Abstract
Objective
HLA-DRB1*1001 (DR1001) is a shared epitope allele associated with rheumatoid arthritis. The objectives of this study were to assess the capacity of DR1001 to accommodate citrulline in its binding pockets and to identify citrullinated T cell epitopes derived from joint associated proteins.
Methods
The binding of peptide derivatives containing citrulline, arginine, and other amino acid substitutions was measured. A prediction algorithm was then developed to identify arginine containing sequences from joint associated proteins that preferentially bind to DR1001 upon citrullination. Unmodified and citrullinated versions of these sequences were synthesized and utilized to stimulate CD4+ T cells from healthy subjects and rheumatoid arthritis patients. Responses were measured by MHC class II tetramer staining and confirmed by isolating CD4+ T cell clones.
Results
DR1001 accepted citrulline, but not arginine in three of its anchoring pockets. The prediction algorithm identified sequences that preferentially bound to DR1001 with arginine replaced by citrulline. Three of these sequences elicited CD4+ T cell responses. T cell clones specific for these sequences proliferated only in response to citrullinated peptides.
Conclusions
Conversion of arginine to citrulline generates ‘altered-self’ peptides that can be bound and presented by DR1001. Responses to these peptides implicate the corresponding proteins (fibrinogen α, fibrinogen β and cartilage intermediate layer protein) as relevant antigens. Preferential responses to citrullinated sequences suggests that altered peptide binding affinity due to this post-translational modification may be an important factor in the initiation or progression of RA. As such, measuring responsiveness to these peptides may be useful for immune monitoring.
Keywords: Human, MHC, T cells, Antigens/Peptides/Epitopes, Antigen Presentation
Introduction
Rheumatoid arthritis (RA) is a chronic disease characterized by inflammation and autoimmune-mediated destruction of joints and surrounding tissue (1). RA is differentiated from other forms of arthritis by important immunological hallmarks, including rheumatoid factor and anti-citrulline antibodies (2). The appearance of these auto-antibodies implies a breakdown of both T and B cell tolerance. The risk of developing RA (and its immune markers) is linked to a subset of MHC class II haplotypes containing the shared epitope (residues 70–74) within their third hypervariable region (3). It is established that these residues dictate the peptide binding preferences for pocket 4 of the MHC class II peptide binding groove and perhaps some aspects of TCR recognition (4). Several mechanisms have been proposed for the contribution of the shared epitope to the disease process, including direct triggering by the five-amino acid shared epitope sequence leading to NO production (5), biased selection of autoreactive TCR (6–7), ability to bind to heat shock proteins (8), and the ability to present citrullinated peptides (9). However, the precise effects of the shared epitope remain unresolved.
The process of citrullination is a deimination of arginine catalyzed by peptidyl arginine deiminases (PADs), which convert the side chain from basic to polar. Notably, PAD2 and PAD4 are expressed at increased levels within joint tissues during inflammation (10). The latter of these PAD isoforms has been associated with RA susceptibility (11). Due to the activity of these enzymes, joint-associated proteins such as fillagrin typically contain citrulline, thereby increasing their antigenicity (12). In addition, PAD expression has been shown to increase due to tissue inflammation or environmental insults such as smoking (13). As a result, additional joint associated proteins such as fibrin, fibrinogen, and vimentin can be citrullinated during inflammation and cell death. Other than a few notable exceptions, it has been previously demonstrated that arginine is poorly tolerated in most anchor residue positions, particularly for position 4 of the MHC class II proteins that comprise the shared epitope alleles (14–15). These differences in pocket 4 binding preference correlate with susceptibility to autoimmune disease (16). Furthermore, it has been demonstrated that a joint associated epitope (vimentin 66–77) binds to shared epitope alleles (DRB1*0101 and DRB1*0401) with appreciable affinity only when residue 70 is changed to citrulline (9). Therefore, it is plausible that the citrullination generates “altered-self” epitopes that can be presented only when key arginine residues are converted by PAD enzymes.
Among the shared epitope alleles, HLA-DRB1*1001 (DR1001) is strongly associated with RA in Spanish and Hungarian populations (16–17) and has been reported as one of the alleles most strongly associated with anti-citrulline antibodies (17). However, DR1001 is among the least studied shared epitope alleles. For example, relatively few DR1001 restricted epitopes are known (18). One recent paper inferred a binding motif for DR1001 by aligning the sequences of eluted peptides (19), but there has been no study of citrulline binding to DR1001. For this current work we hypothesized that DR1001 accepts citrulline at some of its class II MHC anchor positions. Accordingly, conversion of arginine to citrulline by PAD would increase the binding affinity of ‘altered-self' peptides. T cells which recognize these peptides would be expected to escape selection, creating a latent pool of autoreactive cells.
Materials and Methods
Peptides and MHC Class II Protein
Panels of 20-mer peptides with overlapping sequences spanning the Influenza A/Puerto Rico/8/34 Nucleoprotein (NP), Influenza A/Puerto Rico/8/34 Matrix protein (MP), Influenza B/Hong Kong/330/2001 Hemagglutinin (Flu B HA), Influenza A/Panama/2007/99 Hemagglutinin (HA Pan), Influenza A/New Caledonia/20/99 Hemagglutinin (HA NC), tetanus toxin heavy chain (TT), and anthrax protective antigen (PA) proteins were synthesized on polyethylene pins with 9-fluorenylmethoxycarbonyl chemistry by Mimotopes (Clayton, Australia). The biotinylated reference tetanus toxoid peptide (TT560–571) was synthesized using an Applied Biosystems 433A Peptide Synthesizer (Foster City, CA). For flexibility, two Fmoc-6-aminohexanoic acid spacers were added between the N-terminal biotin label and the remainder of the peptide sequence. Panels of 12-mer peptides, 37 with sequences based on the TT560–571 sequence and 20 with sequences based on the HA217–236 sequence (modified for increased solubility) were synthesized on polyethylene pins with 9-fluorenylmethoxycarbonyl chemistry by MIMOTOPES (Clayton, Australia). Each peptide was dissolved in DMSO at 20 mg/ml and subsequently diluted as needed.
Recombinant DR1001 protein was produced as previously described (20). Briefly, soluble DR1001 was purified from insect cell culture supernatants by affinity chromatography and dialyzed against phosphate storage buffer (100 mM, pH 6.0).
DR1001 Tetramer Reagents
For the preparation of MHC class II tetramers, DR1001 protein was biotinylated at a sequence-specific site using biotin ligase (Avidity, Denver, CO) prior to dialysis into phosphate storage buffer. The biotinylated monomer was loaded with 0.2 mg/ml of peptide by incubating at 37°C for 72 hours in the presence of 2.5 mg/ml n-octyl-β-D-glucopyranoside and 1 mM Pefabloc SC (Sigma-Aldrich, St. Louis, MO). Peptide loaded monomers were subsequently conjugated as tetramers using R-PE streptavidin (Invitrogen) at a molar ratio of 8 to1.
Human Subjects
Samples for this study were obtained from healthy subjects with no personal or family history of RA or autoimmunity and from individuals with rheumatoid arthritis who are participants in the Benaroya Research Institute’s Immune Mediated Disease Registry. This protocol has been approved by the IRB at Benaroya Research Institute. Subjects were defined as having RA based on the diagnosis of RA based on ACR criteria, made by their rheumatologist. Anti-CCP antibody status was based on results obtained from a clinical laboratory. HLA typing was performed by the BRI Translational Core and all subjects were confirmed to have HLA DRB1*1001 haplotypes.
Tetramer-based T cell Assays
The Tetramer guided Epitope Mapping procedure was conducted as previously described (21) for each protein. PBMC were isolated from the blood of vaccinated healthy DR1001 subjects by Ficoll® underlay and CD4+ T cells isolated using the Miltenyi CD4+ T cell isolation kit. Cells from the CD4-fraction were incubated in 48 well plates (3 × 106 cells per well) for 1 hour and then washed, leaving adherent cells as APC. After adding 2 million CD4+ T cells per well, each well was stimulated with a pool of five consecutive peptides (20 amino acids long with a 12 residue overlap). After 14 days, 100 µl of resuspended cells were stained with pooled peptide PE-conjugated tetramers for 60 min at 37°C. Subsequently, cells were stained with CD4-PerCP (BD Biosciences), CD3-FITC and CD25-APC mAbs (eBioscience) and analyzed by flow cytometry. Cells from pools that gave positive staining were analyzed again using the corresponding individual peptide tetramers.
T cell responses to single peptides were conducted in a similar fashion. PBMC were isolated from the blood of healthy subjects and rheumatoid arthritis patients with DR1001 haplotypes. Two million CD4+ T cells were isolated and cultured in 48 well plates in the presence of adherent cells from the CD4- fraction. Each well was stimulated with a single peptide derived from a joint associated protein. After 14 days, 100 µl of resuspended cells were stained with peptide-loaded tetramers for 60 min at 37°C. Subsequently, cells were stained with CD4-APC (eBioscience), CD3-PerCP (BD Biosciences) and CD25-FITC mAbs (eBioscience) and analyzed by flow cytometry.
T-cell cloning and proliferation assays
CD4+ tetramer-positive cells were single-cell sorted using a FACS Vantage (Becton Dickinson) into 96-well plates containing T-cell medium expanded by adding 2 µg/ml phytohemagglutinin and 200,000 irradiated PBMCs plus IL-2. Expanded cells were stained with tetramers and analyzed on a FACSCalibur (Becton Dickinson). To assess proliferation, 104 T cells/well were plated in T-cell medium with 105 irradiated PBMCs from a DRB1*1001 donor and 0 or 10 µg/ml peptide (in triplicate), incubated at 37°C for 48 hours, pulsed with [3H]thymidine (1 µCi/well) and harvested 18 hours later, and [3H]thymidine measured with a scintillation counter. For blocking experiments, anti-DR (L243) or anti-DQ (SPVL3) antibody was added at 20 µg/ml.
Peptide Binding Competition
Various concentrations of each test peptide were incubated in competition with 0.01 mM biotinylated TT560–571 peptide in wells coated with HLA-DR1001 protein as previously described (22). After washing, residual biotin-Tetanus peptide was labeled using europium-conjugated streptavidin (Perkin Elmer, Waltham, Massachusetts, USA) and quantified using a Victor2 D time resolved fluorometer (Perkin Elmer, Waltham, Massachusetts, USA). Peptide binding curves were fitted by non-linear regression with a sigmoidal dose response curve model using Prism software (Version 4.03, GraphPad Software Inc.). IC50 binding values (the concentration needed to reduce binding of the biotinylated reference peptide by 50%) and their corresponding error bounds were calculated from the resulting curves using Prism software. Relative binding affinity (RBA) values were calculated as the IC50 value of the substituted peptide divided by the IC50 value of the non-substituted peptide. The ratio of two IC50 values indicates the fold difference in peptide binding affinity.
Molecular Modeling
Models of DR1001 with the modified HA217–236 sequence and several citrullinated variants were prepared on a Silicon Graphics Fuel work station using the program Insight II, version 2005 (Accelrys, San Diego, CA, USA), essentially as previously described (23). Energy minimization at was performed at pH 5.4, the experimental pH used for binding studies. The crystal structure of HLA-DRB1*0401 in complex with the Collagen II peptide (24) was used as the base molecule for all simulation studies. Figures were drawn with the aid of WebLabViewer version 3.5 and DSViewer Pro version 6.0, of Accelrys using previously published formatting and color conventions (25). The pdb coordinates are freely available to interested researchers upon requests to WWK or GKP.
Epitope Prediction
To predict epitopes we adapted the approach from our published work for DRB1*0901 (26). An array of binding coefficients (Cp) was developed for each pocket based on the observed binding of single (at pocket 1, 4, 6, or 9) amino acid substituted versions of the TT560–571 and modified HA217–236 peptide sequence. These Cp values are summarized in Supplementary Table I. Because all possible amino acids were not measured for any of the pockets, missing values in the data set were estimated based on the observed values for chemically similar amino acids. Peptide binding affinities were calculated based on the following formula:
In this formula, each Cp refers to the observed or estimated binding coefficient. To predict RBA values for peptides, Cp values were taken from the table using lookup procedure by scanning across every possible binding register and taking the highest observed RBA value for the sequence. To predict RBA values for entire proteins, Cp values were tabulated for every possible binding register.
Results
Tetramer Guided Epitope Mapping of DR1001 Restricted Epitopes
Because relatively few DR1001 restricted epitopes are known, we used tetramer guided Epitope Mapping to identify DR1001 restricted epitopes within a variety of antigenic proteins as described in Materials and Methods. CD4+ T cells from multiple DR1001 subjects were stimulated with pooled peptides and analyzed by two rounds of staining using pooled peptide tetramers and, for wells with tetramer positive populations, the corresponding individual peptide tetramers (representative results are shown in Supplementary Figure 1). A total of 16 peptides were identified that contained DR1001-restricted epitopes. These results are summarized in the first two columns of Table 1. Among these two sequences, TT560–571 and a modified version of the HA217–236, were chosen as reference peptides to determine the binding preferences of DR1001.
Table 1.
Motif Analysis for Novel and Published DR1001 Epitopes
| Peptide | Sequence* | Pred. RBA |
|---|---|---|
| NP 73-92 | ERRNKYLEEHPSAGKDPKKT | 0.21 |
| NP 305-324 | RLLQNSQVYSLIRPNENPAH | 0.20 |
| MP 41-60 | VLMEWLKTRPILSPLTKGIL | 0.35 |
| MP 97-116 | VKLYRKLKREITFHGAKEIS | 0.02 |
| HA HK 169-188 | ATNSLTIEVPYICTEGEDQI | 0.05 |
| HA Pan 97-116 | CYPYDVPDYASLRSLVASSG | 0.13 |
| HA Pan 297-316 | VNRITYGACPRYVKQNTLKL | 0.07 |
| HA Pan 305-324 | CPRYVKQNTLKLATGMRNVP | 0.07 |
| HA Pan 313-332 | TLKLATGMRNVPEKQTRGIF | 0.01 |
| HA NC 201-220 | IGNQRALYHTENAYVSVVSS | 1.3 |
| HA NC 217-236 | VVSSHYSRRFTPEIAKRPKV | 2.0 |
| TT 482-501 | NSFSEEPFQDEIVSYNTKNK | 0.45 |
| TT 554-573 | NIDDNTIYQYLYAQKSPTTL | 2.0 |
| PA 449-468 | ALNAQDDFSSTPITMNYNQF | 0.05 |
| PA 465-484 | YNQFLELEKTKQLRLDTDQV | 0.04 |
| PA 585-604 | QQTSQNIKNQLAELNATNIY | 0.25 |
| MP1 17-29 | SGPLKAEIAQRLE | 0.18 |
| HA1 255-270 | RGYFKMRTGKSSIMRS | 0.24 |
| HDV 50-65 | WLGNIKGILGKKDKDG | 0.13 |
| PTPRK 670-682 | PYYFAAELPPRNLPEP | 0.52 |
Predicted binding registers indicated in boldface secondary registers underlined
NP: Influenza A/Puerto Rico/8/34 Nucleoprotein (novel epitopes)
MP: Influenza A/Puerto Rico/8/34 Matrix Protein M1 (novel epitopes)
HA HK: Influenza B/Hong Kong/330/2001 Hemagglutinin (novel epitopes)
HA Pan: Influenza A/Panama/2007/99 Hemagglutinin (novel epitopes)
HA NC: Influenza A/New Caledonia/20/99 Hemagglutinin (novel epitopes)
TT: Tetanus Toxin Heavy Chain (novel epitopes)
PA: Antrax protective antigen (novel epitopes)
MP1: Matrix Protein (ref. 28)
HA1: Hemagglutinin (ref. 29)
HDV: Hepatitis Delta Virus (ref. 30)
PTPRK: Melanoma antigen (ref. 31)
Binding of Arginine and Citrulline to DR1001 at MHC class II Anchor Positions
The presence of arginine at MHC class II anchor positions has been previously shown to disrupt peptide binding, in particular for pocket 4 of shared epitope alleles (9, 15). We confirmed this by measuring the binding of arginine substituted versions of the TT560–571 peptide (Supplementary figure 2) to DR1001 protein. Substitutions at residues 561, 564, 566, 567, 569 and 571 blocked peptide binding. These results suggest that arginine is not accepted within the binding pockets of DR1001 and implicate 561Y as the P1 anchor, 564L as the P4 anchor, 566A as the P6 anchor, 567Q as the P7 anchor, and 569S as the P9 anchor. Based on previously published observations for DR0401 (9), the conversion of arginine to citrulline at MHC class II anchor positions at pocket 4 can facilitate peptide binding to multiple shared epitope alleles. To determine whether citrulline is accepted within the binding pockets of DR1001, peptides with single citrulline or arginine substitutions were designed based on the HA217–236 sequence and bound to recombinant DR1001 protein. Arginine was not accepted within the binding pockets of DR1001 (Figure 1). However, as shown in Figure 1, citrulline was accepted at pockets 4 and 9. Citrulline substitution at position 7 also significantly enhanced peptide binding. Therefore, conversion of arginine to citrulline at these three positions would be expected to facilitate peptide binding to DR1001.
Figure 1.
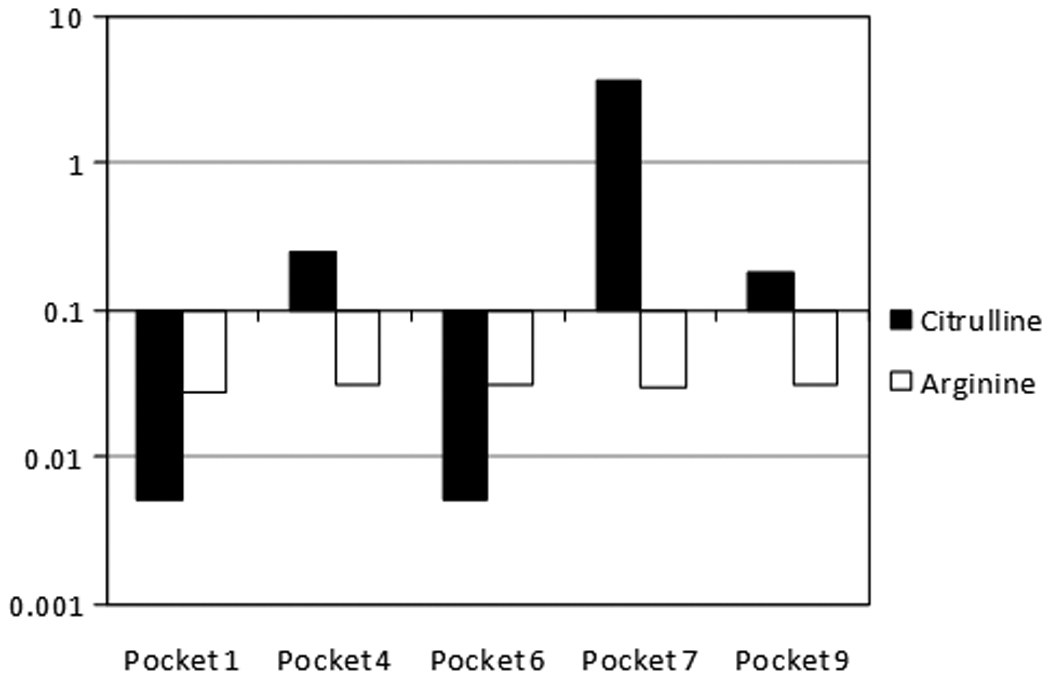
Binding of Citrulline within the Pockets of DR1001. IC50 values for Citrulline (black bars) or Arginine (open bars) substitutions at positions 1, 4, 6, 7, and 9 of the modified HA 217–236 peptide sequence. Values are shown on a logarithmic scale centered on 0.1, the threshold value for binding. Relative binding affinity (RBA) values were calculated as the IC50 value of the unsubstituted peptide divided by the IC50 value of the substituted peptide.
Modeling Analysis of Peptides Bound to DR1001
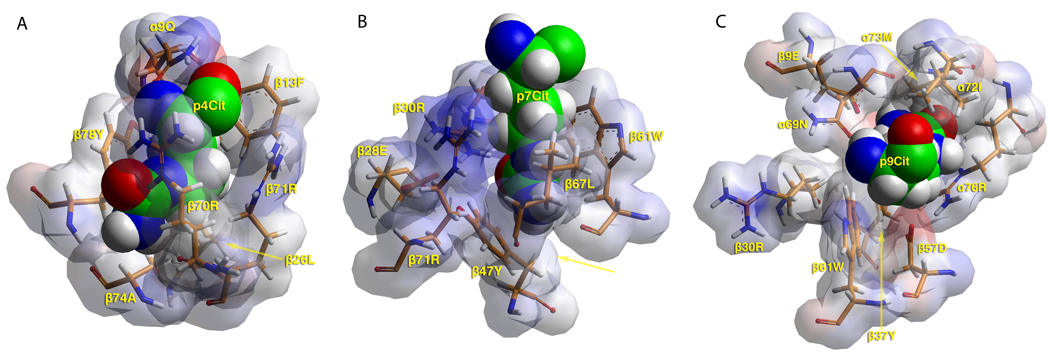
To visualize the binding pockets of DR1001, models of the HA 217–236 peptide (RFTKLIAKRSKV) in complex with DR1001 were created as described in Materials and Methods. Modeling results for the unmodified peptide (Supplementary Figure 3A) suggested high affinity binding with 218F as the P1 anchor. Based on the model, pocket 4 would be expected to favor medium sized aliphatic (such as leucine, depicted in Supplementary Figure 3B) or polar amino acids. As shown in Figure 2A, pocket 4 accepts citrulline due to the formation of three hydrogen bonds and favorable hydrophobic interactions between β13Phe and β26Leu and the anchor’s methylene groups. To allow these interactions the citrulline side chain bends within the pocket. Pocket 6 accepts leucine (despite the presence of α11Glu and α66Asp, depicted in Supplementary Figure 3C) but probably not larger residues. Thus, it is not surprising that citrulline cannot anchor within this pocket. Position 7, previously described as a solvent accessible shelf that accommodates a wide variety of side chains (27), accepts charged residues such as lysine and arginine. However, as depicted in Figure 2B, a variety of interactions (numerous hydrogen bonds and favorable interaction of its methylene groups with β67Leu and β61Trp) strongly favor the binding of citrulline. Pocket 9 accepts serine (depicted in Supplementary Figure 3D) and probably favors small polar and aliphatic residues. As shown in Figure 2C, citrulline is a difficult fit within pocket 9 because of β37Tyr, but favorable interactions with α76Arg and β9Glu promote its binding. In total the modeling results further support the notion that conversion of arginine to citrulline at positions 4, 7, and 9 would facilitate peptide binding to DR1001.
Figure 2.
(A) TCR view of pocket 4 of HLA-DR1001/RFTKCitIAKRSKV complex. Citrulline is able to bind, as its side chain is flexible and uncharged. Its terminus forms hydrogen bonds to β70Arg and β74Ala. Its methylene groups interact with β13Phe and β26Leu. To accommodate these interactions the side chain bends, introducing strain. DR10 depicted in van der Waals surface representation, with atomic charges color-coded (blue, positive; grey, neutral; red, negative). Peptide shown in space filling form (carbon, green; oxygen, red; nitrogen, blue; hydrogen, white; sulfur, yellow). Selected DR10 residues shown in stick form color-coded the same as the peptide except carbon (orange). (B) TCR view of pocket 7 of DR1001/RFTKLIACitRSKV complex. The arrangement of this pocket is favorable to accepting citrulline. Citrulline forms hydrogen bonds with β71Arg and β30Arg. Its methylene groups interact favorably with β67Leu and β61Trp. β47Tyr forms hydrogen bonds to the terminus of Citrulline. Figure rotated −20° with respect to the y-axis. Color and depiction identical to panel A. (C) TCR view of pocket 9 of DR1001 in complex with RFTKLIAKRCitKV peptide. Citrulline barely fits the pocket because of β37Tyr. Yet favorable interactions with α76Arg and β9Glu promote its weak binding. Color and depiction identical to panel A.
Predicting the Binding of Citrullinated Peptides to DR1001
It would be convenient to predict sequences that preferentially bind to DR1001 upon citrullination. To develop such an algorithm, two panels of peptides were designed to empirically measure the binding of various amino acids to the pockets of DR1001. These peptides were bound at various concentrations to DR1001 protein as described in materials and methods. The sequence of each peptide and the measured relative binding affinities (RBA) are summarized in Supplementary Table II. The preferences for Pocket 1, Pocket 4, Pocket 6, and Pocket 9 (Cumulative RBA values, calculated by averaging observed values for both peptide panels) are summarized in Figure 3. These observations were consistent (but not identical) with the findings of a recent study that identified a peptide anchor motif for DR1001 by de novo sequencing of natural DR10–associated peptide ligands (19). Cumulative RBA values were used to develop an array of binding coefficients (Cp, summarized in Supplementary Table I) to predicting peptide binding to DR1001. The effectiveness of this algorithm was evaluated by testing its ability to predict epitopes within the peptides summarized in Table 1. For each of these peptides, relative binding affinities (RBA) for all possible binding registers were calculated as described in Materials and Methods. The predicted RBA of the best core epitope for each peptide is shown in the final column of Table 1. For the majority of these peptides, single core epitopes consistent with the DR1001 binding motif were identified (boldface). A few peptides contained two distinct registers that could be expected to bind DR1001 (for these, the second motif is underlined). Predicting epitopes within two of the peptides (MP 97–116 and HA 313–332) was problematic because even the best registers for these peptides contained suboptimal residues. In a similar manner, predicted epitopes were identified for four previously published peptide sequences, as shown in the lower portion of Table 1 (28–31). In total, the prediction algorithm could predict epitopes within 18 out of 20 peptides.
Figure 3.
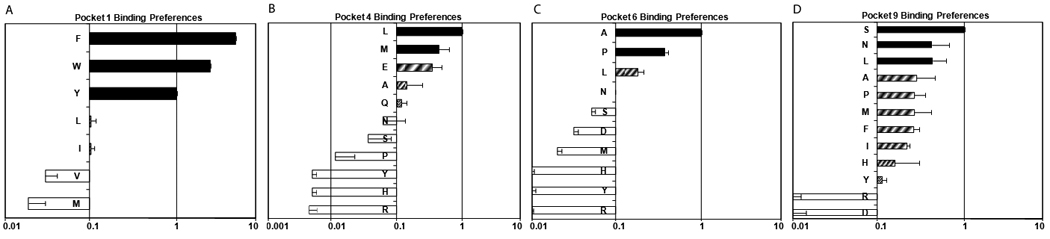
Amino acid preferences for A) pocket 1, B) pocket 4, C) pocket 6, and D) pocket 9 of the DR1001 motif. Black bars represent values within 2.5-fold of the reference peptide (RBA ≥ .4, “preferred”), hatched bars represent values between 4-fold and within 10-fold of the reference peptide (0.4 ≥ RBA ≥ 0.1, “tolerated”), and open bars represent values within less than 10-fold of the reference peptide (RBA ≤ 0.1, “excluded”).
The same prediction algorithm was used to identify arginine containing sequences from within several joint associated proteins: vimentin, fibrinogen alpha (Fib A), figrinogen beta (Fib B), fibrinogen gamma, alpha enolase, filaggrin, and cartilage intermediate layer protein (CILP). The complete results of this analysis are shown in Supplementary Table III. A total of 96 sequences were identified that contained sequences with arginine residues at position 4, 7, or 9 that created a predicted epitope when converted to citrulline. These peptides would be expected to preferentially bind to DR1001 upon citrullination. Twelve of these sequences were selected for further study (with a preference for sequences with aromatic residues in position 1 and citrulline at position 4). Unmodified and citrullinated versions of these peptides were synthesized and binding to DR1001 was measured. The sequences and binding results for these peptides are summarized in Table 2. Five of the 12 citrullinated peptides bound to DR1001, while (with one exception) the unmodified versions were unable to bind. For the remaining sequences, binding may have been influenced by intervening sequences. For example, Fib A 24–38 contains three consecutive glycine residues which could destabilize binding.
Table 2.
In Vitro Binding of Citrullinated Peptides from Joint Associated Proteins
| Peptide | Sequence* | IC50 |
|---|---|---|
| Vimentin 58-72 | GGVYATRSSAVRLRS | >100 |
| GGVYATXSSAVXLRS | 23 | |
| Fib A 24-38 | EGDFLAEGGGVRGPR | >100 |
| EGDFLAEGGGVXGPR | >100 | |
| Fib A 383-397 | TGQWHSESGSFRPDS | >100 |
| TGQWHSESGSFXPDS | >100 | |
| Fib A 506-520 | LDGFRHRHPDEAAFF | >100 |
| LDGFRHXHPDEAAFF | >100 | |
| Fib A 737-751 | YAEYHFRVGSEAEGY | >100 |
| YAEYHFXVGSEAEGY | 39 | |
| Fib B 68-82 | GGGYRARPAKAAATQ | >100 |
| GGGYRAXPAKAAATQ | 14 | |
| Fib G 217-231 | WTVFQKRLDGSVDFK | >100 |
| WTVFQKXLDGSVDFK | >100 | |
| CILP 538-552 | VLTFVDRLQKFVNTT | >100 |
| VLTFVDXLQKFVNTT | >100 | |
| CILP 738-751 | ERRLFNLDVPESRR | 0.4 |
| ERRLFNLDVPESXR | 0.6 | |
| CILP 982-996 | GKLYGIRDVRSTRDR | 13 |
| GKLYGIXDVXSTRDR | 2.8 | |
| Filag 2116-2130 | GSHYDQAQDSSRHSA | >100 |
| GSHYDQAQDSSXHSA | >100 | |
| α Enolase 9-23 | REIFDSRGNPTVEVD | >100 |
| REIFDSXGNPTVEVD | >100 |
Predicted binding register underlined with anchor residues in boldface. X = citrulline
T Cell responses to Citrullinated Peptides
CD4+ T cells from two healthy subjects and two rheumatoid arthritis patients (one CCP+ and one CCP−) were stimulated with citrullinated peptides or the corresponding unmodified sequences (containing arginine) in separate wells. After two weeks in culture these cells were analyzed using tetramers. Results for both of these rheumatoid arthritis patients are shown in Figure 4A. For RA patient #1 (CCP−), two citrullinated peptide sequences (Fib A737–751 and CILP982–996) were positive. For RA patient #2 (CCP+), three citrullinated peptide sequences (Fib A737–751, Fib B68–82, and CILP982–996) were positive. Representative staining results for a healthy control subjects are also shown in Figure 4A. For that control subject, one positive response to CILP982–996 was observed while all other staining was negative. All responses were negative for a second control subject (not shown). Tetramer positive T cell clones specific for these sequences were isolated for each of these specificities (representative clones shown in Figure 4B). As shown in Figure 4C, these T cell clones proliferated only in response to the citrullinated peptide sequences. Blocking experiments (Supplementary figure 4) with anti-DR antibodies and HLA matched antigen presenting cells confirmed that the responses of these clones were restricted by DR1001.
Figure 4.
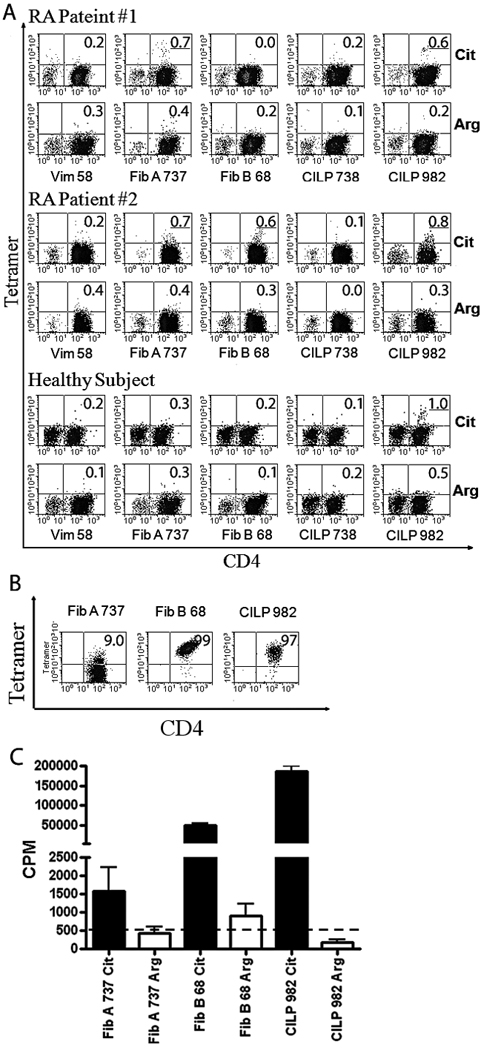
T cell responses to citrullinated peptides. A) Tetramer staining of cultured CD4+ T cells from two RA subjects with DR1001 haplotype and one HLA-matched healthy control subject with citrullinated or unmodified sequences (as labeled). Tetramer positive populations more than 0.5% above background were considered positive (underlined). B) Tetramer staining of representative T cell clones specific for citrullinated sequences from joint associated proteins. C) Proliferation of T cell clones in response to citrullinated (closed bars) or unmodified (open bars) peptide sequences. The dashed line indicates background proliferation (512 CPM).
Discussion
The disease specificity and predictive value of anti-citrulline antibodies (32–33) and the association of these antibodies with severe disease (34) strongly suggest that the recognition of citrullinated epitopes by T and B cells is important in the initiation and progression of rheumatoid arthritis. While a few citrullinated epitopes have been validated, current knowledge is actually quite limited. In this report we demonstrate that citrulline is accepted in multiple pockets (positions 4, 7, and 9) of DR1001, a shared epitope allele that is strongly associated with RA and with anti-citrulline antibodies (16–17, 35). This accommodation of citrulline is consistent with molecular modeling results and with the DR1001 binding motif indicated by our peptide binding results. The acceptance of citrulline at position 4 mirrors a previous study (9), which demonstrated that the conversion of arginine to citrulline at pocket 4 dramatically increases peptide affinity for DRB1*0401 and other shared epitope alleles. These current observations are the first to indicate that citrulline can be accepted at other positions. Among these three pockets, position 4 is likely to be of primary importance because its binding properties are dictated by the shared epitope residues (70–74) of the class II beta chain. However, citrullination at positions 7 and 9 may also play a significant role in the creation of ‘altered-self’ epitopes (for example, position 7 of the CILP 982 epitope was citrullinated).
We also demonstrate an effective approach to predict and validate citrullinated epitopes within RA associated antigens. Our algorithm incorporated information from epitope discovery, peptide binding and molecular modeling to allow the identification of sequences that bind with increased affinity after citrullination. Using this approach we identified numerous sequences from joint-associated proteins that could contain citrullinated epitopes. For this initial study, only a subset of these peptides was included in binding and T cell studies, leading to the validation of three peptides that appear to contain citrullinated epitopes (however, it has not yet been confirmed that these sequences are naturally processed and presented). Based on these findings, it seems likely that other epitopes are present among the peptides that were not studied. The approach applied here for DR1001 should be readily applied to predict citrullinated epitopes for other RA associated alleles once their binding characteristics have been measured. Citrullinated T cell epitopes identified using this approach will provide important tools, aiding our ability to monitor relevant T cell responses during RA progression and to appropriately target antigen based therapies.
While the number of subjects tested is modest, the T cell responses observed in this study have a few interesting implications. It is notable that while T cells specific for these RA associated epitopes clearly recognized citrullinated sequences, the T cell repertoire appeared to be blind to the unmodified sequences (presumably because only the citrullinated sequences could be bound and presented by DR1001). This demonstrates that citrullinated T cell epitopes must be modified by PAD enzymes in order to be recognized, as had been suggested for DR0401 restricted epitopes (9). Previous mouse studies have demonstrated vimentin peptides as naturally processed T cell epitopes (36), the induction of arthritis with citrullinated fibrinogen (37) and epitope spreading to multiple citrullinated antigens (38). Therefore, these T cell specificities are likely to play a role in the induction and/or progression of human RA. Our current data are inadequate to compare the T cell responses of rheumatoid arthritis patients and healthy controls. However, it could be expected based on the findings of other recent studies that RA patients are likely to have a higher frequency of memory T cells specific for citrullinated joint antigens. For example, one recent study reported enhanced cytokine responses to an immunogenic vimentin peptide in RA patients compared to healthy control subjects (39). It might be expected that anti-citrulline T cell responses would be tightly correlated with anti-CCP status. However, our preliminary results do not suggest a tight correlation. Subject #1, who was anti-CCP negative had T cell responses to CILP and Fib A. Subject #2, who was anti-CCP positive had T cell responses to CILP, Fib A, and Fib B. One healthy subject also had a T cell response to CILP. Of course, a lack of tight correlation was not totally unexpected since anti-CCP status reflects only a subset of the anti-citrulline antibody responses that are possible.
Recent findings implicate environmental factors such as smoking (40), apoptosis (41), and inflammation (42) in the upregulation of PAD activity and consequent protein modifications. Under inflammatory conditions, CD4+ T cells that recognize the resulting citrullinated antigens secrete IL-17 (43). Based on these observations, citrulline may provide a crucial link between genetic, immunologic, and environmental factors in rheumatoid arthritis. In this paradigm, “altered-self” epitopes are presented to T cells only when key arginine residues are converted by PAD enzymes. Although one PAD isoform may be expressed in the thymus (44), T cells which recognize these peptides appear to escape thymic selection, creating a latent pool of autoreactive cells. Increased PAD activity, due to one or more environmental factors, could then initiate a cascade of self-reactive T cell responses followed by humoral responses to citrullinated self-proteins and epitope spreading to secondary self-antigens, including PAD enzymes themselves (45). Within this paradigm, indentifying citrullinated T cell epitopes and monitoring responsiveness to these citrullinated antigens may be essential for evaluating the efficacy of therapeutic agents and for targeting antigen based therapies.
Supplementary Material
Acknowledgements
We wish to acknowledge the staff of the Benaroya Research Institute Translational Research program for subject recruitment and sample management. The Silicon Graphics Fuel work station and the accompanying molecular simulation software was obtained via an equipment grant to Epirus Institute of Technology, from the Epirus Regional Development Project of the 3rd Community Support Framework (80% EU funds, 20% Hellenic State funds).
Grant support: Autoimmune prevention centers 5U19 AI050864-07 (JHB) NIH contract HHSN266200400028C (WWK)
References
- 1.Steiner G. Auto-antibodies and autoreactive T-cells in rheumatoid arthritis: pathogenetic players and diagnostic tools. Clin Rev Allergy Immunol. 2007;32:23–36. doi: 10.1007/BF02686079. [DOI] [PubMed] [Google Scholar]
- 2.De Rycke L, Peene I, Hoffman IE, Kruithof E, Union A, Meheus L, et al. Rheumatoid factor and anticitrullinated protein antibodies in rheumatoid arthritis: diagnostic value, associations with radiological progression rate, and extra-articular manifestations. Ann Rheum Dis. 2004;63:1587–1593. doi: 10.1136/ard.2003.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vries N, Tijssen H, van Riel PL, van de Putte LB. Reshaping the shared epitope hypothesis: HLA-associated risk for rheumatoid arthritis is encoded by amino acid substitutions at positions 67–74 of the HLA-DRB1 molecule. Arthritis Rheum. 2002;46:921–928. doi: 10.1002/art.10210. [DOI] [PubMed] [Google Scholar]
- 4.Penzotti JE, Doherty D, Lybrand TP, Nepom GT. A structural model for TCR recognition of the HLA class II shared epitope sequence implicated in susceptibility to rheumatoid arthritis. J Autoimmun. 1996;9:287–293. doi: 10.1006/jaut.1996.0037. [DOI] [PubMed] [Google Scholar]
- 5.Ling S, Li Z, Borschukova O, Xiao L, Pumpens P, Holoshitz J. The rheumatoid arthritis shared epitope increases cellular susceptibility to oxidative stress by antagonizing an adenosine-mediated anti-oxidative pathway. Arthritis Res Ther. 2007;9:R5. doi: 10.1186/ar2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nepom GT. The role of the DR4 shared epitope in selection and commitment of autoreactive T cells in rheumatoid arthritis. Rheum Dis Clin North Am. 2001;27:305–315. doi: 10.1016/s0889-857x(05)70203-9. [DOI] [PubMed] [Google Scholar]
- 7.Taneja V, Behrens M, Basal E, Sparks J, Griffiths MM, Luthra H, et al. Delineating the role of the HLA-DR4 "shared epitope" in susceptibility versus resistance to develop arthritis. J Immunol. 2008;181:2869–2877. doi: 10.4049/jimmunol.181.4.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auger I, Escola JM, Gorvel JP, Roudier J. HLA-DR4 and HLA-DR10 motifs that carry susceptibility to rheumatoid arthritis bind 70-kD heat shock proteins. Nat Med. 1996;2:306–310. doi: 10.1038/nm0396-306. [DOI] [PubMed] [Google Scholar]
- 9.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol. 2003;171:538–541. doi: 10.4049/jimmunol.171.2.538. [DOI] [PubMed] [Google Scholar]
- 10.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Méchin MC, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 12.Yamada R. Peptidylarginine deiminase type 4, anticitrullinated peptide antibodies, and rheumatoid arthritis. Autoimmun Rev. 2005;4:201–206. doi: 10.1016/j.autrev.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Makrygiannakis D, Hermansson M, Ulfgren AK, Nicholas AP, Zendman AJ, Eklund A, et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann Rheum Dis. 2008;67:1488–1492. doi: 10.1136/ard.2007.075192. [DOI] [PubMed] [Google Scholar]
- 14.Fremont DH, Hendrickson WA, Marrack P, Kappler J. Structures of an MHC class II molecule with covalently bound single peptides. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 15.Friede T, Gnau V, Jung G, Keilholz W, Stevanovi S, Rammensee HG. Natural ligand motifs of closely related HLA-DR4 molecules predict features of rheumatoid arthritis associated peptides. BBA - Molecular Basis of Disease. 1996;1316:85–101. doi: 10.1016/0925-4439(96)00010-5. [DOI] [PubMed] [Google Scholar]
- 16.Yelamos J, Garcia-Lozano JR, Moreno I, Aguilera I, Gonzalez MF, Garcia A, et al. Association of HLA-DR4-Dw15 (DRB1*0405) and DR10 with rheumatoid arthritis in a Spanish population. Arthritis Rheum. 1993;36:811–814. doi: 10.1002/art.1780360611. [DOI] [PubMed] [Google Scholar]
- 17.Poór G, Nagy ZB, Schmidt Z, Brózik M, Merétey K, Gergely P., Jr Genetic background of anticyclic citrullinated peptide autoantibody production in Hungarian patients with rheumatoid arthritis. Ann N Y Acad Sci. 2007;1110:23–32. doi: 10.1196/annals.1423.004. [DOI] [PubMed] [Google Scholar]
- 18.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez I, Collado J, Daura X, Colomé N, Rodríguez-García M, Gallart T, et al. The rheumatoid arthritis-associated allele HLA-DR10 (DRB1*1001) shares part of its repertoire with HLA-DR1 (DRB1*0101) and HLA-DR4 (DRB*0401). Arthritis Rheum. 2008;58:1630–1639. doi: 10.1002/art.23503. [DOI] [PubMed] [Google Scholar]
- 20.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104:R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novak EJ, Liu AW, Gebe JA, Falk B, Nepom GT, Koelle DM, Kwok WW. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol. 2001;166:6665–6670. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 22.Ettinger RA, Kwok WW. A peptide binding motif for HLA-DQA1*0102/DQB1*0602, the class II MHC molecule associated with dominant protection in insulin-dependent diabetes mellitus. J Immunol. 1998;160:2365–2373. [PubMed] [Google Scholar]
- 23.Masewicz SA, Papadopoulos GK, Swanson E, Moriarity L, Moustakas AK, Nepom GT. Modulation of T cell response to hGAD65 peptide epitopes. Tissue Antigens. 2002;59:101–112. doi: 10.1034/j.1399-0039.2002.590205.x. [DOI] [PubMed] [Google Scholar]
- 24.Dessen A, Lawrence CM, Cupo S, Zaller DM, Wiley DC. X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity. 1997;7:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 25.Reichstetter S, Papadopoulos GK, Moustakas AK, Swanson E, Liu AW, Beheray S, et al. Mutational analysis of critical residues determining antigen presentation and activation of HLA-DQ0602 restricted T-cell clones. Hum Immunol. 2002;63:185–193. doi: 10.1016/s0198-8859(01)00377-9. [DOI] [PubMed] [Google Scholar]
- 26.James EA, Moustakas AK, Bui J, Nouv R, Papadopoulos GK, Kwok WW. The binding of antigenic peptides to HLA-DR is influenced by interactions between pocket 6 and pocket 9. J Immunol. 2009;183:3249–3258. doi: 10.4049/jimmunol.0802228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fremont DH, Monnaie D, Nelson CA, Hendrickson WA, Unanue ER. Crystal structure of I-Ak in complex with a dominant epitope of lysozyme. Immunity. 1998;8:305–317. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 28.Busch R, Strang G, Howland K, Rothbard JB. Degenerate binding of immunogenic peptides to HLA-DR proteins on B cell surfaces. Int Immunol. 1990;2:443–451. doi: 10.1093/intimm/2.5.443. [DOI] [PubMed] [Google Scholar]
- 29.Jones CM, Lake RA, Lamb JR, Faith A. Degeneracy of T cell receptor recognition of an influenza virus hemagglutinin epitope restricted by HLA-DQ and -DR class II molecules. Eur J Immunol. 1994;24:1137–1142. doi: 10.1002/eji.1830240519. [DOI] [PubMed] [Google Scholar]
- 30.Nisini R, Paroli M, Accapezzato D, Bonino F, Rosina F, Santantonio T, et al. Human CD4+ T-cell response to hepatitis delta virus: identification of multiple epitopes and characterization of T-helper cytokine profiles. J Virol. 1997;71:2241–2251. doi: 10.1128/jvi.71.3.2241-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novellino L, Renkvist N, Rini F, Mazzocchi A, Rivoltini L, Greco A, et al. Identification of a mutated receptor-like protein tyrosine phosphatase kappa as a novel, class II HLA-restricted melanoma antigen. J Immunol. 2003;170:6363–6370. doi: 10.4049/jimmunol.170.12.6363. [DOI] [PubMed] [Google Scholar]
- 32.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Gaalen FA, Linn-Rasker SP, van Venrooij WJ, de Jong BA, Breedveld FC, Verweij CL, et al. Autoantibodies to cyclic citrullinated peptides predict progression to rheumatoid arthritis in patients with undifferentiated arthritis: a prospective cohort study. Arthritis Rheum. 2004;50:709–715. doi: 10.1002/art.20044. [DOI] [PubMed] [Google Scholar]
- 34.Kroot EJ, de Jong BA, van Leeuwen MA, Swinkels H, van den Hoogen FH, van't Hof M, et al. The prognostic value of anti-cyclic citrullinated peptide antibody in patients with recent-onset rheumatoid arthritis. Arthritis Rheum. 2000;43:1831–1835. doi: 10.1002/1529-0131(200008)43:8<1831::AID-ANR19>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.van Gaalen FA, van Aken J, Huizinga TW, Schreuder GM, Breedveld FC, Zanelli E, et al. Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum. 2004;50:2113–2121. doi: 10.1002/art.20316. [DOI] [PubMed] [Google Scholar]
- 36.Feitsma AL, van der Voort EI, Franken KL, El Bannoudi H, Elferink BG, Drijfhout JW, et al. Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis Rheum. 2009;62:117–125. doi: 10.1002/art.25059. [DOI] [PubMed] [Google Scholar]
- 37.Hill JA, Bell DA, Brintnell W, Yue D, Wehrli B, Jevnikar AM, et al. Arthritis induced by posttranslationally modified (citrullinated) fibrinogen in DR4-IE transgenic mice. J Exp Med. 2008;205:967–979. doi: 10.1084/jem.20072051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidd BA, Ho PP, Sharpe O, Zhao X, Tomooka BH, Kanter JL, et al. Epitope spreading to citrullinated antigens in mouse models of autoimmune arthritis and demyelination. Arthritis Res Ther. 2008;10:R119. doi: 10.1186/ar2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feitsma AL, van der Voort EI, Franken KL, el Bannoudi H, Elferink BG, Drijfhout JW, et al. Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis Rheum. 2010;62:117–125. doi: 10.1002/art.25059. [DOI] [PubMed] [Google Scholar]
- 40.Klareskog L, Stolt P, Lundberg K, Källberg H, Bengtsson C, Grunewald J, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 41.Liu GY, Liao YF, Chang WH, Liu CC, Hsieh MC, Hsu PC, et al. Overexpression of peptidylarginine deiminase IV features in apoptosis of haematopoietic cells. Apoptosis. 2006;11:183–196. doi: 10.1007/s10495-006-3715-4. [DOI] [PubMed] [Google Scholar]
- 42.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Méchin MC, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 43.von Delwig A, Locke J, Robinson JH, Ng WF. Response of Th17 cells to a citrullinated arthritogenic aggrecan peptide in patients with rheumatoid arthritis. Arthritis Rheum. 2009;62:143–149. doi: 10.1002/art.25064. [DOI] [PubMed] [Google Scholar]
- 44.Guerrin M, Ishigami A, Méchin MC, Nachat R, Valmary S, Sebbag M, et al. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem J. 2003;370:167–174. doi: 10.1042/BJ20020870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halvorsen EH, Pollmann S, Gilboe IM, van der Heijde D, Landewé R, Ødegård S, et al. Serum IgG antibodies to peptidylarginine deiminase 4 in rheumatoid arthritis and associations with disease severity. Ann Rheum Dis. 2008;67:414–417. doi: 10.1136/ard.2007.080267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.