 and the E. coli analog, RecFOR36, which show a strict specificity to act at a 3’ ssDNA overhang, stimulation of DNA strand exchange by human BRCA2 was the same for 3’ versus 5’ tailed DNA (Fig. 4b, compare lanes 3–8 and 11–16, and 4c). This result was consistent with our observation above (Fig. 2a & b) that the binding affinity of BRCA2 for 3’ tailed DNA and 5’ tailed DNA is the same. A bias was also not apparent at sub-stoichiometric concentrations of RAD51 (data not shown) where reduced filament occupancy revealed junction specificity for Brh217. However, we did observe a consistent 2-fold preference for both 3’ and 5’ tailed DNA substrates over ssDNA, indicating that stimulation by BRCA2 is modestly greater for a DNA substrate containing a junction of ssDNA with dsDNA (Fig. 4c), also consistent with our binding results. These findings demonstrate that BRCA2 possesses the capacity to broadly stimulate RAD51 assembly onto ssDNA with or without a dsDNA junction, and to uniquely promote the assembly onto either 3’ or 5’ ssDNA tails.
BRCA2 did not stimulate RecA (Fig. 4d, and Supplementary Fig. 11b, left panel), consistent with the failure to pull-down RecA. When RPA was replaced by SSB, BRCA2 could still stimulate DNA strand exchange by RAD51 (Fig. 4d, and Supplementary Fig. 11b, right panel), implying that neither BRCA2 nor RAD51 need to interact directly with the ssDNA binding proteins,. This idea is further bolstered by lack of interaction between BRCA2 and either SSB or RPA in the pull-down assays (Fig. 1c, d). This finding is consistent with the behavior of a human BRCA2 polypeptide fusion15 but distinct from that of the bacterial RecFOR and fungal Brh2, which require their cognate ssDNA-binding proteins for stimulation17,36. Thus, it appears that direct interactions between human BRCA2, RAD51, and DNA are sufficient to stimulate the ability of RAD51 to gain access to the RPA- or SSB-coated ssDNA and to then displace them as the ensuing nucleoprotein filament is formed and extended.
Our previous work on the BRC repeats demonstrated that they stabilize ssDNA-RAD51 complexes by blocking the ATPase activity of RAD5121. To gain insight into the mechanism by which BRCA2 stimulates presynaptic complex formation, we measured its effect on the ATPase activity of RAD51. BRCA2 inhibited the ssDNA-dependent ATPase activity of RAD51 in a concentration dependent manner to the level seen in the absence of DNA (Fig. 4e). These results suggest that the same mechanism for RAD51 nucleoprotein filament stabilization ascribed to the BRC repeats applies to BRCA221: namely, full length BRCA2 stabilizes the RAD51 bound to the ssDNA substrate by down-regulating its ATPase activity, an activity which is used to inactivate and turnover the RAD51 protein; however, quantitatively, this control is achieved at much lower concentrations of BRCA2 than the BRC repeats.
BRCA2 cannot anneal RPA-ssDNA complexes
The capability of BRCA2 to accelerate displacement of RPA by RAD51 without the need for a ssDNA/dsDNA junction has some parallels to yeast Rad52 (yRad52)37–39, but human RAD52 lacks this ability to stimulate RPA replacement40. However, because yeast lack a known BRCA2 homologue whereas mammals possess both BRCA2 and RAD52, it is possible that evolutionary changes separated the functions of yRad52 into several mammalian proteins. Also another important role of the multi-functional yRad52, and bacterial RecO, is the annealing of complementary ssDNA that is bound by the cognate ssDNA-binding protein41,42; both the U. maydis and C. elegans BRCA2 orthologues can anneal ssDNA in the presence of RPA23,43. Consequently, we investigated whether BRCA2 or human RAD52 possess a similar capacity. Complementary ssDNA substrates, with or without saturating human RPA, were incubated with BRCA2 or RAD52 and then mixed (Fig. 5a). Figure 5b shows that in the absence of proteins, spontaneous annealing occurred over time (lanes 2–5; quantification in Fig. 5c). BRCA2 marginally increased (lanes 6–9), and RAD52 clearly increased the rate of annealing (lanes 10–13). When RPA was added (lanes 15–18), spontaneous annealing was completely blocked. Unlike the U. maydis and C. elegans orthologues23,43, human BRCA2 was unable to overcome this inhibition (lanes 19–22), but RAD52 readily annealed the RPA-ssDNA complexes (lanes 23–26). In contrast, when substituted for BRCA2, neither human RAD52 nor yeast Rad52 stimulated DNA strand exchange by RAD51 when the ssDNA was complexed with RPA (Supplementary Fig. 12). Taken together, these data show that BRCA2 and human RAD52 have assumed divergent roles in mammalian cells. BRCA2 has taken on the functions that stimulate joint molecule formation and DNA strand exchange, whereas RAD52 provides the ssDNA annealing functions of recombination44.
Figure 5. BRCA2 does not anneal ssDNA complexed with RPA.
(a) Schematic of DNA annealing assays. (b) Autoradiogram: absence of RPA, plus indicated protein (left); presence of RPA first, plus indicated protein (right). Lanes 1 and 14, radio-labeled 40-mer. (c) Quantification of (b): No Protein (grey); BRCA2 (blue); RAD52 (green); RPA (orange); RPA plus BRCA2 (yellow); RPA plus RAD52 (purple). Error bars, s.d. (n=3).
Discussion
Our results reveal the biochemical functions of full length human BRCA2, and they establish that BRCA2 augments the functions of RAD51 that are essential for recombinational repair of DNA breaks (Supplementary Fig. 1). Stimulation by BRCA2 is a consequence of several mutually reinforcing effects; BRCA2: 1) enforces binding of RAD51 to ssDNA; 2) accelerates the rate of RPA-displacement from ssDNA by RAD51; 3) inhibits the ATPase activity of RAD51; and 4) limits binding to dsDNA. BRCA2 focuses the assembly of RAD51 onto ssDNA and facilitates the RAD51-mediated displacement of RPA from ssDNA, which is a key regulatory step of DNA pairing. In support of this general concept, the promotion of RAD51 filament formation onto RPA-coated ssDNA was also demonstrated by Liu et al. (submitted) using a different full-length BRCA2 protein expression construct and preparation. By enabling formation of the presynaptic complex, BRCA2 permits progression to the subsequent DNA pairing phase of recombinational DNA repair. Furthermore, by inhibiting the ssDNA-dependent ATP hydrolysis of RAD51, BRCA2 preserves the active and most stable form of RAD51, the ATP-RAD51-ssDNA complex21,22. Because the rate-limiting step in RAD51 nucleoprotein filament assembly is nucleation of the first several monomers of the filament45–47, BRCA2 can act catalytically to stabilize a nucleus by blocking RAD51 self-inactivation and dissociation via its ATPase activity. If the RAD51 molecules bound to BRCA2 do indeed comprise the nucleus, then BRCA2 can stabilize a nascent filament of up to 4–6 RAD51 molecules. Inhibition of ATPase activity was also observed for the C. elegans proteins (RAD51 and BRC-2)23, but not for the U. maydis orthologues (Rad51 and Brh2)17 or the E. coli analogues (RecA and RecFOR)36, suggesting that this mechanism of stimulating RAD51 function is a late adaption of multicellular organisms. In addition, our results show that BRCA2 prevents or slows the assembly of RAD51 onto duplex DNA, an aspect of RAD51 filament assembly that impairs recombination reactions. Based on previous single-molecule studies with the BRC repeats21, we propose that interaction with full-length BRCA2 slows nucleation of RAD51 onto dsDNA.
An unanticipated feature of human BRCA2 is its ability to bind and stimulate RAD51-mediated DNA strand exchange at regions of ssDNA as well as at ssDNA/dsDNA junctions of either polarity. This capability permits the BRCA2-facilitated loading of RAD51 in both the 5’→3’ and 3’→5’ directions. Although most models for BRCA2 function have focused on DSB repair, we note that these characteristics are also consistent and suggestive of a role for BRCA2 in the recombinational repair of DNA gaps that occur during DNA replication due to damage in the template (Supplementary Fig. 1b). Given that BRCA2 facilitates growth of the filament in either polarity from internal ssDNA regions or from either junction, BRCA2 could readily contribute to daughter strand DNA gap repair as well as DSB repair. It is notable that the lack of polarity in human BRCA2 function differs from analogs such as RecFOR and Brh2, which load RecA and U. maydis Rad51, respectively, specifically onto the 3’ overhanging ssDNA17,36. The absence of a bias for pairing 3’-ends by human RAD51 may reflect its underlying intrinsic capability to assemble in both directions48. But because resection of a DSB in vivo produces 3’ tailed ssDNA, there is no compelling need for polarity enforcement by BRCA2.
Another key difference between the BRCA2 orthologues of U. maydis and C. elegans 23,43 is that human BRCA2 is unable to anneal ssDNA complexed with RPA, the physiological intermediate of recombination. However, human RAD52 does manifest this capability, implying that in mammalian cells this function is assumed by RAD52. Yet RAD52 cannot stimulate the assembly of RAD51 onto ssDNA complexed with RPA40 (Supplementary Fig. 12), but BRCA2 can. This behavior of RAD52 is distinct from yRad52 which displays both of these species-specific capabilities37–39,41, explaining the essential role of yRad52 in all recombinational DNA repair processes. Thus, in vertebrates, these functions have separated: BRCA2 targets RAD51 to ssDNA to mediate DNA strand invasion into a duplex donor to produce joint molecules by a distinctive mechanism, whereas RAD52 anneals RPA-ssDNA complexes in steps or pathways of recombinational repair that could include second-end capture in DSB repair, single-strand annealing (SSA), and synthesis-dependent strand annealing. In support of this idea, error prone repair by SSA is increased in cells lacking functional BRCA2, whereas RAD52 complementation in RAD52−/− mouse ES cells augments the SSA pathway44,49. Both in vivo and in vitro studies clarified the role that BRCA2 plays in catalyzing the delivery of RAD51 to sites of DNA damage. Our work has confirmed and extended prior expectations that the intact human BRCA2 protein mediates the rapid and ordered assembly of the RAD51 protein onto ssDNA, and helps to explain why cells lacking functional BRCA2 would be severely impaired for formation of this critical intermediate in recombinational repair. As a consequence of BRCA2 loss, DNA break repair mediated through template-directed repair from homologous sequences within an intact homolog or sister chromosome would be disrupted, leading to error prone repair and potential chromosomal instability. The ability to now purify full length human BRCA2, a protein directly responsible for genetically predisposing individuals to substantially high risks for cancer, should open a whole new venue for understanding this very large and complex protein.
METHODS SUMMARY
Expression and Purification of full length BRCA2
Human BRCA2 cDNA was cloned into phCMV1 with two repeats of (MBP) at the N-terminus. 293TD cells were transfected and harvested 31 hours post-transfection. Extracts were batch bound to Amylose resin. The protein was eluted with 10 mM maltose, loaded onto HiTrap Q, and step eluted at 450 mM NaCl.
DNA Substrates
Oligonucleotides were PAGE purified. The 3’ tailed, 5’ tailed, and dsDNA were annealed at 1:1 molar ratio, and 32P-labeled at the 5’-end.
DNA Strand Exchange Assays
Unless otherwise indicated, reactions were at 37 °C for 30 minutes with RAD51 (0.22 µM); RPA (0.1 µM); 3’ tailed, 5’ tailed, or ssDNA (4 nM molecules); and dsDNA (4 nM molecules). Reactions were terminated with Proteinase K-SDS, and analyzed by electrophoresis (6% polyacrylamide) and phosphor-imaging.
Supplementary Material
Acknowledgements
We thank Dave Shin for BRCA2 cDNA clones; Malgorzata Zdzienicka for VC8 cells; Alex Mazin for RAD52; Amitabh Nimonkar for DMC1; Wolf Heyer and Kowalczykowski laboratory for comments. Supported by grants from NIH (NIH GM41347) and DOD-Breast Cancer Research Program (BC085223) to S.C.K., American Cancer Society Postdoctoral Fellowship to R.B.J. (PF-05-225-01-GMC), and Postdoctoral Fellowship from Ministerio de Educación y Ciencia (Spain) to A.C.
Footnotes
Author Contributions
R.B.J. and S.C.K. conceived the general ideas for this study. R.B.J., A.C., and S.C.K. planned experiments and interpreted data; R.B.J. and A.C. performed the experiments. R.B.J. and S.C.K. wrote the manuscript and all authors provided editorial input.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Yu VP, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 3.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 4.Phelan CM, et al. Mutation analysis of the BRCA2 gene in 49 site-specific breast cancer families. Nat. Genet. 1996;13:120–122. doi: 10.1038/ng0596-120. [DOI] [PubMed] [Google Scholar]
- 5.Wong AKC, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 6.Bignell G, Micklem G, Stratton MR, Ashworth A, Wooster R. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum. Mol. Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- 7.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 8.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 9.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 10.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 11.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 12.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 14.Godthelp BC, Artwert F, Joenje H, Zdzienicka MZ. Impaired DNA damage-induced nuclear Rad51 foci formation uniquely characterizes Fanconi anemia group D1. Oncogene. 2002;21:5002–5005. doi: 10.1038/sj.onc.1205656. [DOI] [PubMed] [Google Scholar]
- 15.San Filippo J, et al. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 18.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 19.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 20.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carreira A, Kowalczykowski SC. BRCA2: Shining light on the regulation of DNA-binding selectivity by RAD51. Cell Cycle. 2009;8:3445–3447. doi: 10.4161/cc.8.21.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petalcorin MI, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J. Mol. Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 26.Chen PL, et al. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorslund T, Esashi F, West SC. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinohara A, et al. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat. Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimura Y, Morita T, Yamamoto A, Matsushiro A. Cloning and sequence of the human RecA-like gene cDNA. Nucleic Acids Res. 1993;21:1665. doi: 10.1093/nar/21.7.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong JM, Ionescu D, Ingles CJ. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene. 2003;22:28–33. doi: 10.1038/sj.onc.1206071. [DOI] [PubMed] [Google Scholar]
- 31.Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol. Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 34.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 35.Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- 36.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 37.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 38.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 39.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 40.McIlwraith MJ, et al. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J. Mol. Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- 41.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miné J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baumann P, West SC. Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 49.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.